Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing (original) (raw)
Abstract
Most patients with acute myeloid leukaemia (AML) die from progressive disease after relapse, which is associated with clonal evolution at the cytogenetic level1,2 . To determine the mutational spectrum associated with relapse, we sequenced the primary tumour and relapse genomes from eight AML patients, and validated hundreds of somatic mutations using deep sequencing; this allowed us to define clonality and clonal evolution patterns precisely at relapse. In addition to discovering novel, recurrently mutated genes (for example, WAC, SMC3, DIS3, DDX41 and DAXX) in AML, we also found two major clonal evolution patterns during AML relapse: (1) the founding clone in the primary tumour gained mutations and evolved into the relapse clone, or (2) a subclone of the founding clone survived initial therapy, gained additional mutations and expanded at relapse. In all cases, chemotherapy failed to eradicate the founding clone. The comparison of relapse-specific versus primary tumour mutations in all eight cases revealed an increase in transversions, probably due to DNA damage caused by cytotoxic chemotherapy. These data demonstrate that AML relapse is associated with the addition of new mutations and clonal evolution, which is shaped, in part, by the chemotherapy that the patients receive to establish and maintain remissions.
Similar content being viewed by others
Main
To investigate the genetic changes associated with AML relapse, and to determine whether clonal evolution contributes to relapse, we performed whole-genome sequencing of primary tumour–relapse pairs and matched skin samples from eight patients, including unique patient identifier (UPN) 933124, whose primary tumour mutations were previously reported3. Informed consent explicit for whole-genome sequencing was obtained for all patients on a protocol approved by the Washington University Medical School Institutional Review Board. We obtained >25× haploid coverage and >97% diploid coverage for each sample (Supplementary Table 1 and Supplementary Information). These patients were from five different French–American–British haematological subtypes, with elapsed times of 235–961 days between initial diagnosis and relapse (Supplementary Table 2a, b).
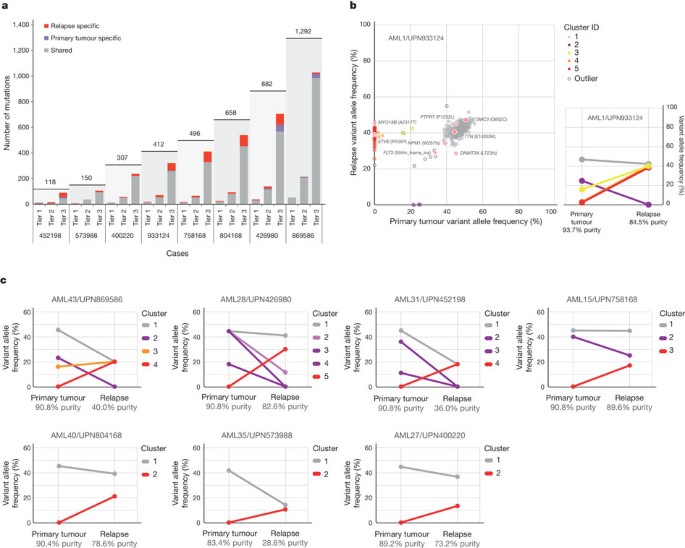
Candidate somatic events in the primary tumour and relapse genomes were identified4,5 and selected for hybridization capture-based validation using methods described in Supplementary Information. Deep sequencing of the captured target DNAs from skin (the matched normal tissue), primary tumour and relapse tumour specimens6 (Supplementary Table 3) yielded a median of 590-fold coverage per site. The average number of mutations and structural variants was 539 (range 118–1,292) per case (Fig. 1a).
