global health – NIH Director's Blog (original) (raw)
3D Animation Captures Viral Infection in Action
Posted on August 1st, 2023 by Lawrence Tabak, D.D.S., Ph.D.
With the summer holiday season now in full swing, the blog will also swing into its annual August series. For most of the month, I will share with you just a small sampling of the colorful videos and snapshots of life captured in a select few of the hundreds of NIH-supported research labs around the country.
To get us started, let’s turn to the study of viruses. Researchers now can generate vast amounts of data relatively quickly on a virus of interest. But data are often displayed as numbers or two-dimensional digital images on a computer screen. For most virologists, it’s extremely helpful to see a virus and its data streaming in three dimensions. To do so, they turn to a technological tool that we all know so well: animation.
This research animation features the chikungunya virus, a sometimes debilitating, mosquito-borne pathogen transmitted mainly in developing countries in Africa, Asia and the Americas. The animation illustrates large amounts of research data to show how the chikungunya virus infects our cells and uses its specialized machinery to release its genetic material into the cell and seed future infections. Let’s take a look.
In the opening seconds, you see how receptor binding glycoproteins (light blue), which are proteins with a carbohydrate attached on the viral surface, dock with protein receptors (yellow) on a host cell. At five seconds, the virus is drawn inside the cell. The change in the color of the chikungunya particle shows that it’s coated in a vesicle, which helps the virus make its way unhindered through the cytoplasm.
At 10 seconds, the virus then enters an endosome, ubiquitous bubble-like compartments that transport material from outside the cell into the cytosol, the fluid part of the cytoplasm. Once inside the endosome, the acidic environment makes other glycoproteins (red, blue, yellow) on the viral surface change shape and become more flexible and dynamic. These glycoproteins serve as machinery that enables them to reach out and grab onto the surrounding endosome membrane, which ultimately will be fused with the virus’s own membrane.
As more of those fusion glycoproteins grab on, fold back on themselves, and form into hairpin-like shapes, they pull the membranes together. The animation illustrates not only the changes in protein organization, but the resulting effects on the integrity of the membrane structures as this dynamic process proceeds. At 53 seconds, the viral protein shell, or capsid (green), which contains the virus’ genetic instructions, is released back out into the cell where it will ultimately go on to make more virus.
This remarkable animation comes from Margot Riggi and Janet Iwasa, experts in visualizing biology at the University of Utah’s Animation Lab, Salt Lake City. Their data source was researcher Kelly Lee, University of Washington, Seattle, who collaborated closely with Riggi and Iwasa on this project. The final product was considered so outstanding that it took the top prize for short videos in the 2022 BioArt Awards competition, sponsored by the Federation of American Societies for Experimental Biology (FASEB).
The Lee lab uses various research methods to understand the specific shape-shifting changes that chikungunya and other viruses perform as they invade and infect cells. One of the lab’s key visual tools is cryo-electron microscopy (Cryo-EM), specifically cryo-electron tomography (cryo-ET). Cryto-ET enables complex 3D structures, including the intermediate state of biological reactions, to be captured and imaged in remarkably fine detail.
In a study in the journal Nature Communications [1] last year, Lee’s team used cryo-ET to reveal how the chikungunya virus invades and delivers its genetic cargo into human cells to initiate a new infection. While Lee’s cryo-ET data revealed stages of the virus entry process and fine structural details of changes to the virus as it enters a cell and starts an infection, it still represented a series of snapshots with missing steps in between. So, Lee’s lab teamed up with The Animation Lab to help beautifully fill in the gaps.
Visualizing chikungunya and similar viruses in action not only makes for informative animations, it helps researchers discover better potential targets to intervene in this process. This basic research continues to make progress, and so do ongoing efforts to develop a chikungunya vaccine [2] and specific treatments that would help give millions of people relief from the aches, pains, and rashes associated with this still-untreatable infection.
References:
[1] Visualization of conformational changes and membrane remodeling leading to genome delivery by viral class-II fusion machinery. Mangala Prasad V, Blijleven JS, Smit JM, Lee KK. Nat Commun. 2022 Aug 15;13(1):4772. doi: 10.1038/s41467-022-32431-9. PMID: 35970990; PMCID: PMC9378758.
[2] Experimental chikungunya vaccine is safe and well-tolerated in early trial, National Institute of Allergy and Infectious Diseases news release, April 27, 2020.
Links:
Chikungunya Virus (Centers for Disease Control and Prevention, Atlanta)
Global Arbovirus Initiative (World Health Organization, Geneva, Switzerland)
The Animation Lab (University of Utah, Salt Lake City)
Video: Janet Iwasa (TED Speaker)
Lee Lab (University of Washington, Seattle)
BioArt Awards (Federation of American Societies for Experimental Biology, Rockville, MD)
NIH Support: National Institute of General Medical Sciences; National Institute of Allergy and Infectious Diseases
Posted In: Cool Videos
Tags: 2022 BioArt Awards, animation, chikungunya, chikungunya vaccine, cryo-electron tomography, cryo-EM, cryo-ET, cytoplasm, cytosol, endosome, FASEB, global health, glycoprotein, imaging, infection, mosquito-borne illnesses, structural biology, vesicle, virology, virus
Case Study Unlocks Clues to Rare Resilience to Alzheimer’s Disease
Posted on May 30th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
Caption: Newly discovered Reelin-COLBOS gene variation may delay or prevent Alzheimer’s disease. Credit: Donny Bliss, NIH
Biomedical breakthroughs most often involve slow and steady research in studies involving large numbers of people. But sometimes careful study of even just one truly remarkable person can lead the way to fascinating discoveries with far-reaching implications.
An NIH-funded case study published recently in the journal Nature Medicine falls into this far-reaching category [1]. The report highlights the world’s second person known to have an extreme resilience to a rare genetic form of early onset Alzheimer’s disease. These latest findings in a single man follow a 2019 report of a woman with similar resilience to developing symptoms of Alzheimer’s despite having the same strong genetic predisposition for the disease [2].
The new findings raise important new ideas about the series of steps that may lead to Alzheimer’s and its dementia. They’re also pointing the way to key parts of the brain for cognitive resilience—and potentially new treatment targets—that may one day help to delay or even stop progression of Alzheimer’s.
The man in question is a member of a well-studied extended family from the country of Colombia. This group of related individuals, or kindred, is the largest in the world with a genetic variant called the “Paisa” mutation (or Presenilin-1 E280A). This Paisa variant follows an autosomal dominant pattern of inheritance, meaning that those with a single altered copy of the rare variant passed down from one parent usually develop mild cognitive impairment around the age of 44. They typically advance to full-blown dementia around the age of 50 and rarely live past the age of 60. This contrasts with the most common form of Alzheimer’s, which usually begins after age 65.
The new findings come from a team led by Yakeel Quiroz, Massachusetts General Hospital, Boston; Joseph Arboleda-Velasquez, Massachusetts Eye and Ear, Boston; Diego Sepulveda-Falla, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; and Francisco Lopera, University of Antioquia, Medellín, Colombia. Lopera first identified this family more than 30 years ago and has been studying them ever since.
