Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution (original) (raw)
- Letter
- Open access
- Published: 08 October 2009
- Ryan D. Morin3 na1,
- Jaswinder Khattra1,
- Leah Prentice1,
- Trevor Pugh3,
- Angela Burleigh1,
- Allen Delaney3,
- Karen Gelmon4,
- Ryan Guliany1,
- Janine Senz2,
- Christian Steidl2,5,
- Robert A. Holt3,
- Steven Jones3,
- Mark Sun1,
- Gillian Leung1,
- Richard Moore3,
- Tesa Severson3,
- Greg A. Taylor3,
- Andrew E. Teschendorff6,
- Kane Tse1,
- Gulisa Turashvili1,
- Richard Varhol3,
- René L. Warren3,
- Peter Watson7,
- Yongjun Zhao3,
- Carlos Caldas6,
- David Huntsman2,5,
- Martin Hirst3,
- Marco A. Marra3 &
- …
- Samuel Aparicio1,2,5
Nature volume 461, pages 809–813 (2009)Cite this article
- 19k Accesses
- 883 Citations
- 82 Altmetric
- Metrics details
Abstract
Recent advances in next generation sequencing1,2,3,4 have made it possible to precisely characterize all somatic coding mutations that occur during the development and progression of individual cancers. Here we used these approaches to sequence the genomes (>43-fold coverage) and transcriptomes of an oestrogen-receptor-α-positive metastatic lobular breast cancer at depth. We found 32 somatic non-synonymous coding mutations present in the metastasis, and measured the frequency of these somatic mutations in DNA from the primary tumour of the same patient, which arose 9 years earlier. Five of the 32 mutations (in ABCB11, HAUS3, SLC24A4, SNX4 and PALB2) were prevalent in the DNA of the primary tumour removed at diagnosis 9 years earlier, six (in KIF1C, USP28, MYH8, MORC1, KIAA1468 and RNASEH2A) were present at lower frequencies (1–13%), 19 were not detected in the primary tumour, and two were undetermined. The combined analysis of genome and transcriptome data revealed two new RNA-editing events that recode the amino acid sequence of SRP9 and COG3. Taken together, our data show that single nucleotide mutational heterogeneity can be a property of low or intermediate grade primary breast cancers and that significant evolution can occur with disease progression.
Similar content being viewed by others
Main
Lobular breast cancer is an oestrogen-receptor-positive (ER+, also known as ESR1+) subtype of breast cancer (approximately 15% of all breast cancers). It is usually of low-intermediate histological grade and can recur many years after initial diagnosis. To interrogate the genomic landscape of this class of tumour, we re-sequenced1,2,3,4 the DNA from a metastatic lobular breast cancer specimen (89% tumour cellularity; Supplementary Fig. 1) at approximately 43.1-fold aligned, haploid reference genome coverage (120.7 gigabases (Gb) aligned paired-end sequence; Supplementary Fig. 2, Table 1 and Supplementary Methods). Deep high-throughput transcriptome sequencing (RNA-seq)5 performed on the same sample generated 160.9-million reads that could be aligned (Supplementary Table 1, see also Supplementary Fig. 2 and Supplementary Methods). The saturation of the genome (Table 1) and RNA-seq (Supplementary Table 1) libraries for single nucleotide variant (SNV) detection is discussed in Supplementary Information. The aligned (hg18) reads were used to identify (Supplementary Fig. 2) the presence of genomic aberrations, including SNVs (Supplementary Table 2), insertions/deletions (indels), gene fusions, translocations, inversions and copy number alterations (Supplementary Methods). We examined predicted coding indels and predicted inversions (coding or non-coding; Supplementary Methods); however, all of the events that were validated by Sanger re-sequencing were also present in the germ line (Supplementary Tables 3 and 4). None of the 12 predicted gene fusions revalidated. We also computed the segmental copy number (Supplementary Methods and Supplementary Table 5a) from aligned reads, and revalidated high level amplicons by fluorescence in situ hybridization (FISH) (Supplementary Table 5b), revealing the presence of a new low-level amplicon in the INSR locus (Supplementary Fig. 3).
Table 1 Summary of sequence library coverage
We identified coding SNVs from aligned reads, using a Binomial mixture model, SNVMix (Supplementary Table 2, Methods and Supplementary Appendix 1). From the RNA-seq (WTSS-PE) and genome (WGSS-PE) libraries we predicted 1,456 new coding non-synonymous SNVMix variants (Supplementary Table 2). After the removal of pseudogene and HLA sequences (1,178 positions remaining) and after primer design, we re-sequenced (Sanger amplicons) 1,120 non-synonymous coding SNV positions in the tumour DNA and normal lymphocyte DNA. Some 437 positions (268 unique to WGSS-PE, 15 unique to WTSS-PE, and 154 in common) were confirmed as non-synonymous coding variants. Of these, 405 were new germline alleles and 32 were revealed as non-synonymous coding somatic point mutations (Table 2). Of the 32 somatic mutations, 30 were present in WGSS-PE and/or WTSS-PE, whereas two were detected from the WTSS library sequence alone (Table 2). None of the 32 genes were found in common with the CAN breast genes6, which were discovered from ER- cell lines. Eleven genes appear in the current release of COSMIC7 (CHD3, SP1, PALB2, ERBB2, USP28, KLHL4, CDC6, KIAA1468, RNF220, COL1A1 and SNX4) but with mutations at different positions. We examined the population frequency of the somatic mutation positions for PALB2, ERBB2, USP28, CDC6, CHD3, HAUS3 (previously known as C4orf15), SP1, KIAA1468 and DLG4 in a further 192 breast cancers (Supplementary Methods; 112 lobular, 80 ductal). None of these 192 breast cancers showed identical mutations to those described here; however, 3 out of 192 cases (2 lobular, 1 ductal) contained neighbouring non-synonymous variants/deletions affecting the ERBB2 kinase domain (Supplementary Fig. 4). Interestingly, 2 out of 192 cases (both lobular) contained two different heterozygous truncating variants in HAUS3: chr4:2203685 G>T on minus strand, GAG>TAG (Glu>stop), and chr4:2203483 C>G on minus strand, TCA>TGA (Ser>stop) (Supplementary Fig. 5). Notably, HAUS3 is a member of the recently described8,9,10 multiprotein augmin complex, the function of which is required for genome stability mediated by appropriate kinetochore attachment and centrosome morphogenesis.
Table 2 Somatic coding sequence SNVs validated by Sanger sequencing
To determine how many of the somatic non-synonymous coding sequence mutations were already present at diagnosis 9 years earlier, we next examined genomic DNA from the primary tumour directly, by a single molecule frequency counting experiment (Supplementary Methods)4. Twenty-eight of the 32 mutations yielded amplicons compatible with Illumina sequencing (Supplementary Methods), and two extra mutations were sampled by Sanger sequencing (Supplementary Fig. 5). As controls we selected 36 heterozygous germline SNVs at random. The PCR amplicons for known germline and somatic mutations were sequenced on an Illumina device. After alignment, the observed counts of reference and non-reference bases at the target position were compared using the Binomial exact test. To calibrate the expected mean of the Binomial distribution, we used the non-reference allele frequency from positions -5 to +5 surrounding (but not including) the target position (Supplementary Table 6a, b), where only reference bases should be called. Unequal segmental amplification/deletion in the genome may contribute to a departure from the theoretical ratio of 0.5 for a heterozygous allele. As a result, amplicons from heterozygous germline alleles showed occasional measured frequencies of between 0.2 and 0.8 in both the primary and metastatic tumour DNA (Table 3 and Supplementary Table 7), but with a modal frequency around 0.5, as expected. In the metastatic genomic DNA the somatic mutations showed frequencies of between 0.2 and 0.79 (Table 3). Notably, the somatic coding mutation positions examined in the primary tumour showed three patterns of abundance: prevalent, rare and undetectable (Table 3). Mutations in ABCB11, PALB2 and SLC24A4 were detected at prevalent frequencies for heterozygous mutations (≥0.2, the lowest value seen for known germline alleles) given a 73% tumour content. The frequency of the mutation in HAUS3 was 0.79, consistent with it being a prevalent homozygous mutation, also confirmed by Sanger sequencing (Supplementary Fig. 5). Sanger amplicon sequencing showed that the SNX4 somatic mutation was also present in the primary tumour, whereas the KIAA1772 (also known as GREB1L) mutation was not. Six mutations (KIF1C, USP28, MORC1, MYH8, KIAA1468 and RNASEH2A) showed statistically significant (P < 0.01, Binomial exact test) intermediate frequencies of between 1% and 13% (Table 3), suggesting that these mutations were restricted to minor subclones of tumour cells. The remaining 19 out of 30 of the somatic coding mutations were not detected in the primary tumour DNA. Thus, significant heterogeneity in tumour somatic mutation content existed in the primary tumour at diagnosis. In contrast with the recently reported sequence of cytogenetically normal acute myeloid leukaemia (AML) tumour4, significant evolution of coding mutational content occurred between primary and metastasis. It is unknown whether the 19 mutations present in the metastasis, but not detected in the primary, were a consequence of radiation therapy or innate tumour progression.
Table 3 Frequency of germline and somatic alleles in the metastatic and primary genomes
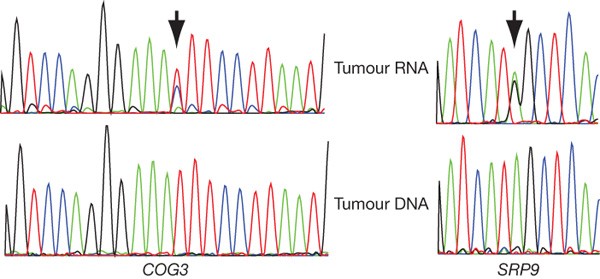
We also examined how the transfer of information from the nuclear genome to proteins was modified by alternative splicing (Supplementary Table 8 and Supplementary Fig. 6), biased allelic expression (Supplementary Table 9) and RNA editing. At the single nucleotide level, RNA-editing enzymes (which can be regulated by oestrogens11) may also recode transcripts resulting in a proteome divergent from the genome12,13,14,15. Interestingly, the ADAR enzyme—one of the principal RNA-editing enzymes that mediates A→I(G) edits—was one of the top 5% of genes expressed (145.6 reads per base, Supplementary Table 10), and the only editing enzyme expressed at a high level. We searched for potential editing events (Methods) and found 3,122 candidate edits in 1,637 gene loci (Supplementary Table 11). Some 526 out of 3,122 candidate edits are non-synonymous changes and 232 are synonymous changes (with the remainder affecting untranslated regions). We revalidated independently (Supplementary Methods) by Sanger sequencing 75 editing events in 12 gene loci from the lobular metastasis (Supplementary Table 12 and see trace data at http://molonc.bccrc.ca/). Two genes, COG3 and SRP9 (Fig. 1), showed confirmed high frequency non-synonymous transcript editing, resulting in variant protein sequences. These observations emphasize the importance of integrating RNA-seq data with tumour genomes in assessing protein variation.
Figure 1: RNA editing in COG3 and SRP9.
Sanger sequence traces from the non-synonymous editing positions in COG3 and SRP9. The editing position is arrowed. Top trace is tumour RNA, bottom trace tumour DNA. The editing positions were confirmed with reverse strand reads (not shown).
The coding mutation landscape of breast cancers has, so far, been mostly determined from ER- metastatic cell lines/samples6,16, and has suggested the presence of large numbers of passenger events as well as drivers. Our results show the importance of sequencing samples of tumour cell populations early as well as late in the evolution of tumours, and of estimating allele frequency in tumour genomes. Our observations suggest that the sequencing of primary breast cancers and pre-invasive malignancy may reveal significantly fewer candidates for tumour initiating mutations.
Methods Summary
Paired-end reads were assigned quality scores and aligned to the reference genome (hg18) using Maq17 (Supplementary Methods and Supplementary Fig. 2). For identification of SNVs we used a simple Binomial mixture model, SNVMix (Supplementary Appendix 1), which assigns a probability to each base position as homozygous reference (aa), heterozygous non-reference (ab) and homozygous non-reference (bb), based on the occurrence of reference (hg18) and non-reference bases at each aligned position. This model was calibrated initially, using high confidence allele calls from Affymetrix SNP6.0 hybridization of tumour and normal DNA. We estimated the receiver operating characteristic (ROC) performance (Supplementary Fig. 8) and determined that an SNVMix threshold of P = 0.77 for (ab) or (bb) for a non-reference call would yield a false discovery rate (FDR) of 1%. For the RNA-seq library, a threshold of P = 0.53 was used (Supplementary Fig. 8; FDR = 0.01) to call non-reference positions. Non-reference positions were then filtered for known variants against the sources of germline variation, the single nucleotide polymorphism database (dbSNP) and the completed individual genomes18,19 (Supplementary Table 2). Saturation of the libraries for SNV discovery was determined by random re-sampling (Supplementary Fig. 9 and Supplementary Methods). Segmental copy number was inferred with a hidden Markov model (HMM) method (Supplementary Table 4a, b and Supplementary Methods).
We searched for RNA-editing events by examining all very high confidence (P(ab) + P(bb) > 0.9) SNVMix predictions from the RNA-seq library of the metastatic tumour, that were not found with extreme confidence (P(aa) > 0.99, derived from the SNVMix receiver operating curve at FDR = 0.01) at the same positions in the metastatic tumour genome library.
Accession codes
Data deposits
Genome sequence data have been deposited at the European Genotype Phenotype Archive (http://www.ebi.ac.uk/ega) which is hosted by the EBI, under accession number EGAS00000000054.
References
- Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 40, 722–729 (2008)
Article CAS Google Scholar - Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008)
Article ADS CAS Google Scholar - Morin, R. et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 45, 81–94 (2008)
Article CAS Google Scholar - Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008)
Article ADS CAS Google Scholar - Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcripomes by RNA-Seq. Nature Methods 5, 621–628 (2008)
Article CAS Google Scholar - Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007)
Article ADS CAS Google Scholar - Forbes, S. A. et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr. Protoc. Hum. Genet. Unit 10.11 10.1002/0471142905.hg1011s57 (2008)
- Goshima, G., Mayer, M., Zhang, N., Stuurman, N. & Vale, R. D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 (2008)
Article CAS Google Scholar - Meireles, A. M., Fisher, K. H., Colombie, N., Wakefield, J. G. & Ohkura, H. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184, 777–784 (2009)
Article CAS Google Scholar - Lawo, S. et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 (2009)
Article CAS Google Scholar - Pauklin, S., Sernandez, I. V., Bachmann, G., Ramiro, A. R. & Petersen-Mahrt, S. K. Estrogen directly activates AID transcription and function. J. Exp. Med. 206, 99–111 (2009)
Article CAS Google Scholar - Blow, M., Futreal, P. A., Wooster, R. & Stratton, M. R. A survey of RNA editing in human brain. Genome Res. 14, 2379–2387 (2004)
Article CAS Google Scholar - Athanasiadis, A., Rich, A. & Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2, e391 (2004)
Article Google Scholar - Maas, S., Kawahara, Y., Tamburro, K. M. & Nishikura, K. A-to-I RNA editing and human disease. RNA Biol. 3, 1–9 (2006)
Article CAS Google Scholar - Li, J. B. et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324, 1210–1213 (2009)
Article ADS CAS Google Scholar - Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007)
Article ADS CAS Google Scholar - Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008)
Article CAS Google Scholar - Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008)
Article ADS CAS Google Scholar - Wheeler, D. A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008)
Article ADS CAS Google Scholar
Acknowledgements
We thank C. Eaves and M. Pollak for comments on earlier versions of the manuscript. We thank and acknowledge the patients of the BC Cancer Agency for donations of tumour tissues to the TTR-BREAST tumour banking program. S.A. is supported by a Canada Research Chair in Molecular Oncology, S.P.S., J.K., L.P., A.B. and T.P. are supported by Michael Smith Foundation for Health Research awards. R.D.M. is a Vanier scholar (CIHR). A.B. is also supported by an NSERC award, and L.P. by a CIHR award. We are grateful for platform support from CIHR, Genome Canada, Genome BC, Canada Foundation for Innovation and the Michael Smith Foundation for Health Research. The work was funded by the BC Cancer Foundation and the CBCF BC/Yukon chapter.
Author Contributions S.P.S. and R.D.M.: led the data analysis and wrote the manuscript. M.H.: oversaw the sequencing efforts. J.K., L.P., T.P., J.S., C.S., A.B., R.M. and T.S.: validation of variants. A.D.: primer design. K.G. and P.W.: establishment of TTR-BREAST tumour bank. K.T., R.G., R.A.H., S.J., M.S., G.L., A.E.T., R.V., G.A.T. and R.L.W.: bioinformatic analysis. G.T., D.H. and P.W.: sample selection and histological grading. Y.Z.: Illumina sequencing library preparation. C.C. and D.H.: data analysis and interpretation. S.A. and M.A.M.: conceived and oversaw the study and wrote the manuscript.
Author information
Author notes
- Sohrab P. Shah and Ryan D. Morin: These authors contributed equally to this work.
Authors and Affiliations
- Molecular Oncology,,
Sohrab P. Shah, Jaswinder Khattra, Leah Prentice, Angela Burleigh, Ryan Guliany, Mark Sun, Gillian Leung, Kane Tse, Gulisa Turashvili & Samuel Aparicio - Centre for Translational and Applied Genomics,,
Sohrab P. Shah, Janine Senz, Christian Steidl, David Huntsman & Samuel Aparicio - Michael Smith Genome Sciences Centre, BC Cancer Agency, 675 West 10th Avenue, Vancouver V5Z 1L3, Canada ,
Ryan D. Morin, Trevor Pugh, Allen Delaney, Robert A. Holt, Steven Jones, Richard Moore, Tesa Severson, Greg A. Taylor, Richard Varhol, René L. Warren, Yongjun Zhao, Martin Hirst & Marco A. Marra - Medical Oncology, BC Cancer Agency, 600 West 10th Avenue, Vancouver V5Z 1L3, Canada ,
Karen Gelmon - Department of Pathology, University of British Columbia, G227-2211 Wesbrook Mall, British Columbia, Vancouver V6T 2B5, Canada,
Christian Steidl, David Huntsman & Samuel Aparicio - Cancer Research UK, Cambridge Research Institute, Li Ka Shing Centre, Robinson Way, Cambridge CB2 0RE, UK ,
Andrew E. Teschendorff & Carlos Caldas - Deeley Research Centre, BC Cancer Agency, Victoria V8R 6V5, Canada
Peter Watson
Authors
- Sohrab P. Shah
You can also search for this author inPubMed Google Scholar - Ryan D. Morin
You can also search for this author inPubMed Google Scholar - Jaswinder Khattra
You can also search for this author inPubMed Google Scholar - Leah Prentice
You can also search for this author inPubMed Google Scholar - Trevor Pugh
You can also search for this author inPubMed Google Scholar - Angela Burleigh
You can also search for this author inPubMed Google Scholar - Allen Delaney
You can also search for this author inPubMed Google Scholar - Karen Gelmon
You can also search for this author inPubMed Google Scholar - Ryan Guliany
You can also search for this author inPubMed Google Scholar - Janine Senz
You can also search for this author inPubMed Google Scholar - Christian Steidl
You can also search for this author inPubMed Google Scholar - Robert A. Holt
You can also search for this author inPubMed Google Scholar - Steven Jones
You can also search for this author inPubMed Google Scholar - Mark Sun
You can also search for this author inPubMed Google Scholar - Gillian Leung
You can also search for this author inPubMed Google Scholar - Richard Moore
You can also search for this author inPubMed Google Scholar - Tesa Severson
You can also search for this author inPubMed Google Scholar - Greg A. Taylor
You can also search for this author inPubMed Google Scholar - Andrew E. Teschendorff
You can also search for this author inPubMed Google Scholar - Kane Tse
You can also search for this author inPubMed Google Scholar - Gulisa Turashvili
You can also search for this author inPubMed Google Scholar - Richard Varhol
You can also search for this author inPubMed Google Scholar - René L. Warren
You can also search for this author inPubMed Google Scholar - Peter Watson
You can also search for this author inPubMed Google Scholar - Yongjun Zhao
You can also search for this author inPubMed Google Scholar - Carlos Caldas
You can also search for this author inPubMed Google Scholar - David Huntsman
You can also search for this author inPubMed Google Scholar - Martin Hirst
You can also search for this author inPubMed Google Scholar - Marco A. Marra
You can also search for this author inPubMed Google Scholar - Samuel Aparicio
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toMarco A. Marra or Samuel Aparicio.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and Data and Supplementary References. (PDF 267 kb)
Supplementary Information
This file contains Supplementary Figures S1-S9 with Legends together with the Legends for Supplementary Tables in S1-S12 (see file s3). (PDF 2977 kb)
Supplementary Tables
This files contains Supplementary Tables S1-S12 (see file s2 for Legends). (XLS 6835 kb)
Supplementary Data
This Supplemental Appendix file contains inferring SNVs from aligned reads using a Bayesian mixture model. (PDF 1962 kb)
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This license does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Shah, S., Morin, R., Khattra, J. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution.Nature 461, 809–813 (2009). https://doi.org/10.1038/nature08489
- Received: 04 September 2009
- Accepted: 10 September 2009
- Issue Date: 08 October 2009
- DOI: https://doi.org/10.1038/nature08489
This article is cited by
Editorial Summary
A cancer evolves: heterogeneity and mutational evolution in a breast tumour
Next-generation sequencing approaches have been used to investigate the genomes and transcriptomes of an oestrogen-receptor-α-positive metastatic lobular breast cancer from a patient — rather than from a cell line or xenograft — over a 9-year period between the diagnosis of the primary tumour and the appearance of metastasis. Comparison of the somatic non-synonymous coding mutations in the metastasis and the primary tumour of the same patient and the combined analysis of genome and transcriptome data provided insights into the mutational evolution that can occur with disease progression. The cover shows sequence elements of the HAUS3 locus, one of the genes found to be mutated in the tissue (shown in the background) from the primary lobular cancer used for this work.