Figure 1: Somatic mutations quantified by deep sequencing of capture validation targets in eight acute myeloid leukaemia primary tumour and relapse pairs.
a, Summary of tier 1–3 mutations detected in eight cases (not including translocations). All mutations shown were validated using capture followed by deep sequencing. Shared mutations are in grey, primary tumour-specific mutations in blue and relapse-specific mutations in red. The total number of tier 1–3 mutations for each case is shown above the light-grey rectangle. b, Mutant allele frequency distribution of validated mutations from tier 1–3 in the primary tumour and relapse of case UPN 933124 (left). Mutant allele frequencies for five primary-tumour-specific mutations were obtained from a 454 deep read-count experiment. Four mutation clusters were identified in the primary tumour, and one was found at relapse. Five low-level mutations in both the primary tumour and relapse (including four residing in known copy number variable regions) were excluded from the clustering analysis. Non-synonymous mutations from genes that are recurrently mutated in AML are shown. The change of mutant allele frequencies for mutations from the five clusters is shown (right) between the primary tumour and relapse. The orange and red lines are superimposed. c, The mutation clusters detected in the primary tumour and relapse samples from seven additional AML patients. The relationship between clusters in the primary tumour and relapse samples are indicated by lines linking them.
The general approach for relapse analysis is exemplified by the first sequenced case (UPN 933124). A total of 413 somatic events from tiers 1 to 3 were validated (see ref. 7 for tier designations; Supplementary Fig. 1a and Supplementary Tables 4a and 5). Of these, 78 mutations were relapse-specific (63 point mutations, 1 dinucleotide mutation, 13 indels and 1 translocation; relapse-specific criteria described in Supplementary Information and shown in Supplementary Fig. 1b), 5 point mutations were primary-tumour-specific, and 330 (317 point mutations and 13 indels) were shared between the primary tumour and relapse samples (Fig. 1a, b and Supplementary Fig. 2). The skin sample was contaminated with leukaemic cells for this case (peripheral white blood cell count was 105,000 cells mm−3 when the skin sample was banked), with an estimated tumour content in the skin sample of 29% (Supplementary Information). In addition to the ten somatic non-synonymous mutations originally reported for the primary tumour sample3, we identified one deletion that was not detected in the original analysis (DNMT3A L723fs (ref. 8)) and three mis-sense mutations previously misclassified as germline events (SMC3 G662C, PDXDC1 E421K and TTN E14263K) (Fig. 1b, Table 1 and Supplementary Table 4b).
Table 1 Coding mutations identified in eight primary tumour–relapse pairs
A total of 169 tier 1 coding mutations (approximately 21 per case) were identified in the eight patients (Table 1 and Supplementary Tables 4b and 6), of which 19 were relapse-specific. In addition to mutations in known AML genes such as DNMT3A (ref. 8), FLT3 (ref. 9), NPM1 (ref. 10), IDH1 (ref. 7), IDH2 (ref. 11), WT1 (ref. 12), RUNX1 (refs 13, 14), PTPRT (ref. 3), PHF6 (ref. 15) and ETV6 (ref. 16) in these eight patients, we also discovered novel, recurring mutations in WAC, SMC3, DIS3, DDX41 and DAXX using 200 AML cases whose exomes were sequenced as part of the Cancer Genome Atlas AML project (Table 1, Supplementary Table 4b and Supplementary Fig. 3; T.J.L., R.K.W. and The Cancer Genome Atlas working group on AML, unpublished data). Details regarding the novel, recurrently mutated genes are provided in Table 1, Supplementary Tables 4b and 7 and Supplementary Figs 3 and 4. Structural and functional analyses of structural variants are presented in the Supplementary Information (Supplementary Figs 5–10 and Supplementary Tables 2, 8 and 9).
The generation of high-depth sequencing data allowed us to quantify accurately mutant allele frequencies in all cases, permitting estimation of the size of tumour clonal populations in each AML sample. On the basis of mutation clustering results, we inferred the identity of four clones having distinct sets of mutations (clusters) in the primary tumour of AML1/UPN 933124 (Supplementary Information). The median mutant allele frequencies in the primary tumour for clusters 1 to 4 were 46.86%, 24.89%, 16.00% and 2.39%, respectively (Fig. 1b and Supplementary Table 5c). Clone 1 is the ‘founding’ clone (that is, the other subclones are derived from it), containing the cluster 1 mutations; assuming that nearly all of these mutations are heterozygous, they must be present in virtually all the tumour cells at presentation and at relapse, as the variant frequency of these mutations is ∼40–50%. Clone 2 (with cluster 2 mutations) and clone 3 (with cluster 3 mutations) must be derived from clone 1, because virtually all the cells in the sample contain the cluster 1 mutations (Fig. 2a). It is likely that a single cell from clone 3 gained a set of mutations (cluster 4) to form clone 4: these survived chemotherapy and evolved to become the dominant clone at relapse. We do not know whether any of the cluster 4 mutations conferred chemotherapy resistance; although none had translational consequences, we cannot rule out a relevant regulatory mutation in this cluster.
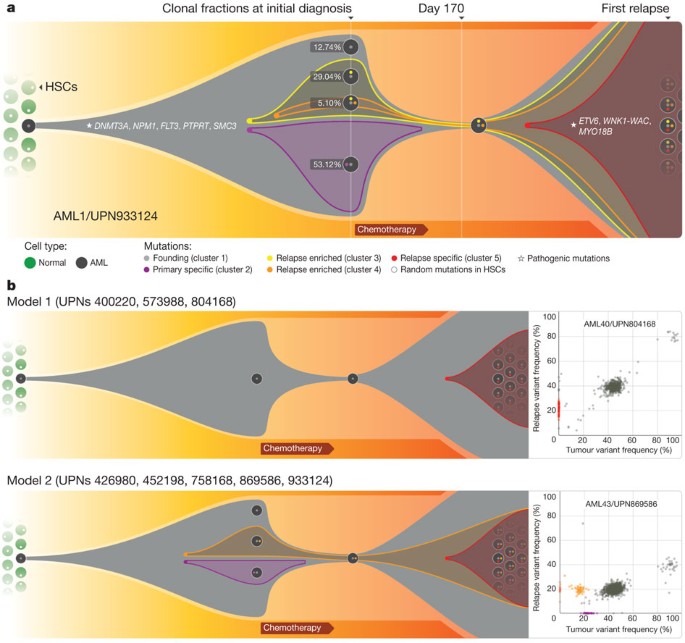
Figure 2: Graphical representation of clonal evolution from the primary tumour to relapse in UPN 933124, and patterns of tumour evolution observed in eight primary tumour and relapse pairs.
a, The founding clone in the primary tumour in UPN 933124 contained somatic mutations in DNMT3A, NPM1, PTPRT, SMC3 and FLT3 that are all recurrent in AML and probably relevant for pathogenesis; one subclone within the founding clone evolved to become the dominant clone at relapse by acquiring additional mutations, including recurrent mutations in ETV6 and MYO18B, and a _WNK1_-WAC fusion gene. HSC, haematopoietic stem cell. b, Examples of the two major patterns of tumour evolution in AML. Model 1 shows the dominant clone in the primary tumour evolving into the relapse clone by gaining relapse-specific mutations; this pattern was identified in three primary tumour and relapse pairs (UPN 804168, UPN 573988 and UPN 400220). Model 2 shows a minor clone carrying the vast majority of the primary tumour mutations survived and expanded at relapse. This pattern was observed in five primary tumour and relapse pairs (UPN 933124, UPN 452198, UPN 758168, UPN 426980 and UPN 869586).
Assuming that all the mutations detected are heterozygous in the primary tumour sample (with a malignant cellular content at 93.72% for the primary bone marrow sample, see Supplementary Information), we were able to calculate the fraction of total malignant cells in each clone. Clone 1 is the founding clone; 12.74% of the tumour cells contain only this set of mutations. Clones 2, 3 and 4 evolved from clone 1. The additional mutations in clones 2 and 3 may have provided a growth or survival advantage, as 53.12% and 29.04% of the tumour cells belonged to these clones, respectively. Only 5.10% of the tumour cells were from clone 4, indicating that it may have arisen last (Fig. 2a). However, the relapse clone evolved from clone 4. A single clone containing all of the cluster 5 mutations was detected in the relapse sample; clone 5 evolved from clone 4, but gained 78 new somatic alterations after sampling at day 170. As all mutations in clone 5 appear to be present in all relapse tumour cells, we suspect that one or more of the mutations in this clone provided a strong selective advantage that contributed to relapse. The ETV6 mutation, the MYO18B mutation, and/or the _WNK1_-WAC fusion are the most likely candidates, as ETV6, MYO18B and WAC are recurrently mutated in AML.
We evaluated the mutation clusters in the seven additional primary tumour–relapse pairs by assessing peaks of allele frequency using kernel density estimation (Supplementary Fig. 11 and Supplementary Information). We thus inferred the numbers and malignant fractions of clones in each primary tumour and relapse sample. Similar to UPN 933124, multiple mutation clusters (2–4) were present in each of the primary tumours from four patients (UPN 869586, UPN 426980, UPN 452198 and UPN 758168). However, only one major cluster was detected in each of the primary tumours from the three other patients (UPN 804168, UPN 573988 and UPN 400220) (Fig. 1c and Supplementary Table 10). Importantly, all eight patients gained relapse-specific mutations, although the number of clusters in the relapse samples varied (Fig. 1).
Two major patterns of clonal evolution were detected at relapse (Fig. 2b and Supplementary Fig. 3): in cases with pattern 1, the dominant clone in the primary tumour gained additional mutations and evolved into the relapse clone (UPN 804168, UPN 573988 and UPN 400220). These patients may simply be inadequately treated (for example, elderly patients who cannot tolerate aggressive consolidation, like UPN 573988), or they may have mutations in their founding clones (or germline variants) that make these cells more resistant to therapy (UPN 804168 and UPN 400220). In patients with pattern 2, a minor subclone carrying the vast majority (but not all) of the primary tumour mutations survived, gained mutations, and expanded at relapse; a subset of primary tumour mutations was often eradicated by therapy, and were not detected at relapse (UPN 758168, UPN 933124, UPN 452198, UPN 426980 and UPN 869586). Specific mutations in a key subclone may contribute to chemotherapy resistance, or the mutations important for relapse may be acquired during tumour evolution, or both. Notably, in cases 426980 and 758168, a second primary tumour clone survived chemotherapy and was also present at relapse (Fig. 1c and Supplementary Fig. 3). Owing to current technical limits in our ability to detect mutations in rare cells (mostly related to currently achievable levels of coverage with whole genome sequencing), our models represent a minimal estimate of the clonal heterogeneity in AML.
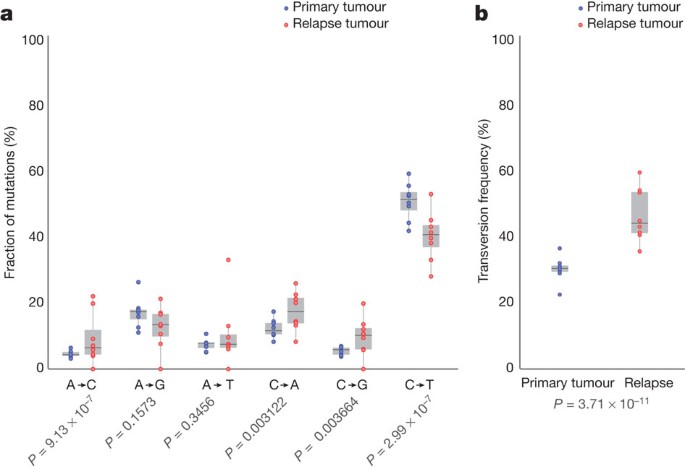
All eight patients received cytarabine and anthracycline for induction therapy, and additional cytotoxic chemotherapy for consolidation; treatment histories are summarized in Supplementary Table 2 and described in Supplementary Information. To investigate the potential impact of treatment on relapse mutation types, we compared the six classes of transition and transversion mutations in the primary tumour with the relapse-specific mutations in all eight patients (Fig. 3a). Although C•G→T•;A transitions are the most common mutations found in both primary and relapse AML genomes, their frequencies are significantly different between the primary tumour mutations (51.1%) and relapse-specific mutations (40.5%) (P = 2.99 × 10−7). Moreover, we observed an average of 4.5%, 5.3% and 4.2% increase in A•T→C•G (P = 9.13 ×10−7), C•G→A•T (P = 0.00312) and C•G→G•C (P = 0.00366) transversions, respectively, in relapse-specific mutations. Notably, an increased A•T→C•G transversion rate has also been observed in cases of chronic lymphocytic leukaemia with mutated immunoglobulin genes17. C•G→A•T transversions are the most common mutation in lung cancer patients who were exposed to tobacco-borne carcinogens18 (Fig. 3b and Supplementary Table 11). We examined the 456 relapse-specific mutations and 3,590 primary tumour point mutations from all eight cases as a group, and found that the transversion frequency is significantly higher for relapse-specific mutations (46%) than for primary tumour mutations (30.7%) (P = 3.71 × 10−11), indicating that chemotherapy has a substantial effect on the mutational spectrum at relapse. Similar results were obtained when we limited the analysis to the 213 mutations that had 0% variant frequency in the primary tumour samples (Supplementary Fig. 1b); the transversion frequency for relapse-specific mutations was 50.4%, versus 31.4% for primary tumour samples (P = 3.89 × 10−9). Very few copy-number alterations were detected in the eight relapse samples, suggesting that the increased transversion rate is not associated with generalized genomic instability (Supplementary Fig. 12).
Figure 3: Comparison of mutational classes between primary tumours and relapse samples.
a, Fraction of the primary tumour and relapse-specific mutations in each of the transition and transversion categories. b, Transversion frequencies of the primary tumour and relapse-specific mutations from eight AML tumour and relapse pairs. 456 relapse-specific mutations and 3,590 primary tumour mutations from eight cases were used for assessing statistical significance using proportion tests.
We first described the use of deep sequencing to define precisely the variant allele frequencies of the mutations in the AML genome of case 933124 (ref. 3), and here have refined and extended this technique to examine clonal evolution at relapse. The analysis of eight primary AML and relapse pairs has revealed unequivocal evidence for a common origin of tumour subpopulations; a dominant mutation cluster representing the founding clone was discovered in the primary tumour sample in all cases. The relationship of the founding clone (and subclones thereof) to the ‘leukaemia initiating cell’ is not yet clear—purification of clonal populations and functional testing would be required to establish this relationship. We observed the loss of primary tumour subclones at relapse in four of eight cases, suggesting that some subclones are indeed eradicated by therapy (Figs 1 and 2 and Supplementary Fig. 3). Some mutations gained at relapse may alter the growth properties of AML cells, or confer resistance to additional chemotherapy. Regardless, each tumour displayed clear evidence of clonal evolution at relapse and a higher frequency of transversions that were probably induced by DNA damage from chemotherapy. Although chemotherapy is required to induce initial remissions in AML patients, our data also raise the possibility that it contributes to relapse by generating new mutations in the founding clone or one of its subclones, which then can undergo selection and clonal expansion. These data demonstrate the critical need to identify the disease-causing mutations for AML, so that targeted therapies can be developed that avoid the use of cytotoxic drugs, many of which are mutagens.
This study extends the findings of previous studies19,20,21, which recently described patterns of clonal evolution in ALL patients using fluorescence in situ hybridization and/or copy number alterations detected by SNP arrays, and it enhances the understanding of genetic changes acquired during disease progression, as previously described for breast and pancreatic cancer metastases22,23,24,25. Our data provide complementary information on clonal evolution in AML, using a much larger set of mutations that were quantified with deep sequencing; this provides an unprecedented number of events that can be used to define precisely clonal size and mutational evolution at relapse. Both ALL and AML share common features of clonal heterogeneity at presentation followed by dynamic clonal evolution at relapse, including the addition of new mutations that may be relevant for relapse pathogenesis. Clonal evolution can also occur after allogeneic transplantation (for example, loss of mismatched HLA alleles via a uniparental disomy mechanism), demonstrating that the type of therapy itself can affect clonal evolution at relapse26,27. Taken together, these data demonstrate that AML cells routinely acquire a small number of additional mutations at relapse, and suggest that some of these mutations may contribute to clonal selection and chemotherapy resistance. The AML genome in an individual patient is clearly a ‘moving target’; eradication of the founding clone and all of its subclones will be required to achieve cures.
Methods Summary
Illumina paired-end reads were aligned to NCBI build36 using BWA 0.5.5 (http://sourceforge.net/projects/bio-bwa/). Somatic mutations were identified using SomaticSniper28 and a modified version of the SAMtools indel caller. Structural variations were identified using BreakDancer5. All predicted non-repetitive somatic SNVs, indels and all structural variants were included on custom sequence capture arrays from Roche Nimblegen. Illumina 2 × 100-bp paired-end sequencing reads were produced after elution from capture arrays. VarScan6 and a read remapping strategy using Crossmatch (P. Green, unpublished data) and BWA were used for determining the validation status of predicted SNVs, indels and structural variants. A complete description of the materials and methods is provided in Supplementary Information. All sequence variants for the AML tumour samples from eight cases have been submitted to dbGaP (accession number phs000159.v4.p2).
Accession codes
Data deposits
All sequence variants for the AML tumour samples from eight cases have been submitted to dbGaP under accession number phs000159.v4.p2.
References
- Testa, J. R., Mintz, U., Rowley, J. D., Vardiman, J. W. & Golomb, H. M. Evolution of karyotypes in acute nonlymphocytic leukemia. Cancer Res. 39, 3619–3627 (1979)
CAS PubMed Google Scholar - Garson, O. M. et al. Cytogenetic studies of 103 patients with acute myelogenous leukemia in relapse. Cancer Genet. Cytogenet. 40, 187–202 (1989)
Article CAS Google Scholar - Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008)
Article ADS CAS Google Scholar - Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Article Google Scholar - Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods 6, 677–681 (2009)
Article CAS Google Scholar - Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009)
Article CAS Google Scholar - Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009)
Article CAS Google Scholar - Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010)
Article CAS Google Scholar - Nakao, M. et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 10, 1911–1918 (1996)
CAS PubMed Google Scholar - Falini, B. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266 (2005)
Article CAS Google Scholar - Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010)
Article CAS Google Scholar - King-Underwood, L., Renshaw, J. & Pritchard-Jones, K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood 87, 2171–2179 (1996)
CAS PubMed Google Scholar - Gao, J. et al. Isolation of a yeast artificial chromosome spanning the 8;21 translocation breakpoint t(8;21)(q22;q22.3) in acute myelogenous leukemia. Proc. Natl Acad. Sci. USA 88, 4882–4886 (1991)
Article ADS CAS Google Scholar - Kirito, K. et al. A novel RUNX1 mutation in familial platelet disorder with propensity to develop myeloid malignancies. Haematologica 93, 155–156 (2008)
Article CAS Google Scholar - Van Vlierberghe, P. et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia 25, 130–134 (2011)
Article CAS Google Scholar - Barjesteh van Waalwijk van Doorn-Khosrovani, S. et al. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 24, 4129–4137 (2005)
Article Google Scholar - Puente, X. S. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011)
Article CAS Google Scholar - Ding, L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008)
Article ADS CAS Google Scholar - Mullighan, C. G. et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380 (2008)
Article ADS CAS Google Scholar - Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011)
Article ADS CAS Google Scholar - Notta, F. et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 469, 362–367 (2011)
Article ADS CAS Google Scholar - Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010)
Article ADS CAS Google Scholar - Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009)
Article ADS CAS Google Scholar - Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010)
Article ADS CAS Google Scholar - Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011)
Article ADS CAS Google Scholar - Vago, L. et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N. Engl. J. Med. 361, 478–488 (2009)
Article CAS Google Scholar - Villalobos, I. B. et al. Relapse of leukemia with loss of mismatched HLA resulting from uniparental disomy after haploidentical hematopoietic stem cell transplantation. Blood 115, 3158–3161 (2010)
Article CAS Google Scholar - Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics (in the press)
Acknowledgements
We thank the Analysis Pipeline group for developing the automated sequence analysis pipelines; the LIMS group for developing tools and software to manage samples and sequencing; the Systems group for providing the IT infrastructure and HPC solutions required for sequencing and analysis; and R. T. Demeter for experimental support. We also thank The Cancer Genome Atlas for allowing us to use unpublished data for this study, and the Washington University Cancer Genome Initiative for their support. This work was funded by grants to R.K.W. and the National Human Genome Research Institute (NHGRI U54 HG003079), and grants to T.J.L. from the National Cancer Institute (PO1 CA101937) and the Barnes-Jewish Hospital Foundation (00335-0505-02).
Author information
Author notes
- Li Ding and Timothy J. Ley: These authors contributed equally to this work.
Authors and Affiliations
- The Genome Institute, Washington University, St Louis, 63108, Missouri, USA
Li Ding, Timothy J. Ley, David E. Larson, Christopher A. Miller, Daniel C. Koboldt, Michael D. McLellan, Joshua F. McMichael, John W. Wallis, Charles Lu, Dong Shen, Christopher C. Harris, David J. Dooling, Robert S. Fulton, Lucinda L. Fulton, Ken Chen, Heather Schmidt, Joelle Kalicki-Veizer, Vincent J. Magrini, Lisa Cook, Sean D. McGrath, Tammi L. Vickery, Michael C. Wendl, Elaine R. Mardis & Richard K. Wilson - Department of Genetics, Washington University, St Louis, 63110, Missouri, USA
Li Ding, John W. Wallis, David J. Dooling, Robert S. Fulton, Lucinda L. Fulton, Ken Chen, Vincent J. Magrini, Michael C. Wendl, Shashikant Kulkarni, Elaine R. Mardis & Richard K. Wilson - Department of Internal Medicine, Division of Oncology, Washington University, St Louis, 63110, Missouri, USA
Timothy J. Ley, John S. Welch, Julie K. Ritchey, Margaret A. Young, Tamara Lamprecht, Sharon Heath, Daniel C. Link, Michael H. Tomasson, Peter Westervelt, Matthew J. Walter, Timothy A. Graubert & John F. DiPersio - Siteman Cancer Center, Washington University, St Louis, 63110, Missouri, USA
Timothy J. Ley, Daniel C. Link, Michael H. Tomasson, Shashikant Kulkarni, Peter Westervelt, Matthew J. Walter, Timothy A. Graubert, Elaine R. Mardis, Richard K. Wilson & John F. DiPersio - Department of Pathology and Immunology, Washington University, St Louis, 63110, Missouri, USA
Mark A. Watson, Jacqueline E. Payton & Shashikant Kulkarni - Division of Biostatistics, Washington University, St Louis, 63110, Missouri, USA
William D. Shannon
Authors
- Li Ding
You can also search for this author inPubMed Google Scholar - Timothy J. Ley
You can also search for this author inPubMed Google Scholar - David E. Larson
You can also search for this author inPubMed Google Scholar - Christopher A. Miller
You can also search for this author inPubMed Google Scholar - Daniel C. Koboldt
You can also search for this author inPubMed Google Scholar - John S. Welch
You can also search for this author inPubMed Google Scholar - Julie K. Ritchey
You can also search for this author inPubMed Google Scholar - Margaret A. Young
You can also search for this author inPubMed Google Scholar - Tamara Lamprecht
You can also search for this author inPubMed Google Scholar - Michael D. McLellan
You can also search for this author inPubMed Google Scholar - Joshua F. McMichael
You can also search for this author inPubMed Google Scholar - John W. Wallis
You can also search for this author inPubMed Google Scholar - Charles Lu
You can also search for this author inPubMed Google Scholar - Dong Shen
You can also search for this author inPubMed Google Scholar - Christopher C. Harris
You can also search for this author inPubMed Google Scholar - David J. Dooling
You can also search for this author inPubMed Google Scholar - Robert S. Fulton
You can also search for this author inPubMed Google Scholar - Lucinda L. Fulton
You can also search for this author inPubMed Google Scholar - Ken Chen
You can also search for this author inPubMed Google Scholar - Heather Schmidt
You can also search for this author inPubMed Google Scholar - Joelle Kalicki-Veizer
You can also search for this author inPubMed Google Scholar - Vincent J. Magrini
You can also search for this author inPubMed Google Scholar - Lisa Cook
You can also search for this author inPubMed Google Scholar - Sean D. McGrath
You can also search for this author inPubMed Google Scholar - Tammi L. Vickery
You can also search for this author inPubMed Google Scholar - Michael C. Wendl
You can also search for this author inPubMed Google Scholar - Sharon Heath
You can also search for this author inPubMed Google Scholar - Mark A. Watson
You can also search for this author inPubMed Google Scholar - Daniel C. Link
You can also search for this author inPubMed Google Scholar - Michael H. Tomasson
You can also search for this author inPubMed Google Scholar - William D. Shannon
You can also search for this author inPubMed Google Scholar - Jacqueline E. Payton
You can also search for this author inPubMed Google Scholar - Shashikant Kulkarni
You can also search for this author inPubMed Google Scholar - Peter Westervelt
You can also search for this author inPubMed Google Scholar - Matthew J. Walter
You can also search for this author inPubMed Google Scholar - Timothy A. Graubert
You can also search for this author inPubMed Google Scholar - Elaine R. Mardis
You can also search for this author inPubMed Google Scholar - Richard K. Wilson
You can also search for this author inPubMed Google Scholar - John F. DiPersio
You can also search for this author inPubMed Google Scholar
Contributions
T.J.L., L.D., J.F.D., E.R.M. and R.K.W. designed the experiments. L.D. and T.J.L. led data analysis. L.D., D.E.L., C.A.M., D.C.K., J.S.W., M.D.M., J.W.W., C.L., D.S., C.C.H., K.C., H.S., J.K.-V., M.C.W., M.A.W., W.D.S., J.E.P. and S.K. performed data analysis. J.F.M., M.D.M. and L.D. prepared figures and tables. J.S.W., J.K.R., M.A.Y., T.L., R.S.F., L.L.F., V.J.M., L.S., L.C., S.D.M. and T.L.V. performed laboratory experiments. S.H. and P.W. provided samples and clinical data. D.J.D. provided informatics support. T.J.L., D.C.L., M.H.T., E.R.M., R.K.W. and J.F.D. developed project concept. L.D., T.J.L., M.J.W., T.A.G. and J.F.D. wrote the manuscript.
Corresponding author
Correspondence toTimothy J. Ley.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Materials and Methods, Supplementary Figures 1-12 with legends, Supplementary Tables 1-12 and additional references. (PDF 26517 kb)
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Ding, L., Ley, T., Larson, D. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing.Nature 481, 506–510 (2012). https://doi.org/10.1038/nature10738
- Received: 29 March 2011
- Accepted: 29 November 2011
- Published: 11 January 2012
- Issue Date: 26 January 2012
- DOI: https://doi.org/10.1038/nature10738