In the new case report, the researchers identified a Colombian man who’d been married with two children and retired from his job as a mechanic in his early 60s. Despite carrying the Paisa mutation, his first cognitive assessment at age 67 showed he was cognitively intact, having limited difficulties with verbal learning skills or language. It wasn’t until he turned 70 that he was diagnosed with mild cognitive impairment—more than 20 years later than the expected age for this family—showing some decline in short-term memory and verbal fluency.
At age 73, he enrolled in the Colombia-Boston biomarker research study (COLBOS). This study is a collaborative project between the University of Antioquia and Massachusetts General Hospital involving approximately 6,000 individuals from the Paisa kindred. About 1,500 of those in the study carry the mutation that sets them up for early Alzheimer’s. As a member of the COLBOS study, the man underwent thorough neuroimaging tests to look for amyloid plaques and tau tangles, both of which are hallmarks of Alzheimer’s.
While this man died at age 74 with Alzheimer’s, the big question is: how did he stave off dementia for so long despite his poor genetic odds? The COLBOS study earlier identified a woman with a similar resilience to Alzheimer’s, which they traced to two copies of a rare, protective genetic variant called Christchurch. This variant affects a gene called apolipoprotein E (APOE3), which is well known for its influence on Alzheimer’s risk. However, the man didn’t carry this same protective variant.
The researchers still thought they’d find an answer in his genome and kept looking. While they found several variants of possible interest, they zeroed in on a single gene variant that they’ve named Reelin-COLBOS. What helped them to narrow it down to this variant is the man also had a sister with the Paisa mutation who only progressed to advanced dementia at age 72. It turned out, in addition to the Paisa variant, the siblings also shared an altered copy of the newly discovered Reelin-COLBOS variant.
This Reelin-COLBOS gene is known to encode a protein that controls signals to chemically modify tau proteins, which form tangles that build up over time in the Alzheimer’s brain and have been linked to memory loss. Reelin is also functionally related to APOE, the gene that was altered in the woman with extreme Alzheimer’s protection. Reelin and APOE both interact with common protein receptors in neurons. Together, the findings add to evidence that signaling pathways influencing tau play an important role in Alzheimer’s pathology and protection.
The neuroimaging exams conducted when the man was age 73 have offered further intriguing clues. They showed that his brain had extensive amyloid plaques. He also had tau tangles in some parts of his brain. But one brain region, called the entorhinal cortex, was notable for having a very minimal amount of those hallmark tau tangles.
The entorhinal cortex is a hub for memory, navigation, and the perception of time. Its degeneration also leads to cognitive impairment and dementia. Studies of the newly identified Reelin-COLBOS variant in Alzheimer’s mouse models also help to confirm that the variant offers its protection by diminishing the pathological modifications of tau.
Overall, the findings in this one individual and his sister highlight the Reelin pathway and brain region as promising targets for future study and development of Alzheimer’s treatments. Quiroz and her colleagues report that they are actively exploring treatment approaches inspired by the Christchurch and Reelin-COLBOS discoveries.
Of course, there’s surely more to discover from continued study of these few individuals and others like them. Other as yet undescribed genetic and environmental factors are likely at play. But the current findings certainly offer some encouraging news for those at risk for Alzheimer’s disease—and a reminder of how much can be learned from careful study of remarkable individuals.
References:
[1] Resilience to autosomal dominant Alzheimer’s disease in a Reelin-COLBOS heterozygous man. Lopera F, Marino C, Chandrahas AS, O’Hare M, Reiman EM, Sepulveda-Falla D, Arboleda-Velasquez JF, Quiroz YT, et al. Nat Med. 2023 May;29(5):1243-1252.
[2] Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Tariot PN, Johnson KA, Reiman EM, Quiroz YT et al. Nat Med. 2019 Nov;25(11):1680-1683.
Links:
Alzheimer’s Disease & Related Dementias (National Institute on Aging/NIH)
“NIH Support Spurs Alzheimer’s Research in Colombia,” Global Health Matters, January/February 2014, Fogarty International Center/NIS
“COLBOS Study Reveals Mysteries of Alzheimer’s Disease,” NIH Record, August 19, 2022.
Yakeel Quiroz (Massachusetts General Hospital, Harvard Medical School, Boston)
Joseph Arboleda-Velasquez (Massachusetts Eye and Ear, Harvard Medical School, Boston)
Diego Sepulveda-Falla Lab (University Medical Center Hamburg-Eppendorf, Hamburg, Germany)
Francisco Lopera (University of Antioquia, Medellín, Colombia)
NIH Support: National Institute on Aging; National Eye Institute; National Institute of Neurological Disorders and Stroke; Office of the Director
Posted In: News
Tags: Alzheimer’s disease, APOE3, brain, Christchurch variant, cognitive resilience, Colombia, Colombia-Boston biomarker research study, dementia, genetics, genomics, global health, Paisa mutation, Paisa variant, Presinilin-1, Reelin-COLBOS gene variant, tau, tau protein
This Is Why NIH Invests in Global Health Research
Posted on December 13th, 2022 by Roger I. Glass, M.D., Ph.D., Fogarty International Center
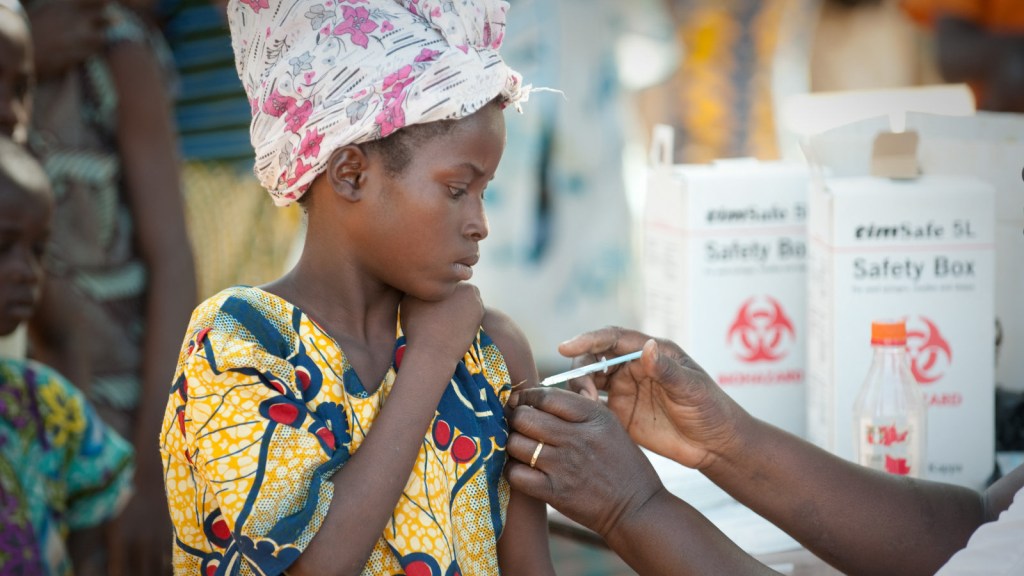
Caption: Global partnerships fostered by NIH’s Fogarty International Center speed translation of scientific discoveries into lifesaving biomedical products. Credit: Gabe Bienczycki, PATH, Seattle
Efforts over the past few years to end the COVID-19 pandemic clearly reveal how global health impacts individual wellbeing and national security. At NIH, the Fogarty International Center helps the other institutes become engaged with global health research, which investigates the dual burden of infectious disease and non-communicable disease.
Global health research also encompasses data science, economics, genetics, climate change science, and many other disciplines. For more than 50 years, Fogarty has been building partnerships among institutions in the U.S. and abroad, while training the next generation of scientists focused on universal health needs.
America’s investment in Fogarty has paid rich dividends
During the pandemic, in particular, we’ve seen researchers trained by our programs make scientific discoveries that contributed to international security. Take Jessica Manning, a former Fogarty fellow who now conducts malaria research in Phnom Penh, Cambodia. Her team at the Ministry of Health sequenced the viral strain of SARS-CoV-2, the cause of COVID-19, infecting the first Cambodian patient and documented early the spread of this novel coronavirus outside of China.
Similarly, Christian Happi, director of the African Centre of Excellence for the Genomics of Infectious Disease, Ede, Nigeria, sequenced the first SARS-CoV-2 genome in Africa. Happi was able to do it by adapting the sequencing and analytical pipelines that he’d created back when he was a Fogarty grantee studying Ebola.
In Botswana, Sikhulile Moyo leveraged the skills he’d acquired while supported by a Fogarty HIV research training grant with Max Essex, Harvard School of Public Health, Cambridge, MA, to track COVID-19 mutations for his country’s Ministry of Health. Last November, he alerted the world of a new Omicron variant. Within six weeks, Omicron became the dominant global strain, challenging the ability of COVID vaccines to control its spread. In the Dominican Republic, William Duke, a national commission member, used what he’d learned as a Fogarty trainee to help create a national COVID-19 intervention plan to prevent and control the disease.
Fogarty’s fostering of global health leaders is one way we advance scientific expertise while ensuring our nation’s biosecurity. Another is by finding effective ways to study abroad the same health conditions that affect our own population.
Research conducted in Colombia, for example, may provide clues for preventing Alzheimer’s disease in the U.S. Fogarty support brought together neuroscientists Kenneth Kosik, University of California, Santa Barbara, and Francisco Lopera, University of Antioquia, Colombia, to study members of the largest-known family with an early-onset, rapidly progressive form of the disease. Over the years, Kosik and Lopera have trained local scientists, explored gene therapy targets, investigated biomarkers to monitor disease progression, and conducted drug trials in search of a cure for Alzheimer’s.
Researchers in other fields also discover unique opportunities to investigate populations with high rates of disease. Siana Nkya, a Fogarty grantee based in Tanzania, has devoted her career to studying the genetic determinants of sickle cell disease, which affects many people around the world, including in the U.S. We hope that US-African partnerships might develop improved, affordable treatments and a cure for all patients with this devastating disease. Similarly, people in the U.S. have access to state-of-the-art HIV treatment studies in places around the globe where incidence rates are higher.
Fogarty has supported many milestone achievements in HIV research over the years. Among them is a study that took place in nine countries. The research, led by Myron Cohen of the University of North Carolina at Chapel Hill, established that antiretroviral therapy can prevent sexual transmission of HIV-1 among couples in which one person is infected and the other is not. In fact, this research informs current HIV treatment recommendations worldwide, including in the U.S.
Americans will also undoubtedly benefit from projects funded by Fogarty’s Global Brain and Nervous System Disorders Research across the Lifespan program. For example, psychologist Tatiana Balachova, University of Oklahoma, Oklahoma City, has designed an intervention for women in Russia to prevent fetal alcohol spectrum disorders. In another project in South Africa, Sandra and Joseph Jacobson, Wayne State University, Detroit, conducted the first-ever prospective longitudinal study of the syndrome. Findings from both projects are ripe for translation within an American context.
Other examples of Global Brain program investigations with broad implications in our own country include studying early psychosis in China; capacity building for schizophrenia research in Macedonia; exploring family consequences from the Zika virus in Brazil; and studying dementia and related health and social challenges in Lebanon.
These are just a few examples of Fogarty’s work and its unique mission. What is most remarkable about Fogarty is that just under 90 percent of our grants are co-funded by at least one other NIH institute, center, or office. Collaboration, both within borders and across them, is Fogarty’s formula for success.
Links:
Fogarty International Center (NIH)
Overview of Brain Disorders: Research Across the Lifespan (Fogarty)
Former Fogarty Scholar Dr Jessica Manning Helps Cambodia Respond to COVID (Fogarty)
Christian Happi: Former Fogarty Grantee Leads COVID-19 Genomics Work in Africa (Fogarty)
Sikhulile Moyo: Fogarty Fellow Recognized for Omicron Discovery (Fogarty)
William Duke: Former Fogarty HIV Trainee Helps Lead Dominican Republic’s COVID Response (Fogarty)
Kenneth Kosic and Francisco Lopera: NIH Support Spurs Alzheimer’s Research in Colombia (Fogarty)
Former Fogarty fellow Siana Nkya Tackles Sickle Cell Disease in Tanzania (Fogarty)
Tatiana Balachova: Researchers Tackle Fetal Alcohol Syndrome in Russia (Fogarty)
Sandra and Joseph Jacobson: Fetal Alcohol Exposure Research Supported by NIAAA in South Africa, Ukraine and Russia Improves Prevention, Outcomes (Fogarty)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 22nd in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Posted In: Generic
Tags: Africa, Alzheimer’s disease, antiretroviral therapy, Botswana, brain, Brazil, Cambodia, China, Colombia, COVID-19, COVID-19 vaccine, dementia, Dominican Republic, early psychosis, early-onset Alzheimer's disease, Ebola, fetal alcohol spectrum disorders, Fogarty, Fogarty International Center, global health, HIV, HIV-1, international security, Lebanon, Macedonia, neuroscience, Nigeria, novel coronavirus, Omicron variant, pandemic, SARS-CoV-2, schizophrenia, sickle cell disease, South Africa, Tanzania, Zika virus
Climate Change and Health Initiative to Expand Research, Build Resiliency
Posted on July 26th, 2022 by Richard Woychik, Ph.D., National Institute of Environmental Health Sciences

Credit: Athawit Ketsak/Shutterstock
Climate change is a global process that affects human health in a variety of complex ways. Wildfires, heat waves, hurricanes, floods, and other climate-related weather events can result in illness, injury, and death. Indirect health threats are cause for concern, too. For example, changes in temperature and rainfall can affect the lifecycle of mosquitoes that transmit diseases such as malaria and dengue fever, thereby paving the way for new outbreaks.
Environmental disruptions worsened by climate change can reduce air quality, diminish water resources, and increase exposure to higher temperatures and pathogens. As a result, we see greater health risks in susceptible individuals such as children, the elderly, the poor, and people with underlying conditions, both in America and around the world.
For decades, the National Institute of Environmental Health Sciences and other NIH institutes and centers (ICs) have advanced important research into how climate change affects health. But expanding knowledge in this area and addressing other key challenges will require much more collaboration. The time is now for an all-hands-on-deck scientific effort—across NIH and the wider biomedical research community—that spans many interconnected disciplines and fields of inquiry.
That is why I am excited to join forces with several other IC directors to launch the NIH Climate Change and Health Initiative. By working together, NIH institutes and centers can harness their technologies, innovative research approaches, and talent to advance the science of climate change and health. Through this timely effort, we will promote resilience in vulnerable communities because our research will help them to understand, prepare for, and recover from climate-related health challenges.
Our Strategic Framework outlines why it is important to go beyond studying the health effects of climate change. We must involve impacted communities in solutions-focused research that empowers them, health care practitioners, and health and social services agencies to reduce climate-related health risks. By generating scientific evidence for public health action, we can use a health equity approach to boost climate resiliency among at-risk groups, whether in the U.S. or low- and middle-income countries.
At the heart of the initiative is a push for transdisciplinary, team-based science that boosts training, research capacity, and community engagement. Our immediate goals are to use existing grant programs to strengthen research infrastructure and enhance communication, internally and externally.
Also, with dedicated support from several ICs and the Office of the Director (OD), NIH is funding a research coordinating center and a community engagement program. The coordinating center will help NIH scientists collaborate and manage data. And the community engagement program will empower underserved populations by encouraging two-way dialogue in which both scientists and community members learn from each other. That inclusive approach will improve research and mitigation efforts and reduce health disparities.
In addition, several Notices of Special Interest are now open for applications. The NIH invites scientists to submit research proposals outlining how they plan either to study the health effects of climate change or develop new technologies to mitigate those effects. Also, with OD support, a Climate and Health Scholars Program will launch later this year. Scientists working on important research will share their expertise and methodologies with the NIH community, spurring opportunities for further collaboration.
Going forward, any additional support from the White House, Congress, and the public will allow NIH to further expand the initiative. For example, we urgently need to test novel interventions for reducing heat stress among agricultural workers and to scale up early-warning systems for climate-related weather events. There is also opportunity to use laboratory-based and clinical methodologies to expand knowledge of how climate factors, such as heat and humidity, affect key cellular systems, including mitochondrial function.
To fill those and other research gaps, we must draw on an array of skill sets and fields of inquiry. Therefore, our Strategic Framework outlines the importance of supporting adaptation research, basic and mechanistic studies, behavioral and social sciences research, data integration, disaster research response, dissemination and implementation science, epidemiology and predictive modeling, exposure and risk assessment, and systems science. Tapping into those areas will help us tackle climate-related health challenges and develop effective solutions.
In recent years, in-depth reports and assessments have provided conclusive evidence that climate change is significantly altering our environment and impacting human health. Although the science of climate change and health has progressed, much work remains. We hope that the Climate Change and Health Initiative expands scientific partnerships and capacity throughout NIH and across the global biomedical and environmental health sciences communities. Greater collaboration will spur new knowledge, interventions, and technologies that help humanity manage the health effects of climate change and strengthen health equity.
(Note: The Initiative’s Executive Committee includes the following IC directors: Richard Woychik, National Institute of Environmental Health Sciences [chair]; Diana Bianchi, Eunice Kennedy Shriver National Institute of Child Health and Human Development; Gary Gibbons, National Heart, Lung, and Blood Institute; Roger Glass, Fogarty International Center; Joshua Gordon, National Institute of Mental Health; Eliseo Pérez-Stable, National Institute on Minority Health and Health Disparities; and Shannon Zenk, National Institute of Nursing Research.)
Links:
Environmental Health Topic: Climate Change (National Institute of Environmental Health Sciences /NIH)
NIH Climate Change and Health Initiative (NIH)
NIH Climate Change and Health Initiative Strategic Framework (NIH)
Research Coordinating Center to Support Climate Change and Health Community of Practice (NIH)
Research Opportunity Announcement: Alliance for Community Engagement—Climate Change and Health (National Heart, Lung, and Blood Institute / NIH)
Notice of Special Interest: Climate Change and Health (NIH)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 14th in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Posted In: Generic
Tags: basic research, climate, climate change, Climate Change and Health Initiative, climate resiliency, climate science, community engagement, environment, global health, mitochondria, NIEHS, Notices of Special Interest, predictive modeling, public health, research capacity, Small Business Innovation Research, social science, Strategic Framework, team science, technology development, underserved communities, weather
Tuberculosis: An Ancient Disease in Need of Modern Scientific Tools
Posted on April 19th, 2022 by Anthony S. Fauci, M.D., National Institute of Allergy and Infectious Diseases

Caption: Here I am with Paul Farmer, who was a strong voice for improving TB prevention and treatments in resource-scarce settings, when he came to NIH in 2007 to deliver my institute’s James C. Hill Memorial Lecture. Credit: NIH
Although COVID-19 has dominated our attention for the past two years, tuberculosis (TB), an ancient scourge, remains a dominating infectious disease globally, with an estimated 10 million new cases and more than 1.3 million deaths in 2020. TB disproportionately afflicts the poor and has long been the leading cause of death in people living with HIV.
Unfortunately, during the global COVID-19 pandemic, recent gains in TB control have been stalled or reversed. We’ve seen a massive drop in new TB diagnoses, reflecting poor access to care and an uptick in deaths in 2020 [1].
We are fighting TB with an armory of old weapons inferior to those we have for COVID-19. The Bacillus Calmette–Guérin (BCG) vaccine, the world’s only licensed TB vaccine, has been in use for more than 100 years. While BCG is somewhat effective at preventing TB meningitis in children, it provides more limited durable protection against pulmonary TB in children and adults. More effective vaccination strategies to prevent infection and disease, decrease relapse rates, and shorten durations of treatment are desperately needed to reduce the terrible global burden of TB.
In this regard, over the past five years, several exciting research advances have generated new optimism in the field of TB vaccinology. Non-human primate studies conducted at my National Institute of Allergy and Infectious Diseases’ (NIAID) Vaccine Research Center and other NIAID-funded laboratories have demonstrated that effective immunity against infection is achievable and that administering BCG intravenously, rather than under the skin as it currently is given, is highly protective [2].
Results from a phase 2 trial testing BCG revaccination in adolescents at high risk of TB infection suggested this approach could help prevent TB [3]. In addition, a phase 2 trial of an experimental TB vaccine based on the recombinant protein M72 and an immune-priming adjuvant, AS01, also showed promise in preventing active TB disease in latently infected adults [4].
Both candidates are now moving on to phase 3 efficacy trials. The encouraging results of these trials, combined with nine other candidates currently in phase 2 or 3 studies [5], offer new hope that improved vaccines may be on the horizon. The NIAID is working with a team of other funders and investigators to analyze the correlates of protection from these studies to inform future TB vaccine development.
Even with these exciting developments, it is critical to accelerate our efforts to enhance and diversify the TB vaccine pipeline by addressing persistent basic and translational research gaps. To this end, NIAID has several new programs. The Immune Protection Against Mtb Centers are taking a multidisciplinary approach to integrate animal and human data to gain a comprehensive understanding of the immune responses required to prevent TB infection and disease.
This spring, NIAID will fund awards under the Innovation for TB Vaccine Discovery program that will focus on the discovery and early evaluation of novel TB vaccine candidates with the goal of diversifying the TB vaccine pipeline. Later this year, the Advancing Vaccine Adjuvant Research for TB program will systematically assess combinations of TB immunogens and adjuvants. Finally, NIAID’s well-established clinical trials networks are planning two new clinical trials of TB vaccine candidates.
