The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X (original) (raw)
- Letters to Nature
- Published: 15 February 2001
- P. Deloukas1,
- A. Dunham1,
- L. French1,
- S. G. Gregory1,
- S. J. Humphray1,
- A. J. Mungall1,
- M. T. Ross1,
- N. P. Carter1,
- I. Dunham1,
- C. E. Scott1,
- K. J. Ashcroft1,
- A. L. Atkinson1,
- K. Aubin1,
- D. M. Beare1,
- G. Bethel1,
- N. Brady1,
- J. C. Brook1,
- D. C. Burford1,
- W. D. Burrill1,
- C. Burrows1,
- A. P. Butler1,
- C. Carder1,
- J. J. Catanese4,
- C. M. Clee1,
- S. M. Clegg1,
- V. Cobley1,
- A. J. Coffey1,
- C. G. Cole1,
- J. E. Collins1,
- J. S. Conquer1,
- R. A. Cooper1,
- K. M. Culley1,
- E. Dawson1,
- F. L. Dearden1,
- R. M. Durbin1,
- P. J. de Jong4,
- P. D. Dhami1,
- M. E. Earthrowl1,
- C. A. Edwards1,
- R. S. Evans1,
- C. J. Gillson1,
- J. Ghori1,
- L. Green1,
- R. Gwilliam1,
- K. S. Halls1,
- S. Hammond1,
- G. L. Harper1,
- R. W. Heathcott1,
- J. L. Holden1,
- E. Holloway1,
- B. L. Hopkins1,
- P. J. Howard1,
- G. R. Howell1,
- E. J. Huckle1,
- J. Hughes1,
- P. J. Hunt1,
- S. E. Hunt1,
- M. Izmajlowicz1,
- C. A. Jones1,
- S. S. Joseph1,
- G. Laird1,
- C. F. Langford1,
- M. H. Lehvaslaiho1,
- M. A. Leversha1,
- O. T. McCann1,
- L. M. McDonald1,
- J. McDowall1,
- G. L. Maslen1,
- D. Mistry1,
- N. K. Moschonas2,
- V. Neocleous3,
- D. M. Pearson1,
- K. J. Phillips1,
- K. M. Porter1,
- S. R. Prathalingam1,
- Y. H. Ramsey1,
- S. A. Ranby1,
- C. M. Rice1,
- J. Rogers1,
- L. J. Rogers1,
- T. Sarafidou2,
- D. J. Scott1,
- G. J. Sharp1,
- C. J. Shaw-Smith1,
- L. J. Smink1,
- C. Soderlund1,
- E. C. Sotheran1,
- H. E. Steingruber1,
- J. E. Sulston1,
- A. Taylor1,
- R. G. Taylor1,
- A. A. Thorpe1,
- E. Tinsley1,
- G. L. Warry1,
- A. Whittaker1,
- P. Whittaker1,
- S. H. Williams1,
- T. E. Wilmer1,
- R. Wooster1 &
- …
- C. L. Wright1
Nature volume 409, pages 942–943 (2001)Cite this article
- 3332 Accesses
- 50 Citations
- 3 Altmetric
- Metrics details
Abstract
We constructed maps for eight chromosomes (1, 6, 9, 10, 13, 20, X and (previously) 22), representing one-third of the genome, by building landmark maps, isolating bacterial clones and assembling contigs. By this approach, we could establish the long-range organization of the maps early in the project, and all contig extension, gap closure and problem-solving was simplified by containment within local regions. The maps currently represent more than 94% of the euchromatic (gene-containing) regions of these chromosomes in 176 contigs, and contain 96% of the chromosome-specific markers in the human gene map. By measuring the remaining gaps, we can assess chromosome length and coverage in sequenced clones.
Main
The task of sequencing the 3,200 megabase (Mb) human genome can be subdivided into individual chromosome projects ranging in size from 263 Mb (chromosome 1)1 to about 35 Mb (chromosomes 21q and 22q)2,3. Our strategy, in common with other groups4,5,6, was to map selected chromosomes individually, and then to combine the results with those of whole-genome mapping studies into a single map of the human genome7. Chromosome maps were constructed as follows (see Supplementary Information). First, we constructed a landmark map for each chromosome. Second, we identified bacterial clones (bacterial- or P1-derived artificial chromosomes (BACs or PACs)) from genomic libraries using the chromosome-specific landmarks, and assembled them into contigs on the basis of shared restriction enzyme fingerprints and landmark content. Third, contigs were extended and joined by chromosome walking. Walking was carried out using BAC end sequences generated in house from clones selected at the ends of contigs, or from the publicly available resources; joins were also made by identification of overlaps using genomic sequence data.
Clones that were representative of each contig were selected regularly for inclusion in the ‘tiling path’ (a set of minimally overlapping clones) for genomic sequencing8. Clones from the tiling path of each chromosome were deposited in the ‘HumanMap’ database at the Genome Sequencing Centre7 (http://genome.wustl.edu/gsc/human/Mapping/), for integration with the data obtained by whole-genome fingerprinting. New clones were identified from this integrated dataset to assist the extension and closure of the chromosome maps (Table 1). A detailed description of tiling paths and the underlying clone contigs is available as Supplementary Information and will continue to be updated at http://www.sanger.ac.uk; all clones are publicly available.
Table 1 Status of chromosome maps
A key question is the extent of coverage of each chromosome in the map. We analysed the coverage of the euchromatic regions on the basis of the assumptions given below and the approximate estimates that were available for chromosome length. Heterochromatic regions, which are estimated to comprise 3–15% of each chromosome (Table 1), are absent from the contigs analysed here. On the basis of the fingerprint bands in the maps (converted to Mb as described in Supplementary Information), 176 contigs represent 927 Mb, or 94% of the estimated total of 981 Mb euchromatin. Half of this (485 Mb) is in fifteen contigs of 22–62 Mb, that illustrates the extent of continuity obtained. We also analysed the representation of human gene markers in the map. The clone map contained 96% of the unique markers that were previously mapped to these chromosomes in GeneMap99 (ref. 9), and 90% were present in the genome sequence. As expected, this figure is higher for individual chromosomes 20 (99%) and 22q (98%), which are nearly or completely finished (Table 1).
Independent corroboration of the coverage of each chromosome required identification of the boundaries between euchromatic sequence and the centromeric, telomeric, and other heterochromatic repeat sequences, as well as measurement of the remaining gaps in the map. We have measured 52 of the 57 remaining gaps in the maps of chromosomes 6, 9, 10, 13 and 20 by fluorescent in situ hybridization (FISH), in addition to the gaps previously measured on chromosome 22. Clones immediately flanking each gap were hybridized to extended DNA fibres, interphase nuclei or metaphase chromosome spreads.
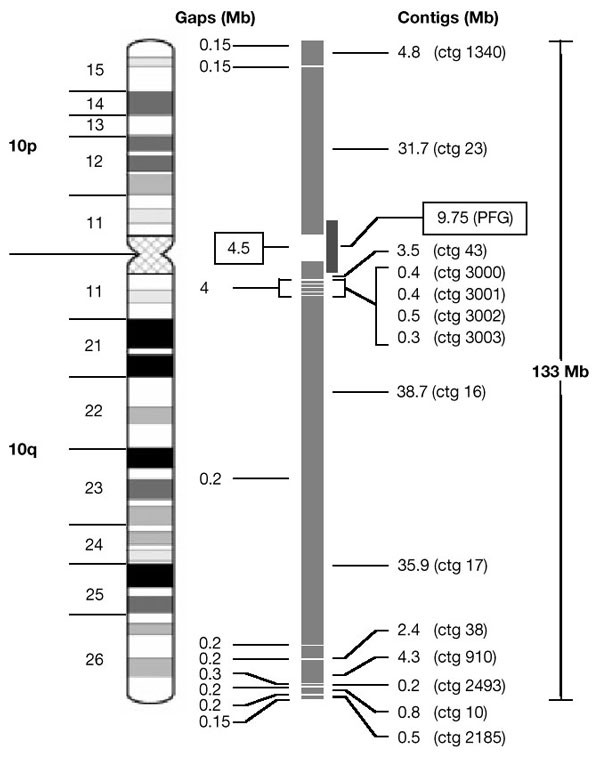
An example of this analysis is given for chromosome 10 (Fig. 1). Fourteen contigs account for 124.4 Mb of DNA. They contain pericentromeric satellite sequences, and also a subtelomeric sequence on the short arm which is <0.1 Mb from the telomere; the most distal clone on the long arm is <0.15 Mb from the telomere10. From FISH analysis, the total extent of euchromatic gaps is ≤ 4.2 Mb. From these measurements, the estimated coverage of euchromatin in the map can be revised from 89% (based on Morton's previous estimate; Table 1) to 96.7% (124.4 of 128.6 Mb). A 9.75-Mb restriction map that spans the centromere has been constructed for chromosome 10 (ref. 11). By anchoring this to the clone maps on both chromosome arms, we estimate that the size of the gap across the centromeric region is around 4.5 Mb. This analysis provides a new estimate of 133 Mb for the total length of the chromosome (Fig. 1), compared with the previous value of 144 Mb. In a similar analysis of chromosome 13, seven contigs span 102 Mb and the remaining gaps in the euchromatic region total a maximum of 5 Mb. This leads to a new, increased estimate of the size of the euchromatic region of 107 Mb, and the current coverage in the map is 95%.
Figure 1: Physical map of chromosome 10.
Clone contigs cover 124.4 Mb, euchromatic gaps cover 4.2 Mb, and the gap across centromeric satellites is 4.5 Mb11. The separation between contigs 43 and 16 was determined as ∼4 Mb on the basis of FISH of metaphase chromosome spreads, from which the sum of contigs 3000–3003 (1.6 Mb) was subtracted for a net gap coverage of 2.4 Mb in this region of 10q11.
Our strategy may form the basis for finishing the map and sequence of every human chromosome. Accurate measurement of all gap sizes assists our continued efforts to bridge the remaining gaps in clones, using other cloning systems (for example yeast artificial chromosomes (YACs), plasmids and bacteriophage lambda), and characterization of the centromeric satellites and the remaining heterochromatic regions, including the short arms of the acrocentric chromosomes (13, 14, 15, 21 and 22), will enable us to determine the true physical extent of DNA in the human genome.
References
- Morton, N. E. Parameters of the human genome. Proc. Natl Acad. Sci. USA 88, 7474–7476 (1991).
Article ADS CAS Google Scholar - The chromosome 21 mapping and sequencing consortium. The DNA sequence of human chromosome 21. Nature 405, 311–319 (2000).
Article Google Scholar - Dunham, I. et al. The DNA sequence of human chromosome 22. Nature 402, 489–495 (1999).
Article ADS CAS Google Scholar - Tilford, C. A. et al. A physical map of the human Y chromosome. Nature 409, 943–945 (2001).
Article ADS CAS Google Scholar - Montgomery, K. T. A high-resolution map of human chromosome 12. Nature 409, 945–946 (2001).
Article ADS CAS Google Scholar - Bruls, T. et al. A physical map of human chromosome 14. Nature 409, 947–948 (2001).
Article ADS CAS Google Scholar - The International Human Genome Mapping Consortium. A physical map of the human genome. Nature 409, 934–941 (2001).
Article ADS Google Scholar - International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Article ADS Google Scholar - Deloukas, P. et al. A physical map of 30,000 human genes. Science 282, 744–746 (1998).
Article ADS CAS Google Scholar - Riethman, H. C. et al. Integration of telomere sequences with the draft human genome sequence. Nature 409, 948–951 (2001).
Article ADS CAS Google Scholar - Jackson, M. S., See, C. G., Mulligan, L. M. & Lauffart, B. F. A 9.75-Mb map across the centromere of human chromosome 10. Genomics 33, 258–270 (1996).
Article CAS Google Scholar
Acknowledgements
We thank M. Sekhon, A. Chinwalla, J. McPherson and staff of the Genome Sequencing Centre, St. Louis for their assistance; the many collaborators who have contributed reagents and information to assist map construction on individual chromosomes; M. Jackson for helpful discussion; T. Cox, R. Pettett and the web team; and the Wellcome Trust for support.
Author information
Authors and Affiliations
- The Sanger Centre, Wellcome Trust Genome Campus, Hinxton, CB10 1SA, Cambridge, UK
D. R. Bentley, P. Deloukas, A. Dunham, L. French, S. G. Gregory, S. J. Humphray, A. J. Mungall, M. T. Ross, N. P. Carter, I. Dunham, C. E. Scott, K. J. Ashcroft, A. L. Atkinson, K. Aubin, D. M. Beare, G. Bethel, N. Brady, J. C. Brook, D. C. Burford, W. D. Burrill, C. Burrows, A. P. Butler, C. Carder, C. M. Clee, S. M. Clegg, V. Cobley, A. J. Coffey, C. G. Cole, J. E. Collins, J. S. Conquer, R. A. Cooper, K. M. Culley, E. Dawson, F. L. Dearden, R. M. Durbin, P. D. Dhami, M. E. Earthrowl, C. A. Edwards, R. S. Evans, C. J. Gillson, J. Ghori, L. Green, R. Gwilliam, K. S. Halls, S. Hammond, G. L. Harper, R. W. Heathcott, J. L. Holden, E. Holloway, B. L. Hopkins, P. J. Howard, G. R. Howell, E. J. Huckle, J. Hughes, P. J. Hunt, S. E. Hunt, M. Izmajlowicz, C. A. Jones, S. S. Joseph, G. Laird, C. F. Langford, M. H. Lehvaslaiho, M. A. Leversha, O. T. McCann, L. M. McDonald, J. McDowall, G. L. Maslen, D. Mistry, D. M. Pearson, K. J. Phillips, K. M. Porter, S. R. Prathalingam, Y. H. Ramsey, S. A. Ranby, C. M. Rice, J. Rogers, L. J. Rogers, D. J. Scott, G. J. Sharp, C. J. Shaw-Smith, L. J. Smink, C. Soderlund, E. C. Sotheran, H. E. Steingruber, J. E. Sulston, A. Taylor, R. G. Taylor, A. A. Thorpe, E. Tinsley, G. L. Warry, A. Whittaker, P. Whittaker, S. H. Williams, T. E. Wilmer, R. Wooster & C. L. Wright - Department of Biology, University of Crete and Institute of Molecular Biology and Biotechnology, PO Box 2208, Heraklion, 71409, Crete, Greece
N. K. Moschonas & T. Sarafidou - Neurogenetic Laboratory, The Cyprus Institute of Neurology and Genetics, 6, International Airport Avenue, PO Box 23462, Nicosia, 1683, Cyprus
V. Neocleous - Children's Hospital-BACPAC Resources, 747 52nd Street, Oakland, 94609, California, USA
J. J. Catanese & P. J. de Jong
Authors
- D. R. Bentley
You can also search for this author inPubMed Google Scholar - P. Deloukas
You can also search for this author inPubMed Google Scholar - A. Dunham
You can also search for this author inPubMed Google Scholar - L. French
You can also search for this author inPubMed Google Scholar - S. G. Gregory
You can also search for this author inPubMed Google Scholar - S. J. Humphray
You can also search for this author inPubMed Google Scholar - A. J. Mungall
You can also search for this author inPubMed Google Scholar - M. T. Ross
You can also search for this author inPubMed Google Scholar - N. P. Carter
You can also search for this author inPubMed Google Scholar - I. Dunham
You can also search for this author inPubMed Google Scholar - C. E. Scott
You can also search for this author inPubMed Google Scholar - K. J. Ashcroft
You can also search for this author inPubMed Google Scholar - A. L. Atkinson
You can also search for this author inPubMed Google Scholar - K. Aubin
You can also search for this author inPubMed Google Scholar - D. M. Beare
You can also search for this author inPubMed Google Scholar - G. Bethel
You can also search for this author inPubMed Google Scholar - N. Brady
You can also search for this author inPubMed Google Scholar - J. C. Brook
You can also search for this author inPubMed Google Scholar - D. C. Burford
You can also search for this author inPubMed Google Scholar - W. D. Burrill
You can also search for this author inPubMed Google Scholar - C. Burrows
You can also search for this author inPubMed Google Scholar - A. P. Butler
You can also search for this author inPubMed Google Scholar - C. Carder
You can also search for this author inPubMed Google Scholar - J. J. Catanese
You can also search for this author inPubMed Google Scholar - C. M. Clee
You can also search for this author inPubMed Google Scholar - S. M. Clegg
You can also search for this author inPubMed Google Scholar - V. Cobley
You can also search for this author inPubMed Google Scholar - A. J. Coffey
You can also search for this author inPubMed Google Scholar - C. G. Cole
You can also search for this author inPubMed Google Scholar - J. E. Collins
You can also search for this author inPubMed Google Scholar - J. S. Conquer
You can also search for this author inPubMed Google Scholar - R. A. Cooper
You can also search for this author inPubMed Google Scholar - K. M. Culley
You can also search for this author inPubMed Google Scholar - E. Dawson
You can also search for this author inPubMed Google Scholar - F. L. Dearden
You can also search for this author inPubMed Google Scholar - R. M. Durbin
You can also search for this author inPubMed Google Scholar - P. J. de Jong
You can also search for this author inPubMed Google Scholar - P. D. Dhami
You can also search for this author inPubMed Google Scholar - M. E. Earthrowl
You can also search for this author inPubMed Google Scholar - C. A. Edwards
You can also search for this author inPubMed Google Scholar - R. S. Evans
You can also search for this author inPubMed Google Scholar - C. J. Gillson
You can also search for this author inPubMed Google Scholar - J. Ghori
You can also search for this author inPubMed Google Scholar - L. Green
You can also search for this author inPubMed Google Scholar - R. Gwilliam
You can also search for this author inPubMed Google Scholar - K. S. Halls
You can also search for this author inPubMed Google Scholar - S. Hammond
You can also search for this author inPubMed Google Scholar - G. L. Harper
You can also search for this author inPubMed Google Scholar - R. W. Heathcott
You can also search for this author inPubMed Google Scholar - J. L. Holden
You can also search for this author inPubMed Google Scholar - E. Holloway
You can also search for this author inPubMed Google Scholar - B. L. Hopkins
You can also search for this author inPubMed Google Scholar - P. J. Howard
You can also search for this author inPubMed Google Scholar - G. R. Howell
You can also search for this author inPubMed Google Scholar - E. J. Huckle
You can also search for this author inPubMed Google Scholar - J. Hughes
You can also search for this author inPubMed Google Scholar - P. J. Hunt
You can also search for this author inPubMed Google Scholar - S. E. Hunt
You can also search for this author inPubMed Google Scholar - M. Izmajlowicz
You can also search for this author inPubMed Google Scholar - C. A. Jones
You can also search for this author inPubMed Google Scholar - S. S. Joseph
You can also search for this author inPubMed Google Scholar - G. Laird
You can also search for this author inPubMed Google Scholar - C. F. Langford
You can also search for this author inPubMed Google Scholar - M. H. Lehvaslaiho
You can also search for this author inPubMed Google Scholar - M. A. Leversha
You can also search for this author inPubMed Google Scholar - O. T. McCann
You can also search for this author inPubMed Google Scholar - L. M. McDonald
You can also search for this author inPubMed Google Scholar - J. McDowall
You can also search for this author inPubMed Google Scholar - G. L. Maslen
You can also search for this author inPubMed Google Scholar - D. Mistry
You can also search for this author inPubMed Google Scholar - N. K. Moschonas
You can also search for this author inPubMed Google Scholar - V. Neocleous
You can also search for this author inPubMed Google Scholar - D. M. Pearson
You can also search for this author inPubMed Google Scholar - K. J. Phillips
You can also search for this author inPubMed Google Scholar - K. M. Porter
You can also search for this author inPubMed Google Scholar - S. R. Prathalingam
You can also search for this author inPubMed Google Scholar - Y. H. Ramsey
You can also search for this author inPubMed Google Scholar - S. A. Ranby
You can also search for this author inPubMed Google Scholar - C. M. Rice
You can also search for this author inPubMed Google Scholar - J. Rogers
You can also search for this author inPubMed Google Scholar - L. J. Rogers
You can also search for this author inPubMed Google Scholar - T. Sarafidou
You can also search for this author inPubMed Google Scholar - D. J. Scott
You can also search for this author inPubMed Google Scholar - G. J. Sharp
You can also search for this author inPubMed Google Scholar - C. J. Shaw-Smith
You can also search for this author inPubMed Google Scholar - L. J. Smink
You can also search for this author inPubMed Google Scholar - C. Soderlund
You can also search for this author inPubMed Google Scholar - E. C. Sotheran
You can also search for this author inPubMed Google Scholar - H. E. Steingruber
You can also search for this author inPubMed Google Scholar - J. E. Sulston
You can also search for this author inPubMed Google Scholar - A. Taylor
You can also search for this author inPubMed Google Scholar - R. G. Taylor
You can also search for this author inPubMed Google Scholar - A. A. Thorpe
You can also search for this author inPubMed Google Scholar - E. Tinsley
You can also search for this author inPubMed Google Scholar - G. L. Warry
You can also search for this author inPubMed Google Scholar - A. Whittaker
You can also search for this author inPubMed Google Scholar - P. Whittaker
You can also search for this author inPubMed Google Scholar - S. H. Williams
You can also search for this author inPubMed Google Scholar - T. E. Wilmer
You can also search for this author inPubMed Google Scholar - R. Wooster
You can also search for this author inPubMed Google Scholar - C. L. Wright
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toD. R. Bentley.
Supplementary information
Rights and permissions
About this article
Cite this article
Bentley, D., Deloukas, P., Dunham, A. et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X.Nature 409, 942–943 (2001). https://doi.org/10.1038/35057165
- Received: 28 November 2000
- Accepted: 21 December 2000
- Issue Date: 15 February 2001
- DOI: https://doi.org/10.1038/35057165