MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma - PubMed (original) (raw)
MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma
Cristian Turato et al. J Clin Med. 2019.
Abstract
The only first-line treatment approved for advanced hepatocellular carcinoma (HCC) is sorafenib. Since many patients experience drug resistance, the discovery of more effective therapeutic strategies represents an unmet clinical need. MicroRNA (MiR)-122 is downregulated in most HCCs, while oncogenic SerpinB3 is upregulated. Here, we assessed the relationship between miR-122 and SerpinB3 and their influence on cell phenotype and sorafenib resistance in HCC. A bioinformatics analysis identified SerpinB3 among hypothetical miR-122 targets. In SerpinB3-overexpressing HepG2 cells, miR-122 transfection decreased SerpinB3 mRNA and protein levels, whereas miR-122 inhibition increased SerpinB3 expression. Luciferase assay demonstrated the interaction between miR-122 and SerpinB3 mRNA. In an HCC rat model, high miR-122 levels were associated with negative SerpinB3 expression, while low miR-122 levels correlated with SerpinB3 positivity. A negative correlation between miR-122 and SerpinB3 or stem cell markers was found in HCC patients. Anti-miR-122 transfection increased cell viability in sorafenib-treated Huh-7 cells, while miR-122 overexpression increased sorafenib sensitivity in treated cells, but not in those overexpressing SerpinB3. In conclusion, we demonstrated that miR-122 targets SerpinB3, and its low levels are associated with SerpinB3 positivity and a stem-like phenotype in HCC. MiR-122 replacement therapy in combination with sorafenib deserves attention as a possible therapeutic strategy in SerpinB3-negative HCCs.
Keywords: hepatocellular carcinoma; micro RNA; molecular targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
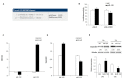
Figure 1
(A) miR-122 binding site in the SerpinB3 3’untranslated region (3′UTR) as reported by the TargetScan algorithm. (B) Dual-luciferase assay in HepG2 cells. The SerpinB3 3’UTR-containing vector was co-transfected with miR-122 or negative control (NC). MiR-122 overexpression determined a decrease of the reporter gene activity in pGL3-SerpinB3 co-transfected HepG2 cells. (C) qPCR analysis of miR-122 levels following miR-122 inhibitor (AM-122) or miR-122 mimic transfection in HepG2 cells stably overexpressing SerpinB3 (HepG2/SerpinB3) with respect to controls. U6RNA was used as housekeeping gene. Y-axis reports 2−ΔΔCt levels expressed in logarithmic form. (D,E) qPCR and Western blot analysis of SerpinB3 in transfected HepG2/SerpinB3 cells. β-Actin was used as housekeeping gene. Y-axis reports 2−ΔΔCt levels. NCi: miRNA inhibitor negative control; NC: miRNA precursor negative control. (* p ˂ 0.05). Data shown are representative results of at least three independent experiments.
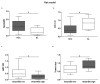
Figure 2
(A) SerpinB3 and (B) miR-122 expression in hepatocellular carcinoma (HCC) nodules and matched surrounding liver (SL) tissues from diethyl nitrosamine (DEN)-induced HCC rats. Y-axis reports 2−ΔΔCt levels from qPCR analysis. The values are represents as box-and-whiskers graph with the minimum, maximum, and median data. (* p ˂ 0.05 Mann–Whitney test). (C) MicroRNA (MiR)-122 expression in tumor tissues from HCC rats in relation to SerpinB3 expression. HCCs were grouped on the basis of SerpinB3 expression as detected by qPCR analysis (* p ˂ 0.05 Mann–Whitney test). Specifically, the high SerpinB3 group included HCC specimens with a cycle threshold (Ct) value lower than 35, whereas the low SerpinB3 group included HCC specimens with a Ct value higher than 35. (D) Box plot graph representing tumor size in HCC rats in relation to SerpinB3 expression (* p ˂ 0.05 Mann–Whitney test). Tumor size was represented by the value of major diameter (cm).
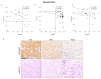
Figure 3
(A) Correlation graphs in human HCC tissues between miR-122 and SerpinB3 mRNA and (B) protein levels, as detected by qPCR and tissue microarray, respectively. Regarding the qPCR analysis, we considered only HCC samples with a Ct value lower than 35, which resulted in 19 out of 35 tested HCC samples. β-Actin was used as housekeeping gene for SerpinB3 normalization; U6RNA was used as housekeeping gene for miR-122 analysis. (C) Correlation graphs in human HCC tissues between SerpinB3 and caspase-3 mRNA levels, as detected by qPCR analysis. (D) Representative images of tissue microarray (TMA) in HCC samples from three patients (Pt2, Pt21 and Pt30) with different expression of miR-122 and SerpinB3 in sequential tissue slides. Scale bar 100 µM.
Figure 4
(A) Correlation graphs between tissue miR-122 and PROM1/CD133 or (B) SerpinB3. (C) Correlation graph between EpCAM/CD326 and SerpinB3 or (D) miR-122 levels in the same HCC patient cohort. The X- and Y-axes report 2−∆∆Ct values from qPCR analysis converted in a log2 form. U6RNA was used as housekeeping gene for miR-122 normalization, whereas β-Actin was used for gene normalization. (E) PROM1/CD133 and EpCAM protein detected by WB analysis in SerpinB3-overexpressing and empty vector-bearing (CTR) HCC cell lines. β-Actin was used as housekeeping gene. (F) PROM1/CD133 and (G) EpCAM mRNA levels in miR-122-overexpressing (miR) or silenced (AM) HCC cell lines. The Y-axis reports relative values with respect to negative controls. (H) Immunophenotype analysis of PROM1/CD133 and EpCAM levels in miR-122 (miR) or anti-miR-122 (AM)-transfected Huh-7 cells. The percentage of positive cells is reported on the top left of the histogram graph. NC: negative control precursor miRNA. NCi: negative control inhibitor miRNA.
Figure 5
(A–C) In each panel, from the top to the bottom: cell viability and caspase 3/7 activity assays and western blot analysis of cleaved caspase-3 in HepG2 and Hep3B cells overexpressing miR-122 (A,B) and in Huh-7 cells transfected with miR-122 inhibitor (AM-122) (C) following vehicle (NT) or sorafenib administration. β-Actin was used as housekeeping gene in western blot analysis and numbers represent fold-change values. NC: miRNA precursor negative control; NCi: miRNA inhibitor’s negative control. (D) Cell viability assay in SerpinB3-overexpressing HepG2 cells following sorafenib treatment (10 and 50 µM for 48 h). (* p ˂ 0.05 Mann–Whitney test). (E) MiR-122 quantification in SerpinB3-overexpressing HepG2 cells following sorafenib treatment. (* p ˂ 0.05 Mann–Whitney test). (F) Western blot analysis of cleaved Poly(ADP-ribose) polymerase 1 (PARP) and cleaved caspase-3 in HepG2 cells overexpressing SerpinB3 (HepG2/SerpinB3) transfected with miR-122 in untreated or sorafenib (10 µM)-treated cells (48 h). β-Actin was used as housekeeping gene in western blot analysis and numbers represent fold-change values. NC: miRNA precursor negative control.
Similar articles
- The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma.
Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, Baldassarre M, Quarta S, Vandewynckel YP, Vandierendonck A, Van Vlierberghe H, Porretti L, Negrini M, Bolondi L, Gramantieri L, Fornari F. Pollutri D, et al. Cell Death Dis. 2018 Jan 5;9(1):4. doi: 10.1038/s41419-017-0076-6. Cell Death Dis. 2018. PMID: 29305580 Free PMC article. - A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma.
Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V, Candy P, Epis M, Wintle L, Kalinowski F, Kopp C, Stuart LM, Yeoh GC, George J, Leedman PJ. Kabir TD, et al. Hepatology. 2018 Jan;67(1):216-231. doi: 10.1002/hep.29478. Epub 2017 Nov 29. Hepatology. 2018. PMID: 28833396 - miR-1226-3p Promotes Sorafenib Sensitivity of Hepatocellular Carcinoma via Downregulation of DUSP4 Expression.
Chen X, Tan W, Li W, Li W, Zhu S, Zhong J, Shang C, Chen Y. Chen X, et al. J Cancer. 2019 Jun 2;10(12):2745-2753. doi: 10.7150/jca.31804. eCollection 2019. J Cancer. 2019. PMID: 31258782 Free PMC article. - MicroRNAs and SerpinB3 in hepatocellular carcinoma.
Turato C, Simonato D, Quarta S, Gatta A, Pontisso P. Turato C, et al. Life Sci. 2014 Mar 28;100(1):9-17. doi: 10.1016/j.lfs.2014.01.073. Epub 2014 Feb 2. Life Sci. 2014. PMID: 24496037 Review. - Role of SERPINB3 in hepatocellular carcinoma.
Pontisso P. Pontisso P. Ann Hepatol. 2014 Nov-Dec;13(6):722-7. Ann Hepatol. 2014. PMID: 25332258 Review.
Cited by
- Novel Molecular Targets for Immune Surveillance of Hepatocellular Carcinoma.
Guerra P, Martini A, Pontisso P, Angeli P. Guerra P, et al. Cancers (Basel). 2023 Jul 15;15(14):3629. doi: 10.3390/cancers15143629. Cancers (Basel). 2023. PMID: 37509293 Free PMC article. Review. - miR-122 and miR-197 expressions in hepatic carcinoma patients before and after chemotherapy and their effect on patient prognosis.
Zhan G, Jiang H, Yang R, Yang K. Zhan G, et al. Am J Transl Res. 2021 Jun 15;13(6):6731-6737. eCollection 2021. Am J Transl Res. 2021. PMID: 34306419 Free PMC article. - CXCL5/NF-_κ_B Pathway as a Therapeutic Target in Hepatocellular Carcinoma Treatment.
Jia X, Wei S, Xiong W. Jia X, et al. J Oncol. 2021 May 31;2021:9919494. doi: 10.1155/2021/9919494. eCollection 2021. J Oncol. 2021. PMID: 34194499 Free PMC article. - Mechanisms Underlying Hepatocellular Carcinoma Progression in Patients with Type 2 Diabetes.
Shi T, Kobara H, Oura K, Masaki T. Shi T, et al. J Hepatocell Carcinoma. 2021 Feb 11;8:45-55. doi: 10.2147/JHC.S274933. eCollection 2021. J Hepatocell Carcinoma. 2021. PMID: 33604315 Free PMC article. Review. - SerpinB3: A Multifaceted Player in Health and Disease-Review and Future Perspectives.
Cagnin S, Pontisso P, Martini A. Cagnin S, et al. Cancers (Basel). 2024 Jul 18;16(14):2579. doi: 10.3390/cancers16142579. Cancers (Basel). 2024. PMID: 39061218 Free PMC article. Review.
References
Grants and funding
- [ 2010-2012/Programma di Ricerca Regione-Università 2010-2012, Regione Emilia-Romagna, Bando "Alessandro Liberati", "Identification of innovative microRNAs-based biomarkers and anticancer strategies for the treatment of Hepatocellular carcinoma" to F.F](/?term=2010-2012%2FProgramma+di+Ricerca+Regione-Universit%C3%A0+2010-2012%2C+Regione+Emilia-Romagna%2C+Bando+%22Alessandro+Liberati%22%2C+%22Identification+of+innovative+microRNAs-based+biomarkers+and+anticancer+strategies+for+the+treatment+of+Hepatocellular+carcinoma%22+to+F.F%5BGrants+and+Funding%5D&sort=date&sort%5Forder=desc "All articles for grant 2010-2012/Programma di Ricerca Regione-Università 2010-2012, Regione Emilia-Romagna, Bando "Alessandro Liberati", "Identification of innovative microRNAs-based biomarkers and anticancer strategies for the treatment of Hepatocellular carcinoma" to F.F")
- [ 2012/Programma di Ricerca Regione-Università 2010-2012, Regione Emilia-Romagna, Bando "Ricerca Innovativa", "Innovative approaches to the diagnosis and pharmacogenetic-based therapies of primary hepatic tumors, peripheral B, and T-cell lymphomas and lymphoblas](/?term=2012%2FProgramma+di+Ricerca+Regione-Universit%C3%A0+2010-2012%2C+Regione+Emilia-Romagna%2C+Bando+%22Ricerca+Innovativa%22%2C+%22Innovative+approaches+to+the+diagnosis+and+pharmacogenetic-based+therapies+of+primary+hepatic+tumors%2C+peripheral+B%2C+and+T-cell+lymphomas+and+lymphoblas%5BGrants+and+Funding%5D&sort=date&sort%5Forder=desc "All articles for grant 2012/Programma di Ricerca Regione-Università 2010-2012, Regione Emilia-Romagna, Bando "Ricerca Innovativa", "Innovative approaches to the diagnosis and pharmacogenetic-based therapies of primary hepatic tumors, peripheral B, and T-cell lymphomas and lymphoblas")
- [ 2014/Progetto Strategico 2011-2014 Ministero Italiano della Ricerca Scientifica e Tecnologica - University of Padova "Nanochemistry and medicine for cancer: from diagnosis to treatment" to PP.](/?term=2014%2FProgetto+Strategico+2011-2014+Ministero+Italiano+della+Ricerca+Scientifica+e+Tecnologica+-+University+of+Padova+%22Nanochemistry+and+medicine+for+cancer%3A+from+diagnosis+to+treatment%22+to+PP.%5BGrants+and+Funding%5D&sort=date&sort%5Forder=desc "All articles for grant 2014/Progetto Strategico 2011-2014 Ministero Italiano della Ricerca Scientifica e Tecnologica - University of Padova "Nanochemistry and medicine for cancer: from diagnosis to treatment" to PP.")
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials