Hypergravity disrupts murine intestinal microbiota - PubMed (original) (raw)
Hypergravity disrupts murine intestinal microbiota
Corentine Alauzet et al. Sci Rep. 2019.
Abstract
During spaceflight, organisms are subjected to various physical stressors including modification of gravity (G) that, associated with lifestyle, could lead to impaired immunity, intestinal dysbiosis and thus potentially predispose astronauts to illness. Whether space travel affects microbiota homeostasis has not been thoroughly investigated. The aim of this study was to evaluate changes in intestinal microbiota and mucosa in a ground-based murine model consisting in a 21-days confinement of mice in a centrifuge running at 2 or 3G. Results revealed an increased α-diversity and a significant change in intracaecal β-diversity observed only at 3G, with profiles characterized by a decrease of the Firmicutes/Bacteroidetes ratio. Compared to 1G microbiota, 12.1% of the taxa were significantly impacted in 3G microbiota, most of them (78%) being enriched. This study shows a G-level-dependent disruption of intracaecal microbiota, without alteration of mucosal integrity. These first data reinforce those recently obtained with in-flight experimentations or microgravity models, and emphasize the critical need for further studies exploring the impact of spaceflight on intestinal microbiota in order to optimize long-term space travel conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures
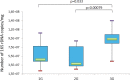
Figure 1
Total bacterial load quantification by qPCR corresponding to the total number of 16S rRNA encoding gene copies per mg of intracaecal content of mice subjected to 2G (n = 10) or 3G (n = 11) hypergravity and 1G control mice (n = 7). Statistical analyses were done using the Mann-Whitney U test. The upper and lower ranges of the box represent the 75% and 25% quartiles, respectively. Error bars reflect standard error of the mean.
Figure 2
Phylogenetic diversity of murine intracaecal microbiomes at 1G (n = 7), 2G (n = 10) or 3G (n = 11). Box plots depict microbiomes diversity differences according to the (A) Shannon index, (B) Evenness index, (C) Simpson’s diversity index and (D) Simpson’s reciprocal index. Statistical analyses were done using the Mann-Whitney U test. The upper and lower ranges of the box represent the 75% and 25% quartiles, respectively. Error bars reflect standard error of the mean.
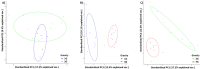
Figure 3
Principal Component Analysis (PCA) of microbiomes of (A) 2G mice versus 1G mice; (B) 3G mice versus 1G mice, and (C) 2G mice versus 3G mice. The variance explained by each of the main two dimensions of the PCA is indicated in parentheses on the axes.
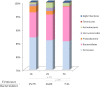
Figure 4
Relative abundance (%) of bacterial phyla and Firmicutes/Bacteroidetes ratio in the caecal content of mice subjected to hypergravity at 2G and 3G compared to control mice (1G). A significant decrease of Verrucomicrobia (p = 0.0095) was noted in 3G samples by comparison to 2G samples. Significant lower levels of Proteobacteria (p = 0.0224 and 0.0095) were noted in 3G samples by comparison to 1G and to 2G samples (p = 0.0224 and p = 0.0095, respectively). Furthermore, higher levels of Deferribacteres (p = 0.078) were noted in 3G samples by comparison to 1G samples. Statistical analyses were done using the Mann-Whitney U test.
Figure 5
Bacterial species significantly enriched or impoverished in 2G samples compared to 1G samples (2G/1G), and in 3G samples compared to 1G samples (3G/1G) and to 2G samples (3G/2G). Taxa names in brackets correspond to species whose classification has to be re-evaluated. Statistical analyses were done using the Mann-Whitney U test.
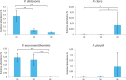
Figure 6
Intracaecal 16S rDNA gene-based real-time PCR quantification of the relative abundance of Parabacteroides distasonis, Paraprevotella clara, Parasutterella excrementihominis, and Flavonifractor plautii. Data are expressed as the relative proportion of the number of 16S rDNA copies of each target to the total number of 16S rDNA copies per mg of intracaecal content of mice subjected to 2G (n = 10) or 3G (n = 11) hypergravity and 1G control mice (n = 7). Statistical analyses were done using the Mann-Whitney U test. *p < 0.05; **p < 0.0005. Error bars reflect standard error of the mean.