As we look to the future, we must apply the lessons learned in the development of the COVID-19 vaccines to longstanding public health challenges such as TB. COVID-19 vaccine development was hugely successful due to the use of novel vaccine platforms, structure-based vaccine design, community engagement for rapid clinical trial enrollment, real-time data sharing with key stakeholders, and innovative trial designs.
However, critical gaps remain in our armamentarium. These include the harnessing the immunology of the tissues that line the respiratory tract to design vaccines more adept at blocking initial infection and transmission, employing thermostable formulations and novel delivery systems for resource-limited settings, and crafting effective messaging around vaccines for different populations.
As we work to develop better ways to prevent, diagnose, and treat TB, we will do well to remember the great public health icon, Paul Farmer, who tragically passed away earlier this year at a much too young age. Paul witnessed firsthand the devastating consequences of TB and its drug resistant forms in Haiti, Peru, and other parts of the world.
In addition to leading efforts to improve how TB is treated, Paul provided direct patient care in underserved communities and demanded that the world do more to meet their needs. As we honor Paul’s legacy, let us accelerate our efforts to find better tools to fight TB and other diseases of global health importance that exact a disproportionate toll among the poor and underserved.
References:
[1] Global tuberculosis report 2021. WHO. October 14, 2021.
[2] Prevention of tuberculosis in macaques after intravenous BCG immunization. Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH,. Hughes TK, Pokkali S, Swanson PA, Grant NL, Rodgers MA, Kamath M, Causgrove CM, Laddy DJ, Bonavia A, Casimiro D, Lin PL, Klein E, White AG, Scanga CA, Shalek AK, Roederer M, Flynn JL, and Seder RA. Nature. 2020 Jan 1; 577: 95–102.
[3] Prevention of M. tuberculosis Infection with H4:IC31 vaccine or BCG revaccination. Nemes E, Geldenhuys H, Rozot V, Rutkowski KT, Ratangee F,Bilek N., Mabwe S, Makhethe L, Erasmus M, Toefy A, Mulenga H, Hanekom WA, et al. N Engl J Med 2018; 379:138-149.
[4] Final analysis of a trial of M72/AS01E vaccine to prevent tuberculosis. Tait DR, Hatherill M, Van Der Meeren O, Ginsberg AM, Van Brakel E, Salaun B, Scriba TJ, Akite EJ, Ayles HM, et al.
[5] Pipeline Report 2021: Tuberculosis Vaccines. TAG. October 2021.
Links:
Tuberculosis (National Institute of Allergy and Infectious Diseases/NIH)
NIAID Strategic Plan for Tuberculosis Research
Immune Mechanisms of Protection Against Mycobacterium tuberculosis Centers (IMPAc-TB) (NIAID)
Partners in Health (Boston, MA)
[Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the seventh in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.]
Posted In: Generic
Tags: Advancing Vaccine Adjuvant Research for TB, Bacillus Calmette-Guerin vaccine, clinical trials, correlates of protection, COVID-19, COVID-19 vaccine, global health, Haiti, Immune Protection Against Mtb Centers, immunology, infectious disease, Innovation for TB Vaccine Discovery, M72, NIAID, Paul Farmer, Peru, public health, pulmonary TB, TB, TB meningitis, TB treatment, translational science, tuberculosis, underserved communities, vaccine
Zooming in on Global Health Research
Posted on July 13th, 2021 by Dr. Francis Collins

In past years, Roger Glass (top left), director of NIH’s Fogarty International Center (FIC), and I have taken an in-person group photo with the FIC fellows and scholars. This year, due to the international health and travel challenges posed by the global COVID-19 pandemic, a Zoom composite of some of the young researchers will have to do! I spoke to the group on the morning of July 13 as part of FIC’s week-long Global Health Program for Fellows and Scholars. The program provides collaborative, mentored global health research training in low- and middle-income countries. Individual students, postdoctoral fellows, or faculty from the U.S. and abroad apply for a 12-month placement at a participating global institution. The meeting has brought together 122 fellows and scholars (US and international), seven Fulbright Fogarty Fellows, 16 alumni, and many others to the event. As you can see in my photo, I had to be out of town this year, and I spoke to everyone buckled up while returning to the Washington, D.C. area. But I didn’t want to miss this opportunity to share my vision for global health research and point to some of the many opportunities available in global health for young academics from the U.S. and other nations.
Genome Data from Africa Reveal Millions of New Variants
Posted on November 10th, 2020 by Dr. Francis Collins

Credit: Human Heredity and Health in Africa Initiative
The first Homo sapiens emerged in Africa hundreds of thousands of years ago. We are all descended from that common pool of ancestors. Put another way, we are all Africans. While it’s not possible to study the DNA of these vanished original human populations, it is possible to study the genetic material of today’s African peoples to learn more about the human genome and its evolution over time. The degree of genetic diversity in Africa is greater than anywhere else in the world.
Progress continues to be made in this important area of genomic research. The latest step forward is a study just published in the journal Nature that analyzes more than 400 complete human genomes, including 50 distinct groups of people from 13 African countries. This work has uncovered about 3.4 million unique gene variants that had never before been described, greatly expanding our knowledge of human genetic variation and its implications for health and disease.
This work is the latest from the Human Heredity and Health in Africa (H3Africa) Initiative , which I helped establish a decade ago. This partnership between NIH, the Wellcome Trust, and the Alliance for Accelerating Excellence in Science in Africa (AESA) seeks to train a new generation of African scientists in genomic science and other disciplines, while conducting state-of-the-art health research on the African continent. The hope is to help these scientists use their new knowledge to improve human health in Africa and to help fill significant gaps in our knowledge of the diversity within human genomes.
The new study was led by Zané Lombard, the University of the Witwatersrand, South Africa; Neil Hanchard, Baylor College of Medicine, Houston; and Adebowale Adeyemo, NIH’s National Human Genome Research Institute, Bethesda, MD. It also included more than 50 other H3Africa data providers and data analysts from across Africa and around the world.
These researchers sequenced and analyzed the genomes of 426 individuals, almost all from studies and countries within the H3Africa Consortium, the network of NIH and Wellcome Trust-funded research sites in Africa. These individuals were carefully selected to provide broad coverage of the diverse landscape of African genomic variation. They also included many populations that hadn’t been studied at the genetic level before. The team focused its attention on single-letter differences, also known as single nucleotide variants (SNVs), located across the 3 billion DNA letters of the human genome.
All told, the researchers observed more than 31 million confirmed SNVs. Of the 3.4 million newly discovered SNVs, most turned up in the genomes of individuals from previously unstudied African ethnic groups with their own distinct languages. Even among SNVs that had been previously reported, several were found much more often than in other populations. That’s important because medical geneticists often include information about frequency in deciding whether a gene variant is a likely cause of rare disease. So, this more complete picture of normal genetic variation will be valuable for diagnosing such genetic conditions around the globe.
The researchers also found more than 100 regions of the genome where the pattern of genetic variation was suggestive of underlying variants that were evolutionarily favored at some time in the past. Sixty-two of those chromosomal locations weren’t previously known to be under such strong natural selection in human populations. Interestingly, those selected regions were found to contain genes associated with viral immunity, DNA repair, reproduction, and metabolism, or occurred close to variants that have been associated with conditions such as uterine fibroids and chronic kidney disease.
The findings suggest that viral infections, such as outbreaks of Ebola, yellow fever, and Lassa fever, may have played an important role over centuries in driving genetic differences on the African continent. The data also point to the possibility of human adaptation to differences across the African continent in local environments and diets, and these adaptations could be relevant to common diseases and traits we see now.
The researchers used the data to help gain insight into past migrations of human populations. The genetic data revealed complex patterns of ancestral mixing within and between groups. It also uncovered how distinct groups likely moved large distances across Africa in the past, going back hundreds to thousands of years. The findings also offered a more complete picture of the timing and extent of the migration of speakers of Africa’s most common language group (Bantu) as they moved from West Africa to the southern and eastern reaches of the continent—a defining event in the genetic history of Africa.
There’s still much more to learn about the diversity of human genomes, and a need for continued studies, including many more individuals representing more distinct groups in Africa. Indeed, H3Africa now consists of 51 projects all across the continent, focused on population-based genomic studies of many common health conditions, from heart disease to tuberculosis. As the cradle of all humanity, Africa has much to offer genomic research in the years ahead that will undoubtedly have far-reaching implications for people living in all parts of our planet.
Reference:
[1] High-depth African genomes inform human migration and health. Choudhury A et al. 2020 Oct;586(7831):741-748.
Links:
Human Heredity and Health in Africa (H3Africa) (NIH)
H3Africa (University of Cape Town, South Africa)
NIH Support: National Human Genome Research Institute; National Institute of Allergy and Infectious Diseases
Posted In: News
Tags: AESA, Africa, Alliance for Accelerating Excellence in Science in Africa, Bantu, chronic kidney disease, Ebola, gene variants, genetic epidemiology, genomics, global health, H3Africa, human diversity, human evolution, Human Heredity and Health in Africa Initiative, human migration, Lassa fever, single nucleotide variants, SNV, uterine fibroids, Wellcome Trust, yellow fever
Public Health Policies Have Prevented Hundreds of Millions of Coronavirus Infections
Posted on June 23rd, 2020 by Dr. Francis Collins

Credit: Stock photo/Juanmonino
The alarming spread of coronavirus disease 2019 (COVID-19) last winter presented a profound threat to nations around the world. Many government leaders responded by shutting down all non-essential activities, implementing policies that public health officials were hopeful could slow the highly infectious SARS-CoV-2, the novel coronavirus that causes COVID-19.
But the shutdown has come at a heavy cost for the U.S. and global economies. It’s also taken a heavy personal toll on many of us, disrupting our daily routines—getting children off to school, commuting to the office or lab, getting together with friends and family, meeting face to face to plan projects, eating out, going to the gym—and causing lots of uncertainty and frustration.
As difficult as the shutdowns have been, new research shows that without these public health measures, things would have been much, much worse. According to a study published recently in Nature [1], the implementation of containment and mitigation strategies across the globe prevented or delayed about 530 million coronavirus infections across six countries—China, South Korea, Iran, Italy, France, and the United States. Take a moment to absorb that number—530 million. Right now, there are 8.8 million cases documented across the globe.
Estimates of the benefits of anti-contagion policies have drawn from epidemiological models that simulate the spread of COVID-19 in various ways, depending on assumptions built into each model. But models are sophisticated ways of guessing. Back when decisions about staying at home had to be made, no one knew for sure if, or how well, such approaches to limit physical contact would work. What’s more, the only real historical precedent was the 1918 Spanish flu pandemic in a very different, much-less interconnected world.
That made it essential to evaluate the pros and cons of these public health strategies within a society. As many people have rightfully asked: are the health benefits really worth the pain?
Recognizing a pressing need to answer this question, an international team of scientists dropped everything that they were doing to find out. Led by Solomon Hsiang, director of the University of California, Berkeley’s Global Policy Laboratory and Chancellor’s Professor at the Goldman School of Public Policy, a research group of 15 researchers from China, France, South Korea, New Zealand, Singapore, and the United States evaluated 1,717 policies implemented in all six countries between January 2020, when the virus began its global rise, and April 6, 2020.
The team relied on econometric methods that use statistics and math to uncover meaningful patterns hiding in mountains of data. As the name implies, these techniques are used routinely by economists to understand, in a before-and-after way, how certain events affect economic growth.
In this look-back study, scientists compare observations before and after an event they couldn’t control, such as a natural disaster or disease outbreak. In the case of COVID-19, these researchers compared public health datasets in multiple localities (e.g., states or cities) within each of the six countries before and several weeks after lockdowns. For each data sample from a given locality, the time period right before a policy deployment was the experimental “control” for the same locality several weeks after it received one or more shutdown policy “treatments.”
Hsiang and his colleagues measured the effects of all the different policies put into place at local, regional, and national levels. These included travel restrictions, business and school closures, shelter-in-place orders, and other actions that didn’t involve any type of medical treatment for COVID-19.
Because SARS-CoV-2 is a new virus, the researchers knew that early in the pandemic, everyone was susceptible, and the outbreak would grow exponentially. The scientists could then use a statistical method designed to estimate how the daily growth rate of infections changed over time within a location after different combinations of large-scale policies were put into place.
The result? Early in the pandemic, coronavirus infection rates grew 38 percent each day, on average, across the six countries: translating to a two-day doubling time. Applying all policies at once slowed the daily COVID-19 infection rate by 31 percentage points! Policies having the clearest benefit were business closures and lockdowns, whereas travel restrictions and bans on social gatherings had mixed results. Without more data, the analysis can’t specify why, but the way different countries enacted those policies might be one reason.
As we continue to try to understand and thwart this new virus and its damage to so many aspects of our personal and professional lives, these new findings add context, comfort, and guidance about the present circumstances. They tell us that individual sacrifices from staying home and canceled events contributed collectively to a huge, positive impact on the world.
Now, as various communities start cautiously to open up, we should continue to practice social distancing, mask wearing, and handwashing. This is not the time to say that the risk has passed. We are all tired of the virus and its consequences for our personal lives, but the virus doesn’t care. It’s still out there. Stay safe, everyone!
Reference:
[1] The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Hsiang S, Allen D, Annan-Phan S, et al. Nature. 2020 June 8 [published online ahead of print].
Links:
Coronavirus (NIH)
Global Policy Lab: Effect of Anti-Contagion Policies (University of California, Berkeley)
Video: How much have policies to slow COVID-19 worked? (UC Berkeley)
Hsiang Lab (UC Berkeley)
“Global Policy Lab Rallies for COVID-19 Research,” COVID-19 News, Goldman School of Public Policy, June 5, 2020.
Posted In: News
Tags: China, COVID-19, disease prevention, econometrics, epidemiological modeling, epidemiology, France, global health, global policy, Iran, Italy, New Zealand, novel coronavirus, pandemic, public health, SARS-CoV-2, shutdown policies, Singapore, social distancing, South Korea, virology
Battling Malaria at the Atomic Level
Posted on February 11th, 2020 by Dr. Francis Collins
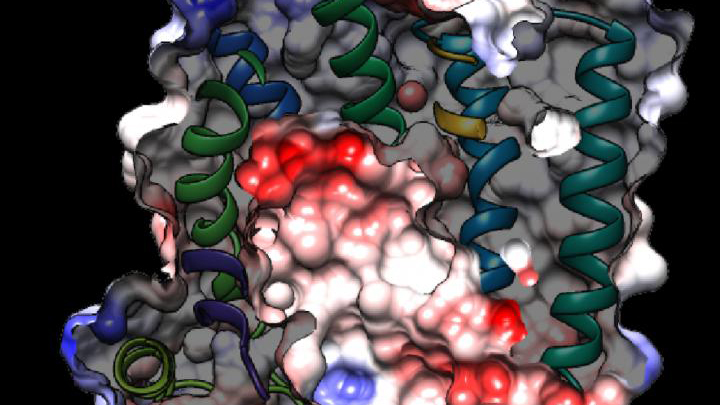
Credit: Columbia University Irving Medical Center, New York
Tropical medicine has its share of wily microbes. Among the most clever is the mosquito-borne protozoan Plasmodium falciparum, which is the cause of the most common—and most lethal—form of malaria. For decades, doctors have used antimalarial drugs against P. falciparum. But just when malaria appeared to be well on its way to eradication, this parasitic protozoan mutated in ways that has enabled it to resist frontline antimalarial drugs. This resistance is a major reason that malaria, one of the world’s oldest diseases, still claims the lives of about 400,000 people each year [1].
This is a situation with which I have personal experience. Thirty years ago before traveling to Nigeria, I followed directions and took chloroquine to prevent malaria. But the resistance to the drug was already widespread, and I came down with malaria anyway. Fortunately, the parasite that a mosquito delivered to me was sensitive to another drug called Fansidar, which acts through another mechanism. I was pretty sick for a few days, but recovered without lasting consequences.
While new drugs are being developed to thwart P. falciparum, some researchers are busy developing tools to predict what mutations are likely to occur next in the parasite’s genome. And that’s what is so exciting about the image above. It presents the unprecedented, 3D atomic-resolution structure of a protein made by P. falciparum that’s been a major source of its resistance: the chloroquine-resistance transporter protein, or PfCRT.
In this cropped density map, you see part of the protein’s biochemical structure. The colorized area displays the long, winding chain of amino acids within the protein as helices in shades of green, blue and gold. These helices enclose a central cavity essential for the function of the protein, whose electrostatic properties are shown here as negative (red), positive (blue), and neutral (white). All this structural information was captured using cryo-electron microscopy (cryo-EM). The technique involves flash-freezing molecules in liquid nitrogen and bombarding them with electrons to capture their images with a special camera.
This groundbreaking work, published recently in Nature, comes from an NIH-supported multidisciplinary research team, led by David Fidock, Matthias Quick, and Filippo Mancia, Columbia University Irving Medical Center, New York [2]. It marks a major feat for structural biology, because PfCRT is on the small side for standard cryo-EM and, as Mancia discovered, the protein is almost featureless.
These two strikes made Mancia and colleagues wonder at first whether they would swing and miss at their attempt to image the protein. With the help of coauthor Anthony Kossiakoff, a researcher at the University of Chicago, the team complexed PfCRT to a bulkier antibody fragment. That doubled the size of their subject, and the fragment helped to draw out PfCRT’s hidden features. One year and a lot of hard work later, they got their homerun.
PfCRT is a transport protein embedded in the surface membrane of what passes for the gut of P. falciparum. Because the gene encoding it is highly mutable, the PfCRT protein modified its structure many years ago, enabling it to pump out and render ineffective several drugs in a major class of antimalarials called 4-aminoquinolines. That includes chloroquine.
Now, with the atomic structure in hand, researchers can map the locations of existing mutations and study how they work. This information will also allow them to model which regions of the protein to be on the lookout for the next adaptive mutations. The hope is this work will help to prolong the effectiveness of today’s antimalarial drugs.
For example, the drug piperaquine, a 4-aminoquinoline agent, is now used in combination with another antimalarial. The combination has proved quite effective. But recent reports show that P. falciparum has acquired resistance to piperaquine, driven by mutations in PfCRT that are spreading rapidly across Southeast Asia [3].
Interestingly, the researchers say they have already pinpointed single mutations that could confer piperaquine resistance to parasites from South America. They’ve also located where new mutations are likely to occur to compromise the drug’s action in Africa, where most malarial infections and deaths occur. So, this atomic structure is already being put to good use.
Researchers also hope that this model will allow drug designers to make structural adjustments to old, less effective malarial drugs and perhaps restore them to their former potency. Perhaps this could even be done by modifying chloroquine, introduced in the 1940s as the first effective antimalarial. It was used worldwide but was largely shelved a few decades later due to resistance—as I experienced three decades ago.
Malaria remains a constant health threat for millions of people living in subtropical areas of the world. Wouldn’t it be great to restore chloroquine to the status of a frontline antimalarial? The drug is inexpensive, taken orally, and safe. Through the power of science, its return is no longer out of the question.
References:
[1] World malaria report 2019. World Health Organization, December 4, 2019
[2] Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, Vendome J, Hagenah LM, Giacometti SI, Warren AL, Nosol K, Roepe PD, Potter CS, Carragher B, Kossiakoff AA, Quick M, Fidock DA, Mancia F. Nature. 2019 Dec;576(7786):315-320.
[3] Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. van der Pluijm RW, Imwong M, Chau NH, Hoa NT, et. al. Lancet Infect Dis. 2019 Sep;19(9):952-961.
Links:
Malaria (National Institute of Allergy and Infectious Diseases/NIH)
Fidock Lab (Columbia University Irving Medical Center, New York)
Video: David Fidock on antimalarial drug resistance (BioMedCentral/YouTube)
Kossiakoff Lab (University of Chicago)
Mancia Lab (Columbia University Irving Medical Center)
Matthias Quick (Columbia University Irving Medical Center)
NIH Support: National Institute of Allergy and Infectious Diseases; National Institute of General Medical Sciences; National Heart, Lung, and Blood Institute
Posted In: Snapshots of Life
Tags: Africa, American Society of Tropical Medicine and Hygiene, antimalarial drugs, atomic structure, chloroquine, chloroquine-resistance transporter, cryo-EM, Fansidar, genomics, global health, imaging, malaria, P. falciparum, P. falciparum mutants, PfCRT, piperaquine, Plasmodium falciparum, Southeast Asia, structural biology, tropical diseases
Celebrating 2019 Biomedical Breakthroughs
Posted on January 2nd, 2020 by Dr. Francis Collins

Happy New Year! As we say goodbye to the Teens, let’s take a look back at 2019 and some of the groundbreaking scientific discoveries that closed out this remarkable decade.
Each December, the reporters and editors at the journal Science select their breakthrough of the year, and the choice for 2019 is nothing less than spectacular: An international network of radio astronomers published the first image of a black hole, the long-theorized cosmic singularity where gravity is so strong that even light cannot escape [1]. This one resides in a galaxy 53 million light-years from Earth! (A light-year equals about 6 trillion miles.)
Though the competition was certainly stiff in 2019, the biomedical sciences were well represented among _Science_’s “runner-up” breakthroughs. They include three breakthroughs that have received NIH support. Let’s take a look at them:
In a first, drug treats most cases of cystic fibrosis: Last October, two international research teams reported the results from phase 3 clinical trials of the triple drug therapy Trikafta to treat cystic fibrosis (CF). Their data showed Trikafta effectively compensates for the effects of a mutation carried by about 90 percent of people born with CF. Upon reviewing these impressive data, the Food and Drug Administration (FDA) approved Trikafta, developed by Vertex Pharmaceuticals.
The approval of Trikafta was a wonderful day for me personally, having co-led the team that isolated the CF gene 30 years ago. A few years later, I wrote a song called “Dare to Dream” imagining that wonderful day when “the story of CF is history.” Though we’ve still got more work to do, we’re getting a lot closer to making that dream come true. Indeed, with the approval of Trikafta, most people with CF have for the first time ever a real chance at managing this genetic disease as a chronic condition over the course of their lives. That’s a tremendous accomplishment considering that few with CF lived beyond their teens as recently as the 1980s.
Such progress has been made possible by decades of work involving a vast number of researchers, many funded by NIH, as well as by more than two decades of visionary and collaborative efforts between the Cystic Fibrosis Foundation and Aurora Biosciences (now, Vertex) that built upon that fundamental knowledge of the responsible gene and its protein product. Not only did this innovative approach serve to accelerate the development of therapies for CF, it established a model that may inform efforts to develop therapies for other rare genetic diseases.
Hope for Ebola patients, at last: It was just six years ago that news of a major Ebola outbreak in West Africa sounded a global health emergency of the highest order. Ebola virus disease was then recognized as an untreatable, rapidly fatal illness for the majority of those who contracted it. Though international control efforts ultimately contained the spread of the virus in West Africa within about two years, over 28,600 cases had been confirmed leading to more than 11,000 deaths—marking the largest known Ebola outbreak in human history. Most recently, another major outbreak continues to wreak havoc in northeastern Democratic Republic of Congo (DRC), where violent civil unrest is greatly challenging public health control efforts.
As troubling as this news remains, 2019 brought a needed breakthrough for the millions of people living in areas susceptible to Ebola outbreaks. A randomized clinical trial in the DRC evaluated four different drugs for treating acutely infected individuals, including an antibody against the virus called mAb114, and a cocktail of anti-Ebola antibodies referred to as REGN-EB3. The trial’s preliminary data showed that about 70 percent of the patients who received either mAb114 or the REGN-EB3 antibody cocktail survived, compared with about half of those given either of the other two medicines.
So compelling were these preliminary results that the trial, co-sponsored by NIH’s National Institute of Allergy and Infectious Diseases (NIAID) and the DRC’s National Institute for Biomedical Research, was halted last August. The results were also promptly made public to help save lives and stem the latest outbreak. All Ebola patients in the DRC treatment centers now are treated with one or the other of these two options. The trial results were recently published.
The NIH-developed mAb114 antibody and the REGN-EB3 cocktail are the first therapeutics to be shown in a scientifically rigorous study to be effective at treating Ebola. This work also demonstrates that ethically sound clinical research can be conducted under difficult conditions in the midst of a disease outbreak. In fact, the halted study was named Pamoja Tulinde Maisha (PALM), which means “together save lives” in Kiswahili.
To top off the life-saving progress in 2019, the FDA just approved the first vaccine for Ebola. Called Ervebo (earlier rVSV-ZEBOV), this single-dose injectable vaccine is a non-infectious version of an animal virus that has been genetically engineered to carry a segment of a gene from the Zaire species of the Ebola virus—the virus responsible for the current DRC outbreak and the West Africa outbreak. Because the vaccine does not contain the whole Zaire virus, it can’t cause Ebola. Results from a large study in Guinea conducted by the WHO indicated that the vaccine offered substantial protection against Ebola virus disease. Ervebo, produced by Merck, has already been given to over 259,000 individuals as part of the response to the DRC outbreak. The NIH has supported numerous clinical trials of the vaccine, including an ongoing study in West Africa.
Microbes combat malnourishment: Researchers discovered a few years ago that abnormal microbial communities, or microbiomes, in the intestine appear to contribute to childhood malnutrition. An NIH-supported research team followed up on this lead with a study of kids in Bangladesh, and it published last July its groundbreaking finding: that foods formulated to repair the “gut microbiome” helped malnourished kids rebuild their health. The researchers were able to identify a network of 15 bacterial species that consistently interact in the gut microbiomes of Bangladeshi children. In this month-long study, this bacterial network helped the researchers characterize a child’s microbiome and/or its relative state of repair.
But a month isn’t long enough to determine how the new foods would help children grow and recover. The researchers are conducting a similar study that is much longer and larger. Globally, malnutrition affects an estimated 238 million children under the age 5, stunting their normal growth, compromising their health, and limiting their mental development. The hope is that these new foods and others adapted for use around the world soon will help many more kids grow up to be healthy adults.
Measles Resurgent: The staff at Science also listed their less-encouraging 2019 Breakdowns of the Year, and unfortunately the biomedical sciences made the cut with the return of measles in the U.S. Prior to 1963, when the measles vaccine was developed, 3 to 4 million Americans were sickened by measles each year. Each year about 500 children would die from measles, and many more would suffer lifelong complications. As more people were vaccinated, the incidence of measles plummeted. By the year 2000, the disease was even declared eliminated from the U.S.
But, as more parents have chosen not to vaccinate their children, driven by the now debunked claim that vaccines are connected to autism, measles has made a very preventable comeback. Last October, the Centers for Disease Control and Prevention (CDC) reported an estimated 1,250 measles cases in the United States at that point in 2019, surpassing the total number of cases reported annually in each of the past 25 years.
The good news is those numbers can be reduced if more people get the vaccine, which has been shown repeatedly in many large and rigorous studies to be safe and effective. The CDC recommends that children should receive their first dose by 12 to 15 months of age and a second dose between the ages of 4 and 6. Older people who’ve been vaccinated or have had the measles previously should consider being re-vaccinated, especially if they live in places with low vaccination rates or will be traveling to countries where measles are endemic.
Despite this public health breakdown, 2019 closed out a memorable decade of scientific discovery. The Twenties will build on discoveries made during the Teens and bring us even closer to an era of precision medicine to improve the lives of millions of Americans. So, onward to 2020—and happy New Year!
Reference:
[1] 2019 Breakthrough of the Year. Science, December 19, 2019.
NIH Support: These breakthroughs represent the culmination of years of research involving many investigators and the support of multiple NIH institutes.
Posted In: News
Tags: 2019 Biomedical Breakdown, 2019 Breakthroughs of the Year, Africa, antibody, Bangladesh, breakdown, breakthroughs, Breakthroughs of 2019, CDC, CF, child health, childhood vaccines, children, clinical trials, Congo, cystic fibrosis, Cystic Fibrosis Foundation, diet, Ebola, Ebola vaccine, Ebola Virus Disease, Ervebo, FDA, gene-based therapy, genetic diseases, global health, Guinea, gut microbiome, hemorrhagic fever, infectious disease, mAB114, malnutrition, measles, measles outbreak, Merck, microbiology, microbiome, PALM, pandemic, precision medicine, public health, rare disease, REGN-EB3, Trikafta, Vertex, Vertex Pharmaceuticals, virus, West Africa
