MAPK1, CPTAC-870 - CPTAC Assay Portal (original) (raw)
Please include the following statement when referencing the CPTAC Assay Portal
We would like to acknowledge the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (assays.cancer.gov) for developing assays and establishing criteria for the assays described in this publication.
Protein Sequence hover to view complete sequence
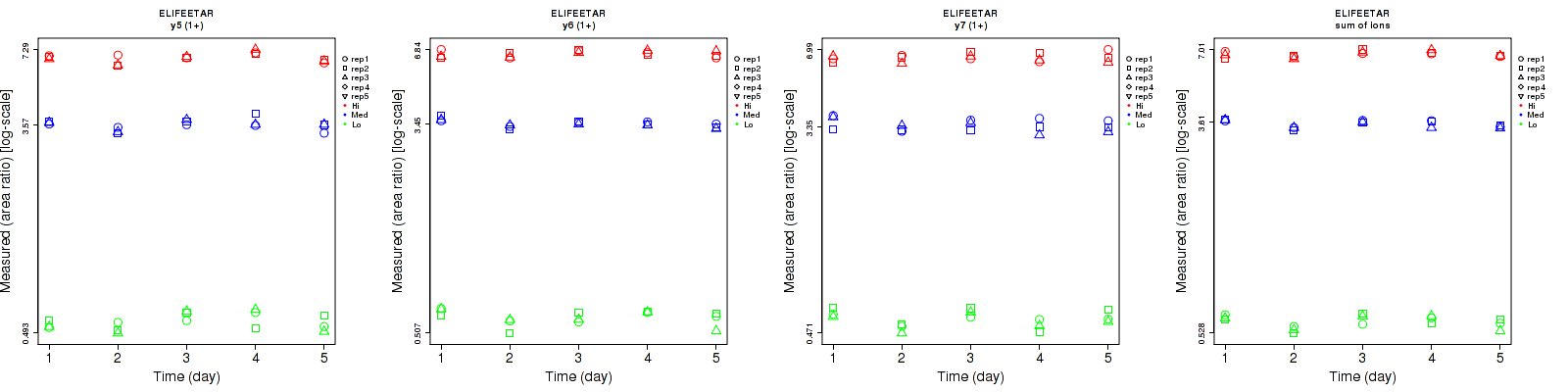
non-CPTAC-6107: View additional ELIFEETAR data
CPTAC-871: View additional ELIFEETAR data
| 10 | 20 | 30 | 40 | 50 |
|---|---|---|---|---|
| MAAAAAAGAG | PEMVRGQVFD | VGPRYTNLSY | IGEGAYGMVC | SAYDNVNKVR |
| 60 | 70 | 80 | 90 | 100 |
| VAIKKISPFE | HQTYCQRTLR | EIKILLRFRH | ENIIGINDII | RAPTIEQMKD |
| 110 | 120 | 130 | 140 | 150 |
| VYIVQDLMET | DLYKLLKTQH | LSNDHICYFL | YQILRGLKYI | HSANVLHRDL |
| 160 | 170 | 180 | 190 | 200 |
| KPSNLLLNTT | CDLKICDFGL | ARVADPDHDH | TGFLTEYVAT | RWYRAPEIML |
| 210 | 220 | 230 | 240 | 250 |
| NSKGYTKSID | IWSVGCILAE | MLSNRPIFPG | KHYLDQLNHI | LGILGSPSQE |
| 260 | 270 | 280 | 290 | 300 |
| DLNCIINLKA | RNYLLSLPHK | NKVPWNRLFP | NADSKALDLL | DKMLTFNPHK |
| 310 | 320 | 330 | 340 | 350 |
| RIEVEQALAH | PYLEQYYDPS | DEPIAEAPFK | FDMELDDLPK | EKLKELIFEE |
| 360 | ||||
| TARFQPGYRS |
Data source: UniProt
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database Entry PhosphoSitePlus ®
Click a point on a node
to view detailed assay information below
All other points link out to UniProt
Phosphorylation Acetylation Ubiquitylation Other
loading

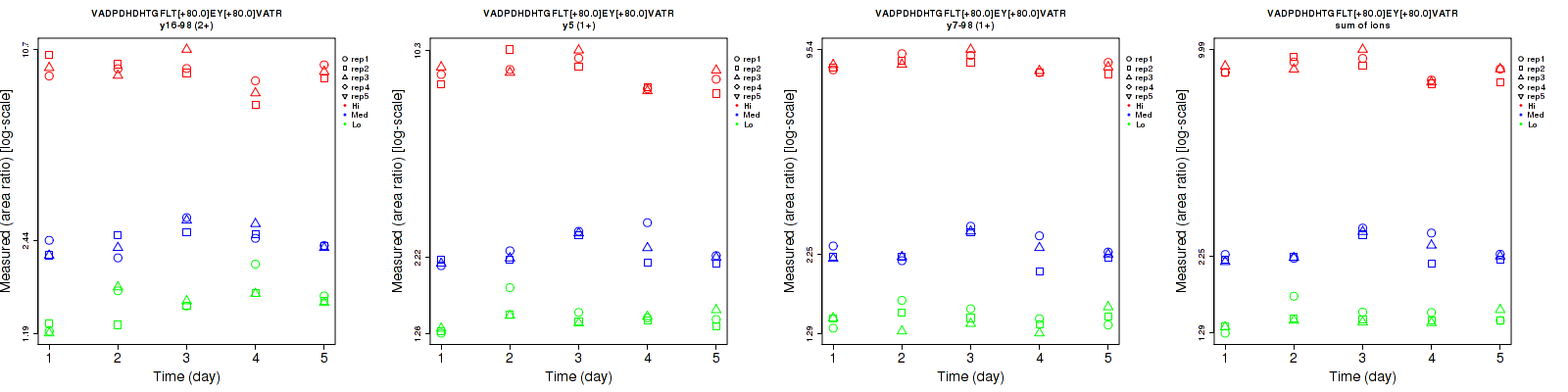
Assay Details for CPTAC-1352 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+80.0]EY[+80.0]VATR
Modification Type
Phospho (ST), Phospho (Y)
Protein - Site of Modification
185, 187
Peptide - Site of Modification
13, 15
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-1352
Peptide Molecular Mass
2,302.9297
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
digested cell lysate
Submitting Laboratory
Broad Institute
Submitting Lab PI
Steven A. Carr
Publication
View Details (opens in a new window)
Mol Cell Proteomics. 2016 May;15(5):1622-41. doi: 10.1074/mcp.M116.058354. Epub 2016 Feb 24. Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Abelin JG1, Patel J1, Lu X1, Feeney CM1, Fagbami L1, Creech AL1, Hu R1, Lam D1, Davison D1, Pino L1, Qiao JW1, Kuhn E1, Officer A1, Li J2, Abbatiello S1, Subramanian A1, Sidman R2, Snyder E3, Carr SA1, Jaffe JD4.
View Details (opens in a new window)
Mol Cell Proteomics. 2014 Jul;13(7):1690-704. doi: 10.1074/mcp.M113.036392. Epub 2014 Apr 9. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mertins P1, Yang F2, Liu T2, Mani DR3, Petyuk VA2, Gillette MA3, Clauser KR3, Qiao JW3, Gritsenko MA2, Moore RJ2, Levine DA4, Townsend R5, Erdmann-Gilmore P5, Snider JE5, Davies SR5, Ruggles KV6, Fenyo D6, Kitchens RT5, Li S5, Olvera N4, Dao F4, Rodriguez H7, Chan DW8, Liebler D9, White F10, Rodland KD2, Mills GB11, Smith RD2, Paulovich AG12, Ellis M5, Carr SA1
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Quantiva
Internal Standard
light stable isotope peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Easy NanoLC1000
Column Packing
Reprosil C18, 1.9um, 200A
Column Dimensions
0.075 x 150 mm
Flow Rate
200 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
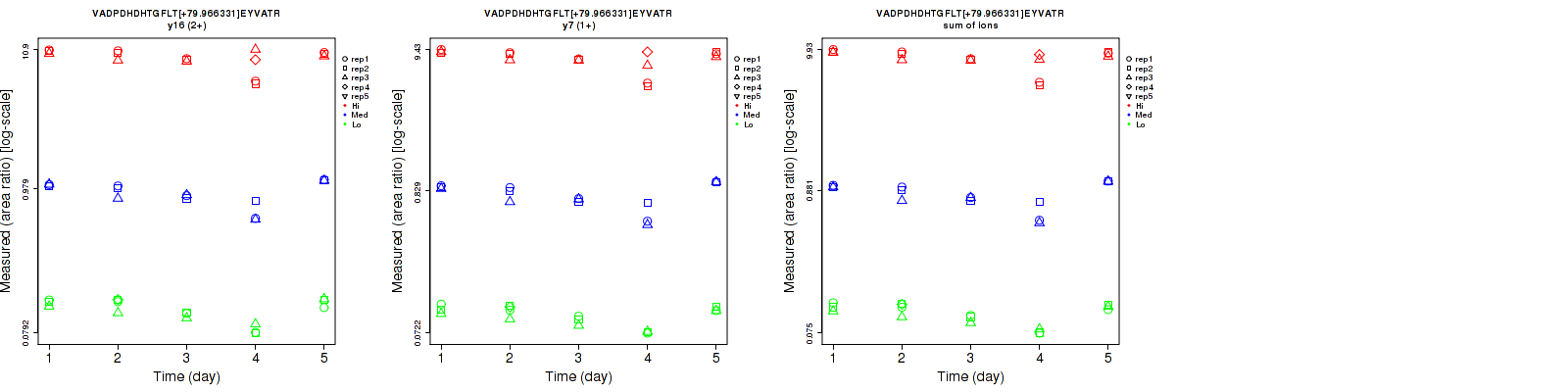
| y16-98 (2+) | 7.6 | 5.7 | 7.5 | 14.2 | 10.5 | 10.4 | 16.1 | 11.9 | 12.8 | 15 | 15 | 15 |
| y5 (1+) | 5.2 | 5 | 6.4 | 7.6 | 10.6 | 11 | 9.2 | 11.7 | 12.7 | 15 | 15 | 15 |
| y7-98 (1+) | 6.3 | 4.6 | 3 | 6.2 | 9.4 | 5.3 | 8.8 | 10.5 | 6.1 | 15 | 15 | 15 |
| sum | 5 | 3.8 | 3.9 | 5.6 | 9.3 | 7.6 | 7.5 | 10 | 8.5 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y16-98 (2+) | 7.6 | 5.7 | 7.5 | 14.2 | 10.5 | 10.4 | 16.1 | 11.9 | 12.8 | 15 | 15 | 15 |
| y5 (1+) | 5.2 | 5 | 6.4 | 7.6 | 10.6 | 11 | 9.2 | 11.7 | 12.7 | 15 | 15 | 15 |
| y7-98 (1+) | 6.3 | 4.6 | 3 | 6.2 | 9.4 | 5.3 | 8.8 | 10.5 | 6.1 | 15 | 15 | 15 |
| sum | 5 | 3.8 | 3.9 | 5.6 | 9.3 | 7.6 | 7.5 | 10 | 8.5 | 15 | 15 | 15 |
Additional Resources and Comments
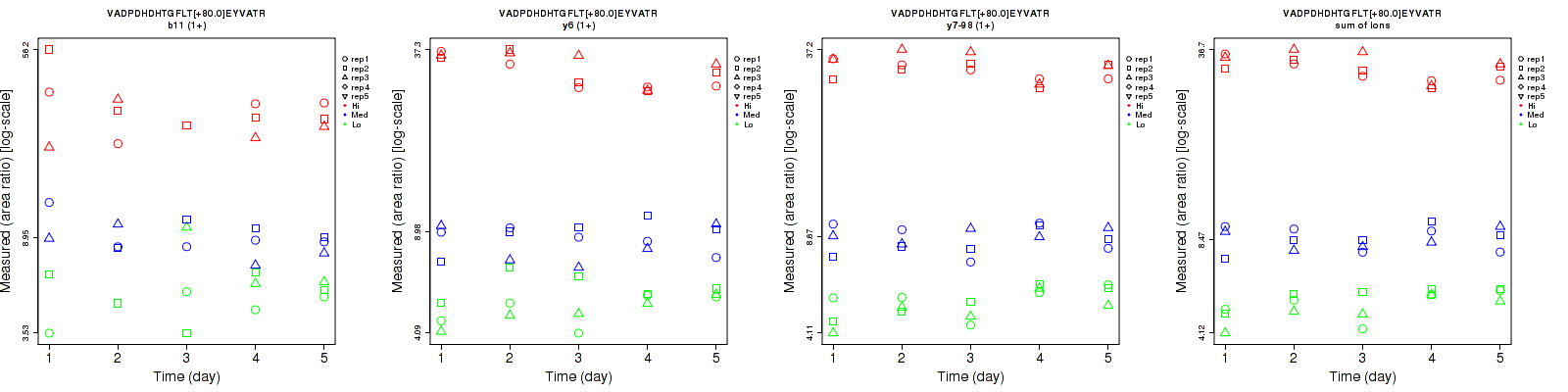
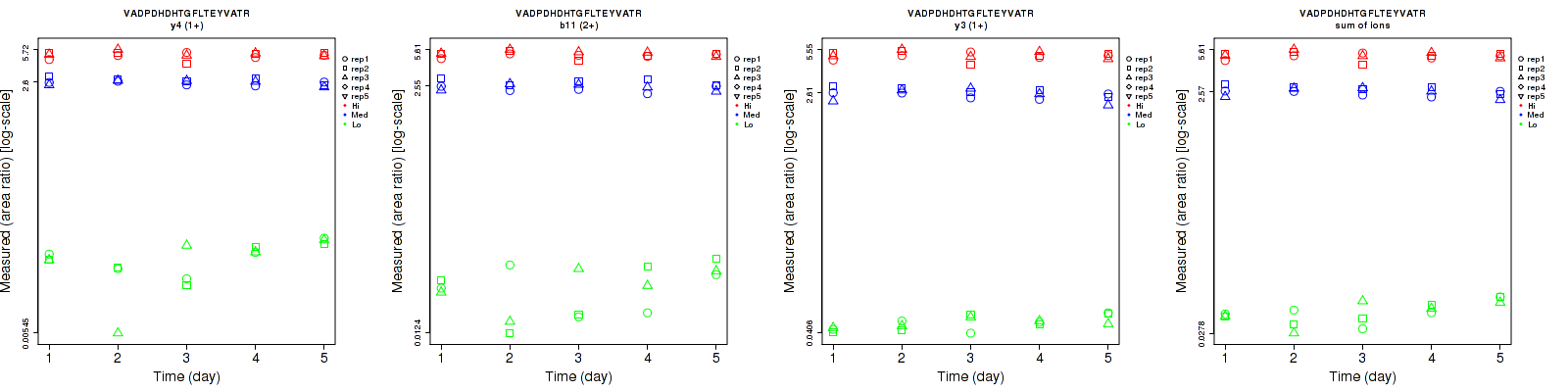
Assay Details for CPTAC-1353 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+80.0]EYVATR
Modification Type
Phospho (ST)
Protein - Site of Modification
185
Peptide - Site of Modification
13
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-1353
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
digested cell lysate
Submitting Laboratory
Broad Institute
Submitting Lab PI
Steven A. Carr
Publication
View Details (opens in a new window)
Mol Cell Proteomics. 2016 May;15(5):1622-41. doi: 10.1074/mcp.M116.058354. Epub 2016 Feb 24. Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Abelin JG1, Patel J1, Lu X1, Feeney CM1, Fagbami L1, Creech AL1, Hu R1, Lam D1, Davison D1, Pino L1, Qiao JW1, Kuhn E1, Officer A1, Li J2, Abbatiello S1, Subramanian A1, Sidman R2, Snyder E3, Carr SA1, Jaffe JD4.
View Details (opens in a new window)
Mol Cell Proteomics. 2014 Jul;13(7):1690-704. doi: 10.1074/mcp.M113.036392. Epub 2014 Apr 9. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mertins P1, Yang F2, Liu T2, Mani DR3, Petyuk VA2, Gillette MA3, Clauser KR3, Qiao JW3, Gritsenko MA2, Moore RJ2, Levine DA4, Townsend R5, Erdmann-Gilmore P5, Snider JE5, Davies SR5, Ruggles KV6, Fenyo D6, Kitchens RT5, Li S5, Olvera N4, Dao F4, Rodriguez H7, Chan DW8, Liebler D9, White F10, Rodland KD2, Mills GB11, Smith RD2, Paulovich AG12, Ellis M5, Carr SA1
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Quantiva
Internal Standard
light stable isotope peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Easy NanoLC1000
Column Packing
Reprosil C18, 1.9um, 200A
Column Dimensions
0.075 x 150 mm
Flow Rate
200 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b11 (1+) | 29.4 | 16.5 | 23.7 | 24.3 | 16.6 | 25.8 | 38.1 | 23.4 | 35 | 12 | 13 | 13 |
| y6 (1+) | 12.4 | 14 | 6.5 | 11.4 | 12.1 | 12.7 | 16.8 | 18.5 | 14.3 | 15 | 15 | 15 |
| y7-98 (1+) | 8.2 | 9.4 | 6.8 | 11.9 | 9.2 | 8.5 | 14.5 | 13.2 | 10.9 | 15 | 15 | 15 |
| sum | 7.7 | 8.8 | 6.2 | 9.9 | 9.3 | 9.3 | 12.5 | 12.8 | 11.2 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b11 (1+) | 29.4 | 16.5 | 23.7 | 24.3 | 16.6 | 25.8 | 38.1 | 23.4 | 35 | 12 | 13 | 13 |
| y6 (1+) | 12.4 | 14 | 6.5 | 11.4 | 12.1 | 12.7 | 16.8 | 18.5 | 14.3 | 15 | 15 | 15 |
| y7-98 (1+) | 8.2 | 9.4 | 6.8 | 11.9 | 9.2 | 8.5 | 14.5 | 13.2 | 10.9 | 15 | 15 | 15 |
| sum | 7.7 | 8.8 | 6.2 | 9.9 | 9.3 | 9.3 | 12.5 | 12.8 | 11.2 | 15 | 15 | 15 |
Additional Resources and Comments
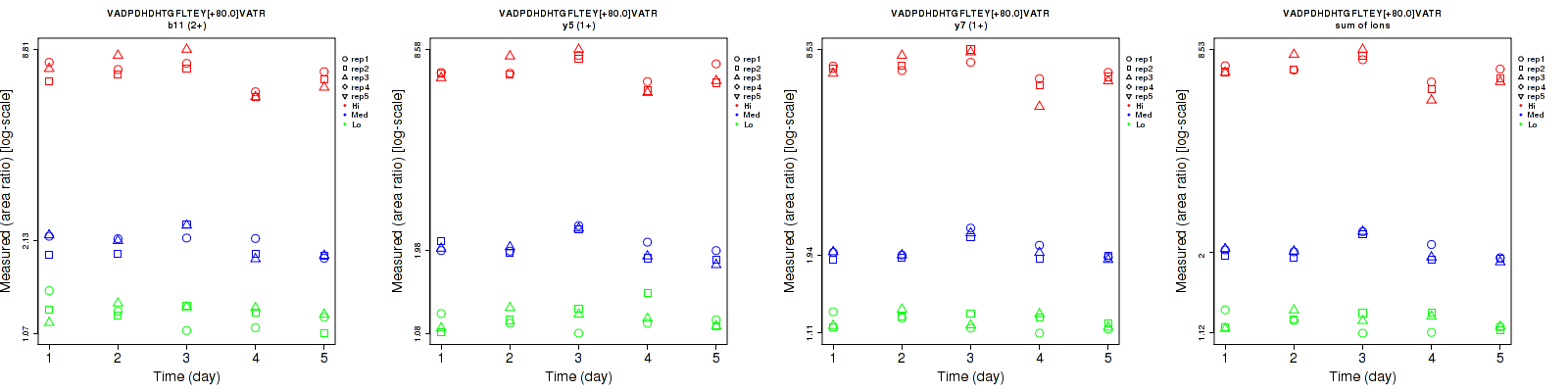
Assay Details for CPTAC-1354 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLTEY[+80.0]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
187
Peptide - Site of Modification
15
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-1354
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
digested cell lysate
Submitting Laboratory
Broad Institute
Submitting Lab PI
Steven A. Carr
Publication
View Details (opens in a new window)
Mol Cell Proteomics. 2016 May;15(5):1622-41. doi: 10.1074/mcp.M116.058354. Epub 2016 Feb 24. Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Abelin JG1, Patel J1, Lu X1, Feeney CM1, Fagbami L1, Creech AL1, Hu R1, Lam D1, Davison D1, Pino L1, Qiao JW1, Kuhn E1, Officer A1, Li J2, Abbatiello S1, Subramanian A1, Sidman R2, Snyder E3, Carr SA1, Jaffe JD4.
View Details (opens in a new window)
Mol Cell Proteomics. 2014 Jul;13(7):1690-704. doi: 10.1074/mcp.M113.036392. Epub 2014 Apr 9. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mertins P1, Yang F2, Liu T2, Mani DR3, Petyuk VA2, Gillette MA3, Clauser KR3, Qiao JW3, Gritsenko MA2, Moore RJ2, Levine DA4, Townsend R5, Erdmann-Gilmore P5, Snider JE5, Davies SR5, Ruggles KV6, Fenyo D6, Kitchens RT5, Li S5, Olvera N4, Dao F4, Rodriguez H7, Chan DW8, Liebler D9, White F10, Rodland KD2, Mills GB11, Smith RD2, Paulovich AG12, Ellis M5, Carr SA1
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Quantiva
Internal Standard
light stable isotope peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Easy NanoLC1000
Column Packing
Reprosil C18, 1.9um, 200A
Column Dimensions
0.075 x 150 mm
Flow Rate
200 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
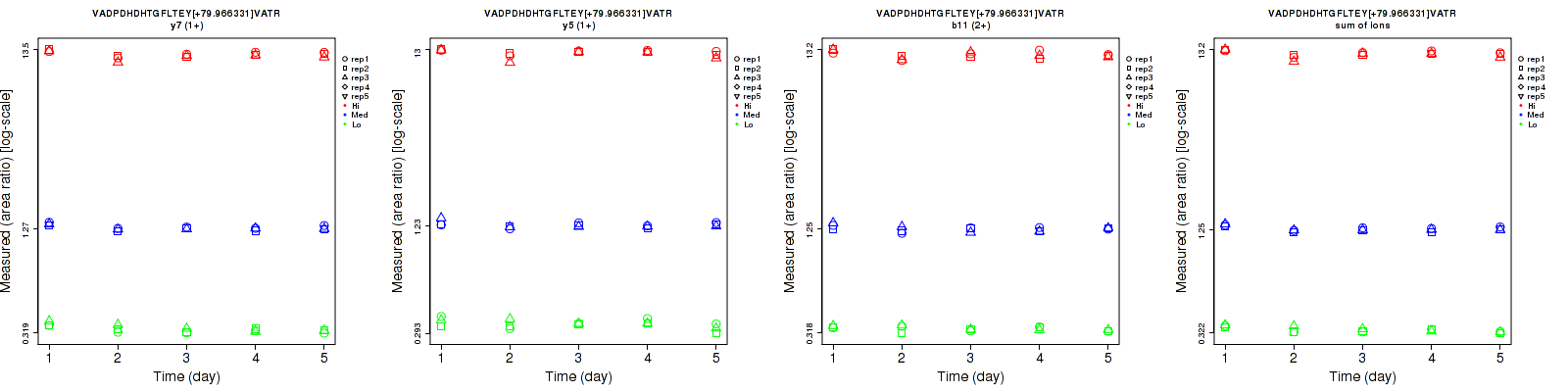
| b11 (2+) | 8.3 | 5.7 | 6 | 8.5 | 9.1 | 10.3 | 11.9 | 10.7 | 11.9 | 15 | 15 | 15 |
| y5 (1+) | 7.5 | 3.8 | 5.1 | 7.6 | 9.3 | 9.7 | 10.7 | 10 | 11 | 15 | 15 | 15 |
| y7 (1+) | 4.9 | 2.8 | 5.3 | 5.5 | 7.8 | 9.8 | 7.4 | 8.3 | 11.1 | 15 | 15 | 15 |
| sum | 5.5 | 2.7 | 4.8 | 6 | 8 | 9.7 | 8.1 | 8.4 | 10.8 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b11 (2+) | 8.3 | 5.7 | 6 | 8.5 | 9.1 | 10.3 | 11.9 | 10.7 | 11.9 | 15 | 15 | 15 |
| y5 (1+) | 7.5 | 3.8 | 5.1 | 7.6 | 9.3 | 9.7 | 10.7 | 10 | 11 | 15 | 15 | 15 |
| y7 (1+) | 4.9 | 2.8 | 5.3 | 5.5 | 7.8 | 9.8 | 7.4 | 8.3 | 11.1 | 15 | 15 | 15 |
| sum | 5.5 | 2.7 | 4.8 | 6 | 8 | 9.7 | 8.1 | 8.4 | 10.8 | 15 | 15 | 15 |
Additional Resources and Comments
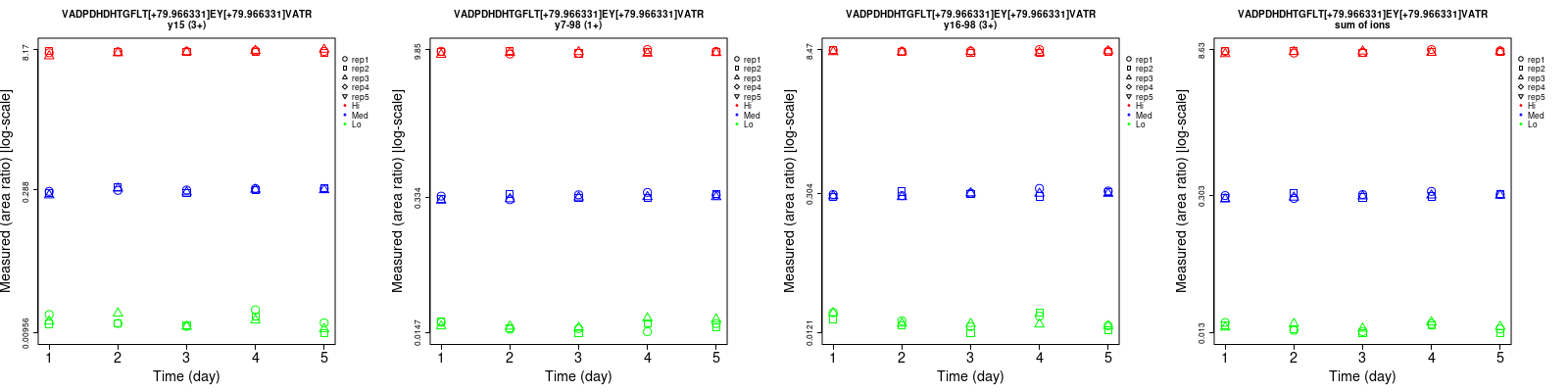
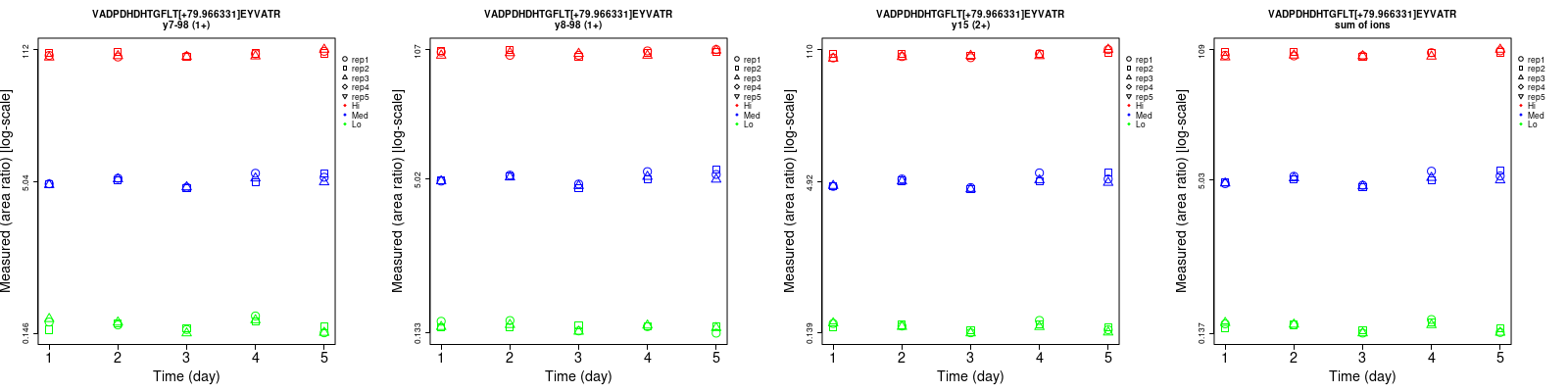
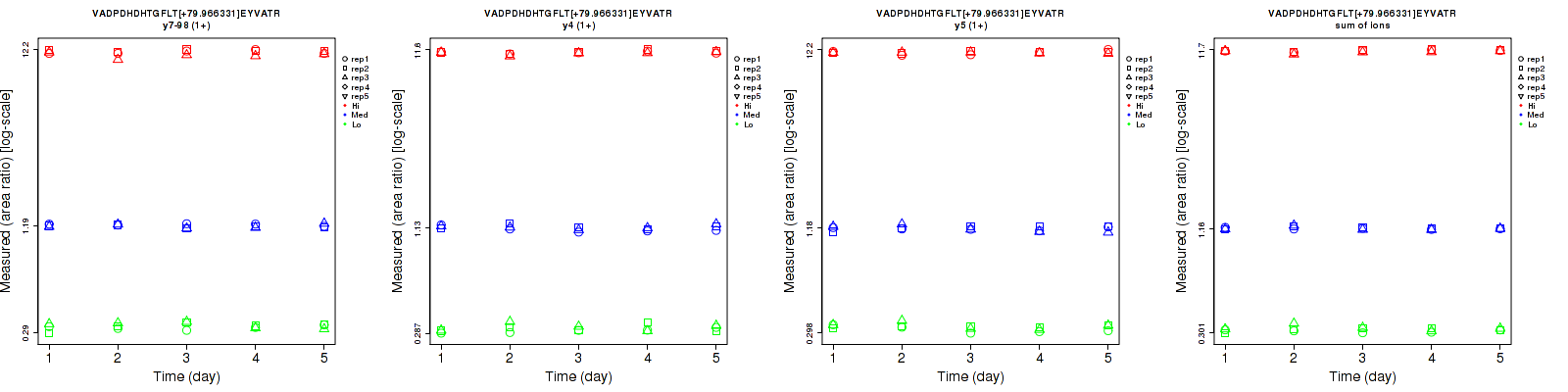
Assay Details for non-CPTAC-6256 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EY[+79.966331]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
8
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-6256
Peptide Molecular Mass
2,302.9297
Species
Human
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
SKBR3 cells
Submitting Laboratory
Cell Signaling Technology
Submitting Lab PI
Yiying Zhu
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Orbitrap Q Exactive (Thermo)
Internal Standard
Heavy isotope-labeled peptides
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
EASY-nLC II (Thermo)
Column Packing
self-packed
Column Dimensions
ID 100um, length 15cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
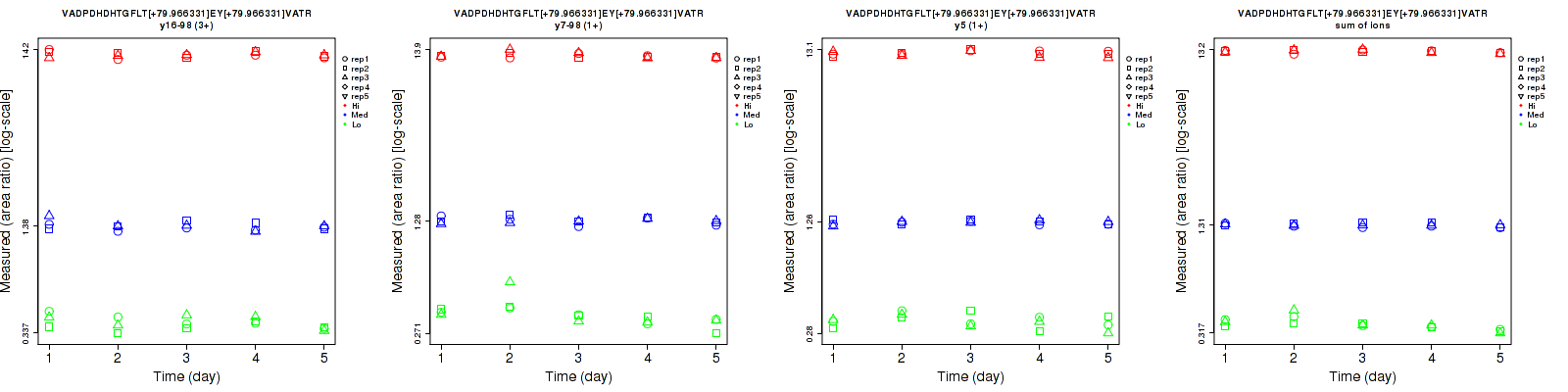
| y7-98 (1+) | 8 | 5.2 | 3.2 | 9.9 | 4.9 | 2.9 | 12.7 | 7.1 | 4.3 | 15 | 15 | 15 |
| y16-98 (2+) | 10 | 5.5 | 3.1 | 11.8 | 5.6 | 2.9 | 15.5 | 7.8 | 4.2 | 15 | 15 | 15 |
| y15 (2+) | 11.7 | 4.8 | 3.2 | 10.9 | 5.5 | 3.2 | 16 | 7.3 | 4.5 | 15 | 15 | 15 |
| y15 (3+) | 10.6 | 3.3 | 2.9 | 15.3 | 5.2 | 3.1 | 18.6 | 6.2 | 4.2 | 15 | 15 | 15 |
| y16-98 (3+) | 8.8 | 4.6 | 2.3 | 15.5 | 5.5 | 2.1 | 17.8 | 7.2 | 3.1 | 15 | 15 | 15 |
| sum | 6 | 4.6 | 2.5 | 8.3 | 4.9 | 2.3 | 10.2 | 6.7 | 3.4 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7-98 (1+) | 8 | 5.2 | 3.2 | 9.9 | 4.9 | 2.9 | 12.7 | 7.1 | 4.3 | 15 | 15 | 15 |
| y16-98 (2+) | 10 | 5.5 | 3.1 | 11.8 | 5.6 | 2.9 | 15.5 | 7.8 | 4.2 | 15 | 15 | 15 |
| y15 (2+) | 11.7 | 4.8 | 3.2 | 10.9 | 5.5 | 3.2 | 16 | 7.3 | 4.5 | 15 | 15 | 15 |
| y15 (3+) | 10.6 | 3.3 | 2.9 | 15.3 | 5.2 | 3.1 | 18.6 | 6.2 | 4.2 | 15 | 15 | 15 |
| y16-98 (3+) | 8.8 | 4.6 | 2.3 | 15.5 | 5.5 | 2.1 | 17.8 | 7.2 | 3.1 | 15 | 15 | 15 |
| sum | 6 | 4.6 | 2.5 | 8.3 | 4.9 | 2.3 | 10.2 | 6.7 | 3.4 | 15 | 15 | 15 |
Additional Resources and Comments
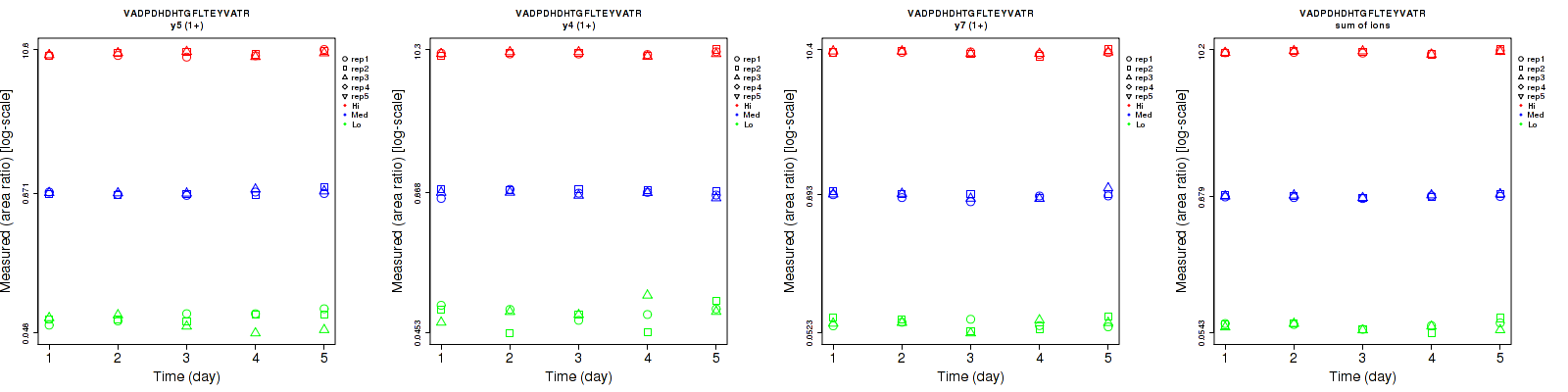
Assay Details for non-CPTAC-6255 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EYVATR
Modification Type
Phospho (ST)
Protein - Site of Modification
N/A
Peptide - Site of Modification
8
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-6255
Peptide Molecular Mass
2,222.9634
Species
Human
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
SKBR3 cells
Submitting Laboratory
Cell Signaling Technology
Submitting Lab PI
Yiying Zhu
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Orbitrap Q Exactive (Thermo)
Internal Standard
Heavy isotope-labeled peptides
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
EASY-nLC II (Thermo)
Column Packing
self-packed
Column Dimensions
ID 100um, length 15cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y8-98 (1+) | 7 | 5.8 | 4.9 | 7.2 | 12 | 5.8 | 10 | 13.3 | 7.6 | 15 | 15 | 15 |
| y7-98 (1+) | 7.5 | 5.1 | 4.3 | 13.2 | 11.7 | 6 | 15.2 | 12.8 | 7.4 | 15 | 15 | 15 |
| y5 (1+) | 5.9 | 7.5 | 4.3 | 10.2 | 12 | 6.2 | 11.8 | 14.2 | 7.5 | 15 | 15 | 15 |
| y16-98 (2+) | 4.7 | 6.5 | 5 | 13 | 12.6 | 5.7 | 13.8 | 14.2 | 7.6 | 15 | 15 | 15 |
| y15 (2+) | 5.1 | 5.7 | 4.1 | 8.9 | 12.5 | 6.5 | 10.3 | 13.7 | 7.7 | 15 | 15 | 15 |
| y16 (2+) | 7.8 | 7.8 | 4.5 | 15.1 | 13.2 | 6.7 | 17 | 15.3 | 8.1 | 15 | 15 | 15 |
| sum | 4.6 | 6 | 4.1 | 10.7 | 12.1 | 5.8 | 11.6 | 13.5 | 7.1 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y8-98 (1+) | 7 | 5.8 | 4.9 | 7.2 | 12 | 5.8 | 10 | 13.3 | 7.6 | 15 | 15 | 15 |
| y7-98 (1+) | 7.5 | 5.1 | 4.3 | 13.2 | 11.7 | 6 | 15.2 | 12.8 | 7.4 | 15 | 15 | 15 |
| y5 (1+) | 5.9 | 7.5 | 4.3 | 10.2 | 12 | 6.2 | 11.8 | 14.2 | 7.5 | 15 | 15 | 15 |
| y16-98 (2+) | 4.7 | 6.5 | 5 | 13 | 12.6 | 5.7 | 13.8 | 14.2 | 7.6 | 15 | 15 | 15 |
| y15 (2+) | 5.1 | 5.7 | 4.1 | 8.9 | 12.5 | 6.5 | 10.3 | 13.7 | 7.7 | 15 | 15 | 15 |
| y16 (2+) | 7.8 | 7.8 | 4.5 | 15.1 | 13.2 | 6.7 | 17 | 15.3 | 8.1 | 15 | 15 | 15 |
| sum | 4.6 | 6 | 4.1 | 10.7 | 12.1 | 5.8 | 11.6 | 13.5 | 7.1 | 15 | 15 | 15 |
Additional Resources and Comments
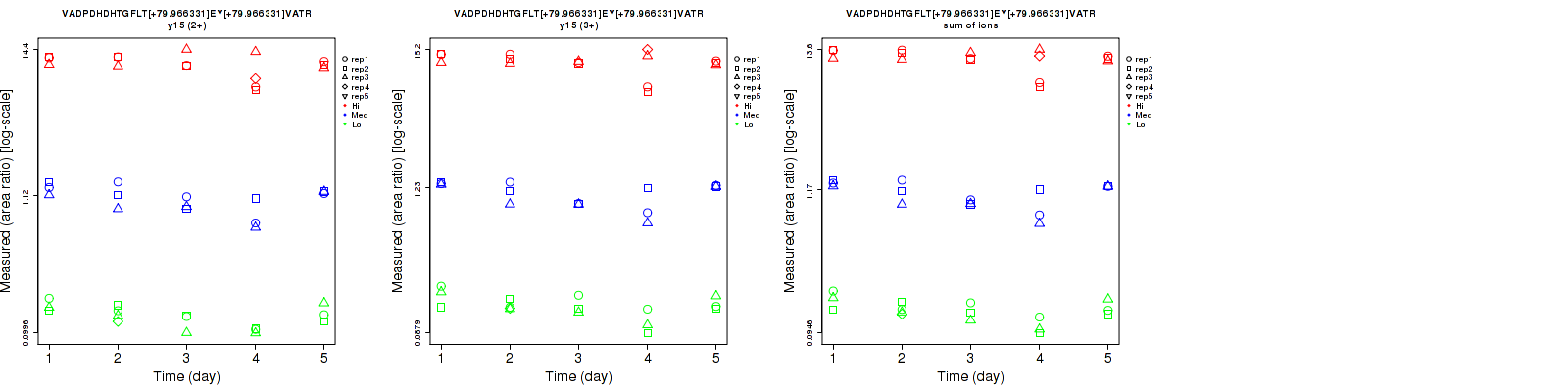
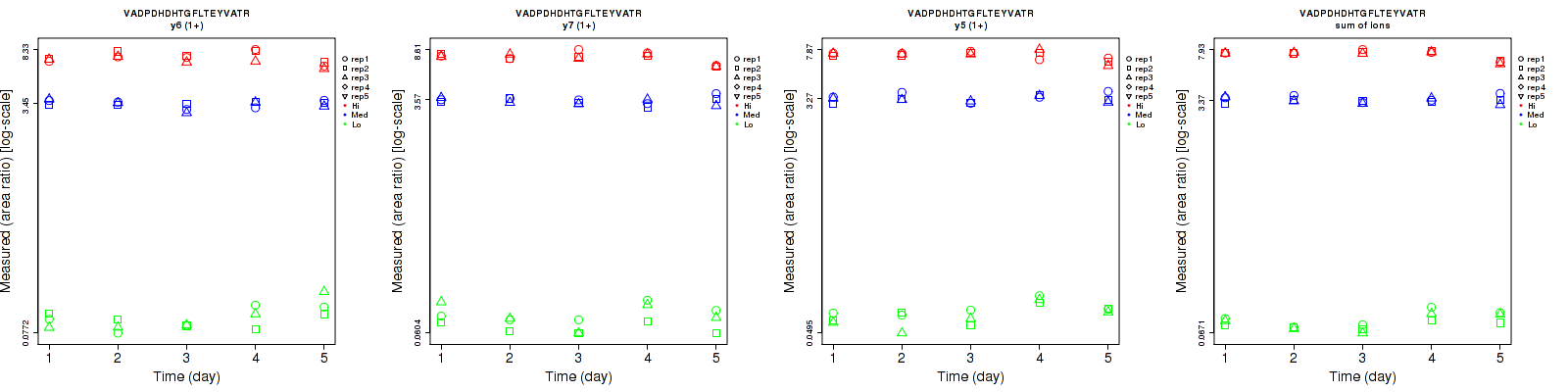
Assay Details for CPTAC-5812 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EY[+79.966331]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-5812
Peptide Molecular Mass
2,302.9297
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y15 (2+) | 12.1 | 15.4 | 14.5 | 19.8 | 21.5 | 18.5 | 23.2 | 26.4 | 23.5 | 16 | 15 | 16 |
| y15 (3+) | 15.8 | 11.6 | 11.9 | 20.9 | 21.5 | 16.8 | 26.2 | 24.4 | 20.6 | 16 | 15 | 16 |
| sum | 13.9 | 12.6 | 11.8 | 19.9 | 21.2 | 17.1 | 24.3 | 24.7 | 20.8 | 16 | 15 | 16 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y15 (2+) | 12.1 | 15.4 | 14.5 | 19.8 | 21.5 | 18.5 | 23.2 | 26.4 | 23.5 | 16 | 15 | 16 |
| y15 (3+) | 15.8 | 11.6 | 11.9 | 20.9 | 21.5 | 16.8 | 26.2 | 24.4 | 20.6 | 16 | 15 | 16 |
| sum | 13.9 | 12.6 | 11.8 | 19.9 | 21.2 | 17.1 | 24.3 | 24.7 | 20.8 | 16 | 15 | 16 |
Additional Resources and Comments
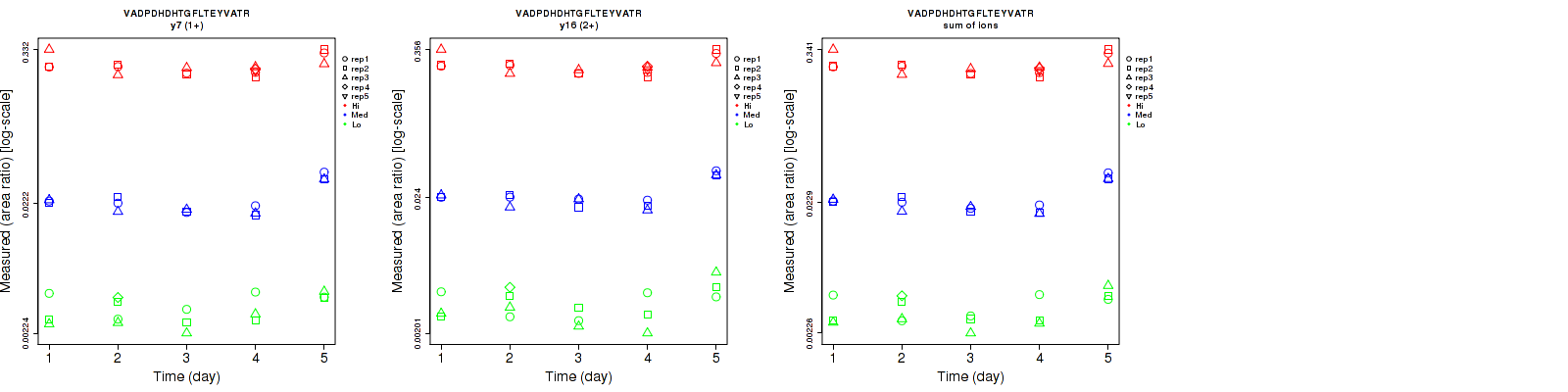
Assay Details for CPTAC-5811 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
VADPDHDHTGFLTEYVATR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-5811
Peptide Molecular Mass
2,142.9970
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 21.4 | 6.9 | 10.7 | 23.8 | 28.6 | 17.6 | 32 | 29.4 | 20.6 | 16 | 15 | 17 |
| y16 (2+) | 25.4 | 7.2 | 10.2 | 32.3 | 26.4 | 17.9 | 41.1 | 27.4 | 20.6 | 16 | 15 | 17 |
| sum | 21.8 | 6.7 | 10.5 | 26.6 | 27.6 | 17.7 | 34.4 | 28.4 | 20.6 | 16 | 15 | 17 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 21.4 | 6.9 | 10.7 | 23.8 | 28.6 | 17.6 | 32 | 29.4 | 20.6 | 16 | 15 | 17 |
| y16 (2+) | 25.4 | 7.2 | 10.2 | 32.3 | 26.4 | 17.9 | 41.1 | 27.4 | 20.6 | 16 | 15 | 17 |
| sum | 21.8 | 6.7 | 10.5 | 26.6 | 27.6 | 17.7 | 34.4 | 28.4 | 20.6 | 16 | 15 | 17 |
Additional Resources and Comments
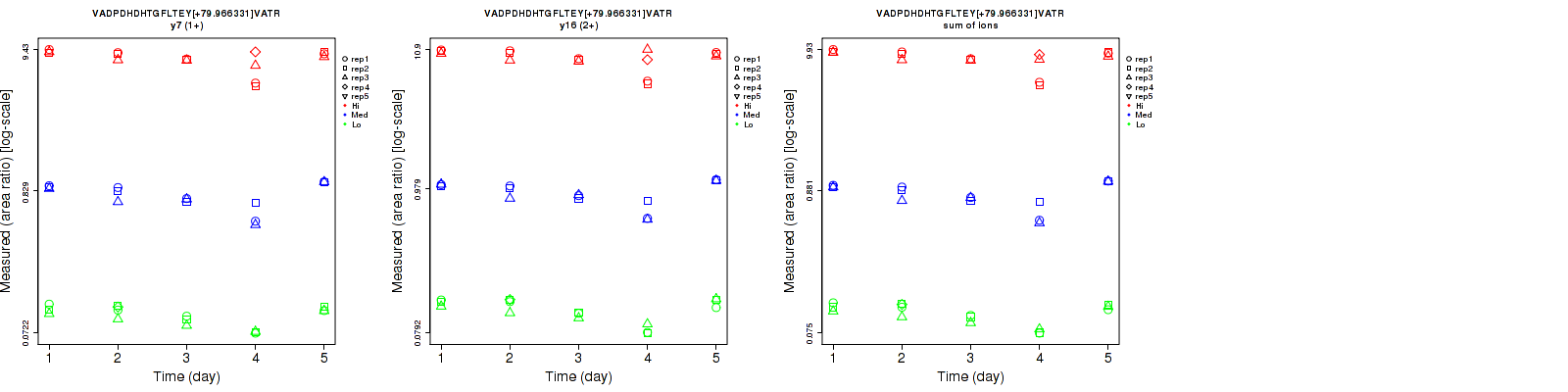
Assay Details for CPTAC-5810 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLTEY[+79.966331]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-5810
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 6.3 | 7.9 | 8.6 | 16.4 | 21.8 | 16.7 | 17.6 | 23.2 | 18.8 | 16 | 15 | 16 |
| y16 (2+) | 7.6 | 7.1 | 9.2 | 19.9 | 20.9 | 16.3 | 21.3 | 22.1 | 18.7 | 16 | 15 | 16 |
| sum | 6.5 | 7.5 | 8.3 | 17.7 | 21.4 | 15.7 | 18.9 | 22.7 | 17.8 | 16 | 15 | 16 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 6.3 | 7.9 | 8.6 | 16.4 | 21.8 | 16.7 | 17.6 | 23.2 | 18.8 | 16 | 15 | 16 |
| y16 (2+) | 7.6 | 7.1 | 9.2 | 19.9 | 20.9 | 16.3 | 21.3 | 22.1 | 18.7 | 16 | 15 | 16 |
| sum | 6.5 | 7.5 | 8.3 | 17.7 | 21.4 | 15.7 | 18.9 | 22.7 | 17.8 | 16 | 15 | 16 |
Additional Resources and Comments
Assay Details for CPTAC-5809 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EYVATR
Modification Type
Phospho (ST)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-5809
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y16 (2+) | 7.6 | 7.1 | 9.2 | 19.9 | 20.9 | 16.3 | 21.3 | 22.1 | 18.7 | 16 | 15 | 16 |
| y7 (1+) | 6.3 | 7.9 | 8.6 | 16.4 | 21.8 | 16.7 | 17.6 | 23.2 | 18.8 | 16 | 15 | 16 |
| sum | 6.5 | 7.5 | 8.3 | 17.7 | 21.4 | 15.7 | 18.9 | 22.7 | 17.8 | 16 | 15 | 16 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y16 (2+) | 7.6 | 7.1 | 9.2 | 19.9 | 20.9 | 16.3 | 21.3 | 22.1 | 18.7 | 16 | 15 | 16 |
| y7 (1+) | 6.3 | 7.9 | 8.6 | 16.4 | 21.8 | 16.7 | 17.6 | 23.2 | 18.8 | 16 | 15 | 16 |
| sum | 6.5 | 7.5 | 8.3 | 17.7 | 21.4 | 15.7 | 18.9 | 22.7 | 17.8 | 16 | 15 | 16 |
Additional Resources and Comments
Assay Details for non-CPTAC-6107 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
ELIFEETAR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
345
Peptide End
353
CPTAC ID
non-CPTAC-6107
Peptide Molecular Mass
1,106.5608
Species
Homo sapiens (Human)
Assay Type
Direct PRM
Matrix
Bovine Serum Albumin
Submitting Laboratory
McGill University
Submitting Lab PI
Christoph H. Borchers, Gerald Batist
Publication
View Details (opens in a new window)
mTORC1-driven protein translation correlates with clinical benefit of capivasertib within a genetically preselected cohort of PIK3CA-altered tumours. Constance A Sobsey, Bjoern C Froehlich, Georgia Mitsa, Sahar Ibrahim, Robert Popp, Rene P Zahedi, Elza C de Bruin, Christoph H Borchers, Gerald Batist
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Agilent 6495B QQQ-MS
Internal Standard
Standard Isotope labeled internal standard peptides with 13Cx, 15Ny (R+10 or K+8)
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Infinity 1290
Column Packing
commercial (Agilent ZORBAX Eclipse Plus C18)
Column Dimensions
4.6 mm × 150 mm, (5 µm)
Flow Rate
0.4 mL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
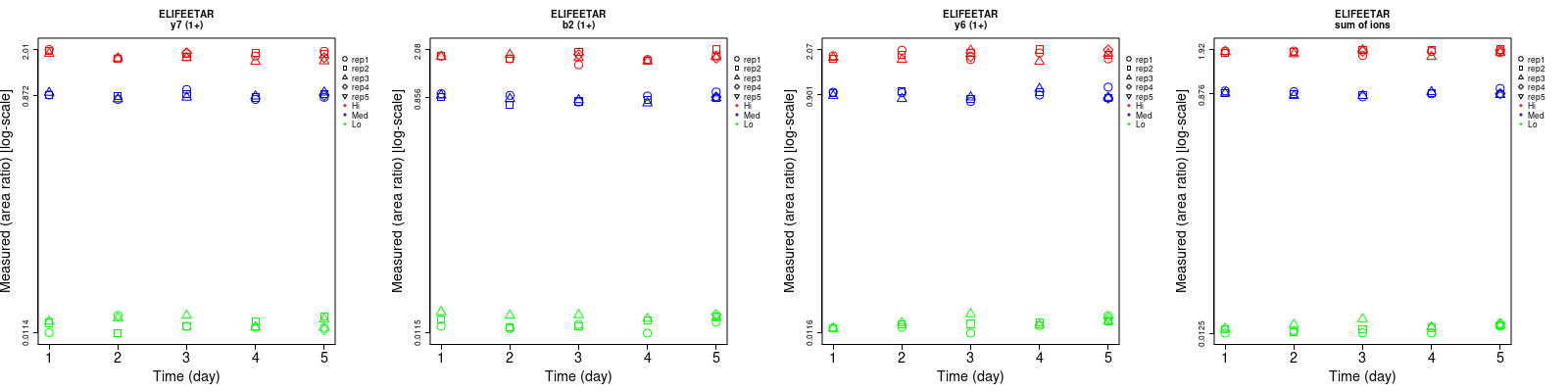
| b2 (1+) | 11.7 | 5.1 | 4.8 | 7.1 | 5.8 | 6.1 | 13.7 | 7.7 | 7.8 | 16 | 16 | 17 |
| y4 (1+) | 11.8 | 7.8 | 5.9 | 12.3 | 7.2 | 5.2 | 17 | 10.6 | 7.9 | 16 | 16 | 17 |
| y6 (1+) | 6.1 | 6.1 | 7.2 | 9 | 7.8 | 7.5 | 10.9 | 9.9 | 10.4 | 16 | 16 | 17 |
| y7 (1+) | 11.4 | 3.9 | 4.9 | 10.4 | 5.3 | 6 | 15.4 | 6.6 | 7.7 | 16 | 16 | 17 |
| sum | 6.5 | 3 | 3.6 | 5.5 | 3.4 | 2.9 | 8.5 | 4.5 | 4.6 | 16 | 16 | 17 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b2 (1+) | 11.7 | 5.1 | 4.8 | 7.1 | 5.8 | 6.1 | 13.7 | 7.7 | 7.8 | 16 | 16 | 17 |
| y4 (1+) | 11.8 | 7.8 | 5.9 | 12.3 | 7.2 | 5.2 | 17 | 10.6 | 7.9 | 16 | 16 | 17 |
| y6 (1+) | 6.1 | 6.1 | 7.2 | 9 | 7.8 | 7.5 | 10.9 | 9.9 | 10.4 | 16 | 16 | 17 |
| y7 (1+) | 11.4 | 3.9 | 4.9 | 10.4 | 5.3 | 6 | 15.4 | 6.6 | 7.7 | 16 | 16 | 17 |
| sum | 6.5 | 3 | 3.6 | 5.5 | 3.4 | 2.9 | 8.5 | 4.5 | 4.6 | 16 | 16 | 17 |
Additional Resources and Comments
Assay Details for non-CPTAC-5722 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLTEY[+79.966331]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-5722
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Thermo Quantiva
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000 RSLCnano
Column Packing
Pepmap 100
Column Dimensions
75um x 25cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y8 (1+) | 5.8 | 3.4 | 3.3 | 4.8 | 4.2 | 4.7 | 7.5 | 5.4 | 5.7 | 15 | 15 | 15 |
| y7 (1+) | 3 | 2 | 2.5 | 4.5 | 3.2 | 4.1 | 5.4 | 3.8 | 4.8 | 15 | 15 | 15 |
| y6 (1+) | 3.5 | 3.5 | 2.5 | 5.2 | 3.9 | 4.6 | 6.3 | 5.2 | 5.2 | 15 | 15 | 15 |
| y5 (1+) | 4.8 | 2.7 | 2.7 | 5.4 | 3.5 | 4.2 | 7.2 | 4.4 | 5 | 15 | 15 | 15 |
| b11 (2+) | 2.2 | 3 | 3.2 | 3 | 3.5 | 4.9 | 3.7 | 4.6 | 5.9 | 15 | 15 | 15 |
| sum | 2.2 | 1.7 | 2.3 | 3.5 | 3 | 4.1 | 4.1 | 3.4 | 4.7 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y8 (1+) | 5.8 | 3.4 | 3.3 | 4.8 | 4.2 | 4.7 | 7.5 | 5.4 | 5.7 | 15 | 15 | 15 |
| y7 (1+) | 3 | 2 | 2.5 | 4.5 | 3.2 | 4.1 | 5.4 | 3.8 | 4.8 | 15 | 15 | 15 |
| y6 (1+) | 3.5 | 3.5 | 2.5 | 5.2 | 3.9 | 4.6 | 6.3 | 5.2 | 5.2 | 15 | 15 | 15 |
| y5 (1+) | 4.8 | 2.7 | 2.7 | 5.4 | 3.5 | 4.2 | 7.2 | 4.4 | 5 | 15 | 15 | 15 |
| b11 (2+) | 2.2 | 3 | 3.2 | 3 | 3.5 | 4.9 | 3.7 | 4.6 | 5.9 | 15 | 15 | 15 |
| sum | 2.2 | 1.7 | 2.3 | 3.5 | 3 | 4.1 | 4.1 | 3.4 | 4.7 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5723 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EY[+79.966331]VATR
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-5723
Peptide Molecular Mass
2,302.9297
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Thermo Quantiva
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000 RSLCnano
Column Packing
Pepmap 100
Column Dimensions
75um x 25cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y16-98 (3+) | 7.3 | 5.2 | 3.3 | 7.7 | 5.4 | 3.9 | 10.6 | 7.5 | 5.1 | 15 | 15 | 15 |
| y7-98 (1+) | 9.2 | 3.9 | 2.5 | 16.4 | 4.7 | 3.5 | 18.8 | 6.1 | 4.3 | 15 | 15 | 15 |
| y5 (1+) | 8.7 | 2.8 | 3 | 9.7 | 2.8 | 3.5 | 13 | 4 | 4.6 | 15 | 15 | 15 |
| sum | 3.7 | 2.3 | 1.7 | 7.5 | 2 | 2.1 | 8.4 | 3 | 2.7 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y16-98 (3+) | 7.3 | 5.2 | 3.3 | 7.7 | 5.4 | 3.9 | 10.6 | 7.5 | 5.1 | 15 | 15 | 15 |
| y7-98 (1+) | 9.2 | 3.9 | 2.5 | 16.4 | 4.7 | 3.5 | 18.8 | 6.1 | 4.3 | 15 | 15 | 15 |
| y5 (1+) | 8.7 | 2.8 | 3 | 9.7 | 2.8 | 3.5 | 13 | 4 | 4.6 | 15 | 15 | 15 |
| sum | 3.7 | 2.3 | 1.7 | 7.5 | 2 | 2.1 | 8.4 | 3 | 2.7 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5721 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Modified Sequence
VADPDHDHTGFLT[+79.966331]EYVATR
Modification Type
Phospho (ST)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-5721
Peptide Molecular Mass
2,222.9634
Species
Homo sapiens (Human)
Assay Type
Enrichment PRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Thermo Quantiva
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000 RSLCnano
Column Packing
Pepmap 100
Column Dimensions
75um x 25cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10-98 (1+) | 9.3 | 4.3 | 5.5 | 11 | 6.8 | 6.1 | 14.4 | 8 | 8.2 | 15 | 15 | 15 |
| y7-98 (1+) | 4.1 | 2.1 | 3.5 | 4 | 2 | 2.4 | 5.7 | 2.9 | 4.2 | 15 | 15 | 15 |
| y6 (1+) | 3.9 | 3.7 | 2.3 | 4 | 4.5 | 3 | 5.6 | 5.8 | 3.8 | 15 | 15 | 15 |
| y5 (1+) | 3.6 | 3.4 | 1.7 | 3.4 | 3.2 | 1.6 | 5 | 4.7 | 2.3 | 15 | 15 | 15 |
| y4 (1+) | 4.3 | 3 | 1.4 | 4.2 | 3.1 | 1.9 | 6 | 4.3 | 2.4 | 15 | 15 | 15 |
| sum | 3 | 1.3 | 1.2 | 2.6 | 1.5 | 1.5 | 4 | 2 | 1.9 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10-98 (1+) | 9.3 | 4.3 | 5.5 | 11 | 6.8 | 6.1 | 14.4 | 8 | 8.2 | 15 | 15 | 15 |
| y7-98 (1+) | 4.1 | 2.1 | 3.5 | 4 | 2 | 2.4 | 5.7 | 2.9 | 4.2 | 15 | 15 | 15 |
| y6 (1+) | 3.9 | 3.7 | 2.3 | 4 | 4.5 | 3 | 5.6 | 5.8 | 3.8 | 15 | 15 | 15 |
| y5 (1+) | 3.6 | 3.4 | 1.7 | 3.4 | 3.2 | 1.6 | 5 | 4.7 | 2.3 | 15 | 15 | 15 |
| y4 (1+) | 4.3 | 3 | 1.4 | 4.2 | 3.1 | 1.9 | 6 | 4.3 | 2.4 | 15 | 15 | 15 |
| sum | 3 | 1.3 | 1.2 | 2.6 | 1.5 | 1.5 | 4 | 2 | 1.9 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5406 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
VADPDHDHTGFLTEYVATR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
173
Peptide End
191
CPTAC ID
non-CPTAC-5406
Peptide Molecular Mass
2,142.9970
Species
Homo sapiens (Human)
Assay Type
Direct PRM
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
RSLCnano and Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
RSLCnano
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nl/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y4 (1+) | 18.2 | 5.4 | 2.5 | 19.1 | 4.3 | 3.5 | 26.4 | 6.9 | 4.3 | 15 | 15 | 15 |
| y5 (1+) | 13 | 3.5 | 3.1 | 11.2 | 4.2 | 4.5 | 17.2 | 5.5 | 5.5 | 15 | 15 | 15 |
| y6 (1+) | 19.9 | 6.2 | 2.1 | 21.3 | 6.3 | 3.2 | 29.1 | 8.8 | 3.8 | 15 | 15 | 15 |
| y7 (1+) | 8.9 | 4.9 | 2 | 9.2 | 5.9 | 2.9 | 12.8 | 7.7 | 3.5 | 15 | 15 | 15 |
| y10 (1+) | 40.9 | 8.3 | 4.7 | 39.6 | 10.3 | 4.9 | 56.9 | 13.2 | 6.8 | 15 | 15 | 15 |
| sum | 4.8 | 1.7 | 1.3 | 6.8 | 2.4 | 2.8 | 8.3 | 2.9 | 3.1 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y4 (1+) | 18.2 | 5.4 | 2.5 | 19.1 | 4.3 | 3.5 | 26.4 | 6.9 | 4.3 | 15 | 15 | 15 |
| y5 (1+) | 13 | 3.5 | 3.1 | 11.2 | 4.2 | 4.5 | 17.2 | 5.5 | 5.5 | 15 | 15 | 15 |
| y6 (1+) | 19.9 | 6.2 | 2.1 | 21.3 | 6.3 | 3.2 | 29.1 | 8.8 | 3.8 | 15 | 15 | 15 |
| y7 (1+) | 8.9 | 4.9 | 2 | 9.2 | 5.9 | 2.9 | 12.8 | 7.7 | 3.5 | 15 | 15 | 15 |
| y10 (1+) | 40.9 | 8.3 | 4.7 | 39.6 | 10.3 | 4.9 | 56.9 | 13.2 | 6.8 | 15 | 15 | 15 |
| sum | 4.8 | 1.7 | 1.3 | 6.8 | 2.4 | 2.8 | 8.3 | 2.9 | 3.1 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-1541 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
VADPDHDHTGFLTEYVATR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-1541
Peptide Molecular Mass
2,142.9970
Species
Homo sapiens (Human)
Assay Type
Direct PRM
Matrix
Ovarian cancer tumor tissue lysate
Submitting Laboratory
Pacific Northwest National Laboratory
Submitting Lab PI
Tao Liu
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
TSQ Vantage
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Waters nanoACQUITY (Part Number 176016000)
Column Packing
Waters BEH C18, 1.7 um 130 Å (Part Number 186007485)
Column Dimensions
100 um x 100 mm
Flow Rate
0.5 uL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10 (1+) | 30.8 | 11.8 | 6.1 | 27.8 | 11 | 11.9 | 41.5 | 16.1 | 13.4 | 15 | 15 | 15 |
| y8 (1+) | 23.2 | 13.2 | 9.2 | 23.1 | 11.3 | 13.4 | 32.7 | 17.4 | 16.3 | 15 | 15 | 15 |
| y7 (1+) | 16.6 | 5.8 | 3.7 | 15.4 | 6.1 | 9.1 | 22.6 | 8.4 | 9.8 | 15 | 15 | 15 |
| y6 (1+) | 12.5 | 5.1 | 5.7 | 19.8 | 5.9 | 8.4 | 23.4 | 7.8 | 10.2 | 15 | 15 | 15 |
| y5 (1+) | 10.2 | 5.6 | 4.6 | 17.6 | 6.4 | 7.6 | 20.3 | 8.5 | 8.9 | 15 | 15 | 15 |
| sum | 6.9 | 5.1 | 1.8 | 11.5 | 4.9 | 7.4 | 13.4 | 7.1 | 7.6 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10 (1+) | 30.8 | 11.8 | 6.1 | 27.8 | 11 | 11.9 | 41.5 | 16.1 | 13.4 | 15 | 15 | 15 |
| y8 (1+) | 23.2 | 13.2 | 9.2 | 23.1 | 11.3 | 13.4 | 32.7 | 17.4 | 16.3 | 15 | 15 | 15 |
| y7 (1+) | 16.6 | 5.8 | 3.7 | 15.4 | 6.1 | 9.1 | 22.6 | 8.4 | 9.8 | 15 | 15 | 15 |
| y6 (1+) | 12.5 | 5.1 | 5.7 | 19.8 | 5.9 | 8.4 | 23.4 | 7.8 | 10.2 | 15 | 15 | 15 |
| y5 (1+) | 10.2 | 5.6 | 4.6 | 17.6 | 6.4 | 7.6 | 20.3 | 8.5 | 8.9 | 15 | 15 | 15 |
| sum | 6.9 | 5.1 | 1.8 | 11.5 | 4.9 | 7.4 | 13.4 | 7.1 | 7.6 | 15 | 15 | 15 |
Additional Resources and Comments
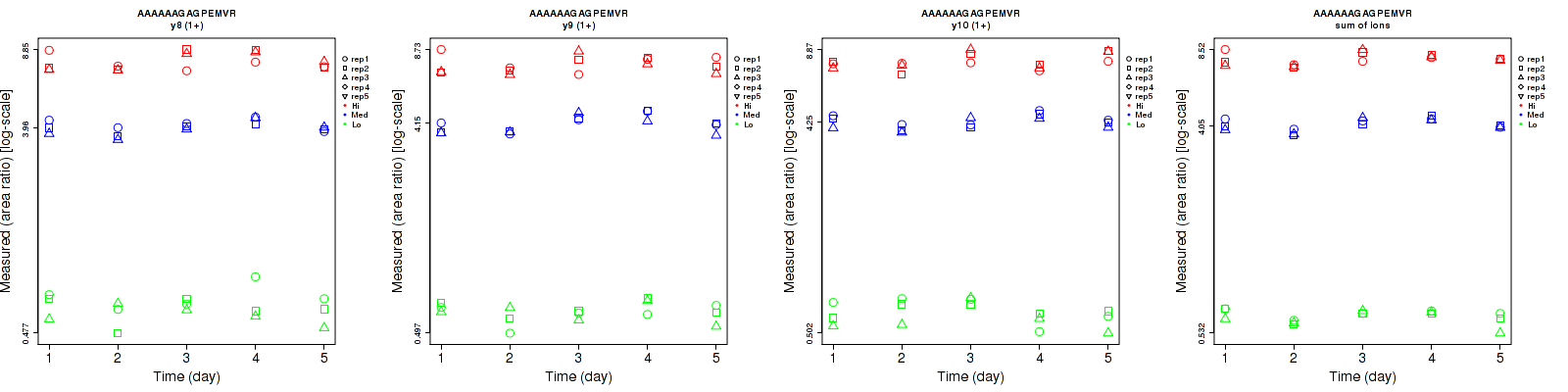
Assay Details for CPTAC-870 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
AAAAAAGAGPEMVR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
2
Peptide End
15
CPTAC ID
CPTAC-870
Peptide Molecular Mass
1,241.6187
Species
Homo sapiens (Human)
Assay Type
Direct PRM
Matrix
Pooled patient derived xenograft breast tumor digest
Submitting Laboratory
Washington University in St. Louis
Submitting Lab PI
Reid Townsend
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
TripleTOF 5600+ (SCIEX)
Internal Standard
Peptide
Peptide Standard Purity
Crude
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
2DPlus nano-LC (Eksigent)
Column Packing
ChromXP C18-CL, 3 µm, 120 Å
Column Dimensions
200 µm x 15 cm
Flow Rate
800 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 22.5 | 14.6 | 10.1 | 28.3 | 11.3 | 9.4 | 36.2 | 18.5 | 13.8 | 15 | 15 | 15 |
| y5 (1+) | 13.4 | 9.9 | 11.9 | 14.1 | 11.2 | 16.3 | 19.5 | 14.9 | 20.2 | 15 | 15 | 15 |
| y6 (1+) | 13.9 | 5 | 5.3 | 16 | 9 | 6.6 | 21.2 | 10.3 | 8.5 | 15 | 15 | 15 |
| y7 (1+) | 10.2 | 9 | 11.5 | 17.4 | 10.8 | 12.4 | 20.2 | 14.1 | 16.9 | 15 | 15 | 15 |
| y8 (1+) | 14.4 | 4.5 | 6.9 | 12.1 | 6.3 | 9.3 | 18.8 | 7.7 | 11.6 | 15 | 15 | 15 |
| y9 (1+) | 8.3 | 4.6 | 8.1 | 9.6 | 9.1 | 8.8 | 12.7 | 10.2 | 12 | 15 | 15 | 15 |
| y10 (1+) | 10 | 4.7 | 4.9 | 11.4 | 6.5 | 7.4 | 15.2 | 8 | 8.9 | 15 | 15 | 15 |
| y11 (1+) | 18.1 | 12 | 10.5 | 21.3 | 13.2 | 11.2 | 28 | 17.8 | 15.4 | 15 | 15 | 15 |
| b6 (2+) | 8.1 | 7.6 | 8.5 | 13.4 | 10.4 | 9.4 | 15.7 | 12.9 | 12.7 | 15 | 15 | 15 |
| b5 (1+) | 22.7 | 13.7 | 15.1 | 26.3 | 16.1 | 17.6 | 34.7 | 21.1 | 23.2 | 15 | 15 | 15 |
| b4 (1+) | 14.6 | 11 | 11.2 | 17.2 | 13 | 11.8 | 22.6 | 17 | 16.3 | 15 | 15 | 15 |
| sum | 4 | 3 | 3.6 | 6.2 | 6 | 6.1 | 7.4 | 6.7 | 7.1 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 22.5 | 14.6 | 10.1 | 28.3 | 11.3 | 9.4 | 36.2 | 18.5 | 13.8 | 15 | 15 | 15 |
| y5 (1+) | 13.4 | 9.9 | 11.9 | 14.1 | 11.2 | 16.3 | 19.5 | 14.9 | 20.2 | 15 | 15 | 15 |
| y6 (1+) | 13.9 | 5 | 5.3 | 16 | 9 | 6.6 | 21.2 | 10.3 | 8.5 | 15 | 15 | 15 |
| y7 (1+) | 10.2 | 9 | 11.5 | 17.4 | 10.8 | 12.4 | 20.2 | 14.1 | 16.9 | 15 | 15 | 15 |
| y8 (1+) | 14.4 | 4.5 | 6.9 | 12.1 | 6.3 | 9.3 | 18.8 | 7.7 | 11.6 | 15 | 15 | 15 |
| y9 (1+) | 8.3 | 4.6 | 8.1 | 9.6 | 9.1 | 8.8 | 12.7 | 10.2 | 12 | 15 | 15 | 15 |
| y10 (1+) | 10 | 4.7 | 4.9 | 11.4 | 6.5 | 7.4 | 15.2 | 8 | 8.9 | 15 | 15 | 15 |
| y11 (1+) | 18.1 | 12 | 10.5 | 21.3 | 13.2 | 11.2 | 28 | 17.8 | 15.4 | 15 | 15 | 15 |
| b6 (2+) | 8.1 | 7.6 | 8.5 | 13.4 | 10.4 | 9.4 | 15.7 | 12.9 | 12.7 | 15 | 15 | 15 |
| b5 (1+) | 22.7 | 13.7 | 15.1 | 26.3 | 16.1 | 17.6 | 34.7 | 21.1 | 23.2 | 15 | 15 | 15 |
| b4 (1+) | 14.6 | 11 | 11.2 | 17.2 | 13 | 11.8 | 22.6 | 17 | 16.3 | 15 | 15 | 15 |
| sum | 4 | 3 | 3.6 | 6.2 | 6 | 6.1 | 7.4 | 6.7 | 7.1 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-871 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
ELIFEETAR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
345
Peptide End
353
CPTAC ID
CPTAC-871
Peptide Molecular Mass
1,106.5608
Species
Homo sapiens (Human)
Assay Type
Direct PRM
Matrix
Pooled patient derived xenograft breast tumor digest
Submitting Laboratory
Washington University in St. Louis
Submitting Lab PI
Reid Townsend
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
TripleTOF 5600+ (SCIEX)
Internal Standard
Peptide
Peptide Standard Purity
Crude
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
2DPlus nano-LC (Eksigent)
Column Packing
ChromXP C18-CL, 3 µm, 120 Å
Column Dimensions
200 µm x 15 cm
Flow Rate
800 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 15 | 6.7 | 6.3 | 14 | 7.2 | 7.1 | 20.5 | 9.8 | 9.5 | 15 | 15 | 15 |
| y4 (1+) | 8.3 | 5.1 | 4.8 | 8.4 | 7.1 | 6.2 | 11.8 | 8.7 | 7.8 | 15 | 15 | 15 |
| y5 (1+) | 6.2 | 3.5 | 2.3 | 8 | 4.8 | 4.4 | 10.1 | 5.9 | 5 | 15 | 15 | 15 |
| y6 (1+) | 4.6 | 1.8 | 2.6 | 6.9 | 3.3 | 3.1 | 8.3 | 3.8 | 4 | 15 | 15 | 15 |
| y7 (1+) | 5 | 5.7 | 4.2 | 7.3 | 4.7 | 3.9 | 8.8 | 7.4 | 5.7 | 15 | 15 | 15 |
| sum | 3.8 | 1.5 | 1.7 | 5.9 | 3.5 | 2.8 | 7 | 3.8 | 3.3 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 15 | 6.7 | 6.3 | 14 | 7.2 | 7.1 | 20.5 | 9.8 | 9.5 | 15 | 15 | 15 |
| y4 (1+) | 8.3 | 5.1 | 4.8 | 8.4 | 7.1 | 6.2 | 11.8 | 8.7 | 7.8 | 15 | 15 | 15 |
| y5 (1+) | 6.2 | 3.5 | 2.3 | 8 | 4.8 | 4.4 | 10.1 | 5.9 | 5 | 15 | 15 | 15 |
| y6 (1+) | 4.6 | 1.8 | 2.6 | 6.9 | 3.3 | 3.1 | 8.3 | 3.8 | 4 | 15 | 15 | 15 |
| y7 (1+) | 5 | 5.7 | 4.2 | 7.3 | 4.7 | 3.9 | 8.8 | 7.4 | 5.7 | 15 | 15 | 15 |
| sum | 3.8 | 1.5 | 1.7 | 5.9 | 3.5 | 2.8 | 7 | 3.8 | 3.3 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-3200 Collapse assay details
Data source: Panorama
Official Gene Symbol
MAPK1
Peptide Sequence
VADPDHDHTGFLTEYVATR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
173
Peptide End
191
CPTAC ID
CPTAC-3200
Peptide Molecular Mass
2,142.9970
Species
Homo Sapiens
Assay Type
Direct PRM
Matrix
Tumor Digest
Submitting Laboratory
Washington University in St. Louis
Submitting Lab PI
Reid Townsend
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
ThermoFisher, Q-Exactive
Internal Standard
25 fmol on column
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Easy-nLC 1000 Thermo Scientific
Column Packing
C18
Column Dimensions
75 µm x 50 cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y4 (1+) | 28.5 | 6.8 | 7.9 | 42.5 | 6.5 | 7.9 | 51.2 | 9.4 | 11.2 | 15 | 15 | 15 |
| b11 (2+) | 45.3 | 10.4 | 5.6 | 47.6 | 6.9 | 5.8 | 65.7 | 12.5 | 8.1 | 15 | 15 | 15 |
| y3 (1+) | 8.3 | 8.6 | 6.3 | 12.1 | 8.2 | 6.8 | 14.7 | 11.9 | 9.3 | 15 | 15 | 15 |
| sum | 13.1 | 8.1 | 6.5 | 21.7 | 7.1 | 6.8 | 25.3 | 10.8 | 9.4 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y4 (1+) | 28.5 | 6.8 | 7.9 | 42.5 | 6.5 | 7.9 | 51.2 | 9.4 | 11.2 | 15 | 15 | 15 |
| b11 (2+) | 45.3 | 10.4 | 5.6 | 47.6 | 6.9 | 5.8 | 65.7 | 12.5 | 8.1 | 15 | 15 | 15 |
| y3 (1+) | 8.3 | 8.6 | 6.3 | 12.1 | 8.2 | 6.8 | 14.7 | 11.9 | 9.3 | 15 | 15 | 15 |
| sum | 13.1 | 8.1 | 6.5 | 21.7 | 7.1 | 6.8 | 25.3 | 10.8 | 9.4 | 15 | 15 | 15 |