Similar articles
- Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota.
Alauzet C, Cunat L, Wack M, Lanfumey L, Legrand-Frossi C, Lozniewski A, Agrinier N, Cailliez-Grimal C, Frippiat JP. Alauzet C, et al. Int J Mol Sci. 2020 Oct 23;21(21):7863. doi: 10.3390/ijms21217863. Int J Mol Sci. 2020. PMID: 33114008 Free PMC article. - Space Environmental Factor Impacts upon Murine Colon Microbiota and Mucosal Homeostasis.
Ritchie LE, Taddeo SS, Weeks BR, Lima F, Bloomfield SA, Azcarate-Peril MA, Zwart SR, Smith SM, Turner ND. Ritchie LE, et al. PLoS One. 2015 Jun 17;10(6):e0125792. doi: 10.1371/journal.pone.0125792. eCollection 2015. PLoS One. 2015. PMID: 26083373 Free PMC article. - Intestinal microbiota contributes to colonic epithelial changes in simulated microgravity mouse model.
Shi J, Wang Y, He J, Li P, Jin R, Wang K, Xu X, Hao J, Zhang Y, Liu H, Chen X, Wu H, Ge Q. Shi J, et al. FASEB J. 2017 Aug;31(8):3695-3709. doi: 10.1096/fj.201700034R. Epub 2017 May 11. FASEB J. 2017. PMID: 28495755 - The human microbiome in space: parallels between Earth-based dysbiosis, implications for long-duration spaceflight, and possible mitigation strategies.
Etlin S, Rose J, Bielski L, Walter C, Kleinman AS, Mason CE. Etlin S, et al. Clin Microbiol Rev. 2024 Sep 12;37(3):e0016322. doi: 10.1128/cmr.00163-22. Epub 2024 Aug 13. Clin Microbiol Rev. 2024. PMID: 39136453 Review. - The brain in micro- and hypergravity: the effects of changing gravity on the brain electrocortical activity.
Marušič U, Meeusen R, Pišot R, Kavcic V. Marušič U, et al. Eur J Sport Sci. 2014;14(8):813-22. doi: 10.1080/17461391.2014.908959. Epub 2014 Apr 15. Eur J Sport Sci. 2014. PMID: 24734884 Review.
Cited by
- Pharmacological Innovations in Space: Challenges and Future Perspectives.
Aksoyalp ZŞ, Temel A, Karpuz M. Aksoyalp ZŞ, et al. Pharm Res. 2024 Nov 12. doi: 10.1007/s11095-024-03788-x. Online ahead of print. Pharm Res. 2024. PMID: 39532779 Review. - Synergistic interplay between radiation and microgravity in spaceflight-related immunological health risks.
Wadhwa A, Moreno-Villanueva M, Crucian B, Wu H. Wadhwa A, et al. Immun Ageing. 2024 Jul 20;21(1):50. doi: 10.1186/s12979-024-00449-w. Immun Ageing. 2024. PMID: 39033285 Free PMC article. Review. - Positive modulation of a new reconstructed human gut microbiota by Maitake extract helpfully boosts the intestinal environment in vitro.
De Giani A, Perillo F, Baeri A, Finazzi M, Facciotti F, Di Gennaro P. De Giani A, et al. PLoS One. 2024 Apr 11;19(4):e0301822. doi: 10.1371/journal.pone.0301822. eCollection 2024. PLoS One. 2024. PMID: 38603764 Free PMC article. - Impact of Microgravity and Other Spaceflight Factors on Retina of Vertebrates and Humans In Vivo and In Vitro.
Grigoryan EN. Grigoryan EN. Life (Basel). 2023 May 26;13(6):1263. doi: 10.3390/life13061263. Life (Basel). 2023. PMID: 37374046 Free PMC article. Review. - Hypergravity Increases Blood-Brain Barrier Permeability to Fluorescent Dextran and Antisense Oligonucleotide in Mice.
Dubayle D, Vanden-Bossche A, Peixoto T, Morel JL. Dubayle D, et al. Cells. 2023 Feb 24;12(5):734. doi: 10.3390/cells12050734. Cells. 2023. PMID: 36899870 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources