Cryopyrin and pyrin activate caspase-1, but not NF-κB, via ASC oligomerization (original) (raw)
Introduction
The pyrin domain (PYD)-containing proteins pyrin (also known as marenostrin) and cryopyrin (also known as Pypaf1, Nalp3), have been implicated in the etiology of a number of inflammatory diseases in humans termed hereditary periodic fever syndromes (PFSs) (reviewed in Gumucio et al.,1 Hull et al.,2 McDermott and Aksentijevich,3 Stehlik and Reed4 and Tschopp5). Mutations in the MEFV gene which encodes pyrin are associated with familial Mediterranean fever (FMF),6, 7 while mutations in the CIAS1 gene which encodes cryopyrin are responsible for three PFSs; familial cold autoinflammatory syndrome (FCAS)/familial cold urticaria (FCU), Muckle–Wells syndrome (MWS), and neonatal-onset multisystem inflammatory disease (NOMID)/chronic infantile neurological cutaneous and articular syndrome (CINCA).8, 9 These diseases are characterized by recurrent episodes of inflammation and fever, and because of the lack of an apparent stimulus and the involvement of autoantibodies and autoreactive T cells, they are defined as autoinflammatory syndromes (reviewed in Gumucio et al.,1 Hull et al.,2 McDermott and Aksentijevich,3 and Stehlik and Reed4).
The human pyrin protein is a 781 amino-acid protein,6, 7 expressed predominantly in neutrophils, monocytes, and eosinophils.10, 11 It is composed of several identifiable conserved domains including an N-terminal PYD, which is a homotypic protein–protein interaction domain belonging to the six-helix bundle death domain (DD)-fold superfamily that includes DDs, death effector domains (DEDs), and caspase-associated recruitment domains (CARDs).12, 13, 14, 15 The PYD is followed by two central B-box zinc-finger and coiled coil domains and a C-terminal B30.2/rfp/SPRY domain. Most FMF-associated mutations are localized in B30.2/rfp/SPRY domain,16 suggesting that this domain is involved in the regulation of the activity of pyrin. Pyrin associates through a PYD–PYD interaction with apoptosis-associated speck-like protein containing a CARD (ASC),17, 18 a 22 kDa adapter protein with an N-terminal PYD and a C-terminal CARD involved in the activation of caspase-1.19, 20, 21 However, the physiological significance of the pyrin–ASC interaction for caspase-1 activation remains unclear because mice with total MEFV gene ablation are not yet available. Nevertheless, the importance of ASC for caspase-1 activation was recently revealed by gene knockout in mice, which showed an increased resistance to endotoxic shock of the ASC_−/− mice and a severe defect in caspase-1 activation and interleukin (IL)-1_β production in macrophages from these mice in response to several pro-inflammatory molecules.22 Pyrin has also been shown to interact with the proline serine threonine phosphatase-interacting protein 1 (PSTPIP1).23 Interestingly, PSTPIP1 is mutated in the autoinflammatory pyogenic arthritis, pyoderma gangrenosum, and acne syndrome,24 suggesting that pyrin and PSTPIP1 function in the same autoinflammatory pathway (reviewed in McDermott 25).
Like pyrin, cryopyrin contains an N-terminal PYD that mediates its interaction with ASC. However, beyond the PYD there are no structural similarities between pyrin and cryopyrin. Cryopyrin belongs to the NACHT-LRR (NLR) protein family, which includes NOD1, NOD2, Ipaf and 14 other family members known as NALPs.26, 5 All family members contain a central nucleotide-binding oligomerization domain (NOD)/domain present in NAIP, CIITA, HET-E and TP1 (NACHT), and a C-terminal leucine-rich repeat (LRR) domain. The disease-associated cryopyrin mutations are located within or near the NOD/NACHT domain,27 suggesting that these mutations might affect oligomerization of cryopyrin. Cryopyrin is primarily expressed in peripheral blood leukocytes.28, 29 Recent reports suggest that cryopyrin is involved in the regulation of NF-κ_B activation,29, 30, 31, 32, 33 and this in turn regulates the expression of genes involved in the immune response and inflammation.34 Cryopyrin also regulates IL-1_β generation by assembling an inflammasome complex with ASC and caspase-1.35 This activity of cryopyrin was recently shown to be regulated by bacterial peptidoglycan possibly through direct interactions with its LRR domain.36
In the present study, we investigated the NF-_κ_B and caspase-1-activating abilities of WT and disease-associated pyrin and cryopyrin proteins using a stable 293T cell-based reconstitution system that expresses physiological amounts of procaspase-1 and the caspase-1 adaptor protein ASC. We demonstrated that expression of either pyrin or cryopyrin or their corresponding disease-associated mutants could induce ASC oligomerization and activation of caspase-1, but not NF-_κ_B. The disease-associated mutants of cryopyrin were more potent than the WT protein in inducing caspase-1 activation because of increased self-oligomerization and interaction with ASC. Our data suggest that both pyrin and cryopyrin are capable of assembling independent inflammasome complexes with ASC and procaspase-1, and activating caspase-1 via ASC oligomerization.
Results
Cryopyrin and pyrin do not activate NF-_κ_B in the presence of physiological amount of ASC
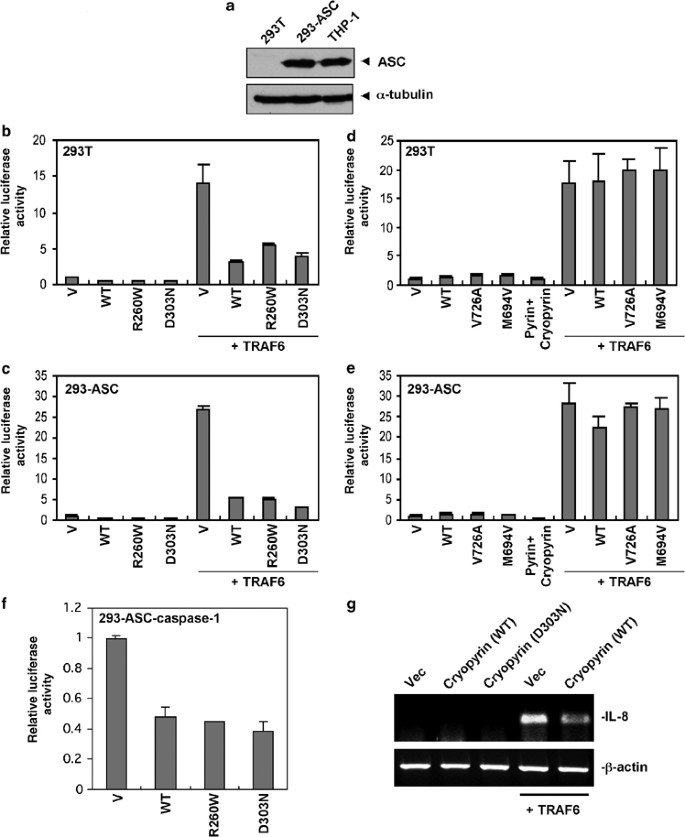
A number of recent studies provided conflicting results on the role of ASC in activation of NF-_κ_B in response to proinflammatory cytokines such as lipopolysaccharide (LPS) and tumor necrosis factor (TNF)-α or expression of cryopyrin or pyrin proteins.29, 30, 31, 32, 33 Since these studies were performed in transfected cells overexpressing ASC, the results of these studies may not reflect the normal physiological function of ASC. This is especially true in light of the results of the ASC knockout mice, which demonstrated that ASC does not appear to play a role in NF-_κ_B activation in response to LPS or TNF-α.22 However, since coexpression of ASC with the disease-associated cryopyrin mutants, but not the WT cryopyrin, was reported to activate NF-_κ_B,31 it is possible that the disease-associated cryopyrin mutants, and perhaps also the disease-associated pyrin mutants, could abnormally recruit ASC to induce NF-_κ_B activation. To test this possibility and to rule out any effect of ASC overexpression, we generated a stable HEK293T cell line expressing physiological amount of ASC (293-ASC cells) comparable to that present in human THP1 monocytes and periferal blood leukocytes (Figure 1a and Figure S1, Supplementary information). We investigated whether expression of WT cryopyrin or two cryopyrin mutants R260W and D303N, commonly found in patients with FCAS/FCU and NOMID/CINCA, respectively, could induce NF-_κ_B in 293-ASC cells by NF-_κ_B reporter gene assay. Surprisingly, neither the WT nor the mutant cryopyrin proteins were able to induce NF-_κ_B activation in these cells (Figure 1c), or in the parental 293T cells (Figure 1b), which lack ASC expression. On the contrary the WT and the mutant cryopyrin proteins reduced the basal NF-_κ_B activity in both the parental 293T cells and the 293-ASC cells. Transfection of the 293T and the 293-ASC cells with TRAF6 induced potent NF-_κ_B activation, ruling out the possibility that these cells contain a defect in NF-_κ_B activation. Interestingly, coexpression of cryopyrin proteins with TRAF6 in 293T and the 293-ASC cells inhibited TRAF6-induced NF-_κ_B activation (Figure 1b and c). Coexpression of cryopyrin proteins with Bcl10 also inhibited NF-_κ_B activation by this protein (data not shown). Our data thus clearly demonstrate that the WT or disease-associated cryopyrin mutants do not induce NF-_κ_B activation alone or in the presence of ASC, but rather they have a dampening effect on this pathway in this 293T system. Moreover, when ASC is expressed at normal levels in 293T cells, it does not collaborate with cryopyrin to induce NF-_κ_B activation. This apparent discrepancy with the reported NF-_κ_B-promoting activity of the disease-associated cryopyrin mutants in 293T cells31 is most likely due to the use of transiently overexpressed ASC in the previous study. The transiently overexpressed ASC has been shown to interact with the I_κ_B kinase (IKK) complex in 293T cells and suppresses its activity.33 Cryopyrin interaction with ASC could displace ASC from the IKK complex, relieving the suppression of the IKK complex, and resulting in artificial NF-_κ_B activation as reported previously.33
Figure 1
Cryopyrin and pyrin do not activate NF-_κ_B. (a) Western blot analysis of total cell extracts from 293T, 293-ASC and THP1 cells with anti-ASC monoclonal antibody. Lower panel, shows the expression of _α_-tubulin in these cells. Note that the expression of ASC in 293-ASC cells is comparable to the expression of endogenous ASC in THP1 cells. (b and c) Effect of cryopyrin expression on NF-_κ_B activity in 293T and 293-ASC cells. 293T (b) or 293-ASC (c) cells were transfected with empty vector (V) or expression constructs encoding wild-type (WT) or mutant R260W or D303N cryopyrins alone or together with TRAF6 as indicated. The NF-_κ_B activity was assayed as described under Materials and Methods. (d and e) Effect of pyrin expression on NF-_κ_B activity in 293T and 293-ASC cells. 293T (d) or 293-ASC (e) cells were transfected with empty vector (V) or expression constructs encoding wild-type (WT) or mutant V726A or M694A pyrins alone or together with TRAF6 as indicated. The NF-_κ_B activity was assayed as in (b) and (c). (f) Effect of cryopyrin expression on NF-_κ_B activity in 293-ASC-caspase-1 cells. 293-ASC-caspase-1 cells were transfected with empty vector (V) or expression constructs encoding wild-type (WT) or mutant R260W or D303N cryopyrins as indicated. The NF-_κ_B activity was assayed as in (b) and (c). (g) Effect of cryopyrin and TRAF6 expression on IL-8 gene in 293-ASC-caspase-1 cells. 293-ASC-caspase-1 cells were transfected with empty vector (Vec, 1st lane), or expression constructs encoding wild-type (WT) cryopyrin (2nd lane), mutant D303N cryopyrin (3rd lane), TRAF6 (4th lane) or WT cryopyrin plus TRAF6 (5th lane) as indicated. The IL-8 gene expression was assayed by RT-PCR with specific human IL-8 gene oligonucleotide primers as described under Materials and Methods. _β_-actin was used as an internal control (lower panel)
Coexpression of WT pyrin with ASC in HEK293T cells, was reported to activate NF-_κ_B.33 In contrast, a second study reported that coexpression of pyrin with ASC and cryopyrin in HEK293T cells inhibited cryopyrin–ASC-mediated NF-_κ_B activation.30 To investigate the role of pyrin in the NF-_κ_B-signaling pathway we expressed the WT pyrin and two FMF-associated pyrin mutants M694V and V726A in the 293-ASC cells and the parental 293T cells. As shown in Figure 1d and e, expression of the different pyrin proteins did not induce significant NF-_κ_B activity in either the parental or the 293-ASC cells. There was also little change in the NF-_κ_B activity when pyrin was coexpressed with cryopyrin in these cells. However, unlike cryopyrin, expression of pyrin or the pyrin mutants with TRAF6 did not inhibit TRAF6-induced NF-_κ_B activation indicating that the inhibitory effect observed with cryopyrin is specific to the cryopyrin protein. Taken together, our data indicate that neither cryopyrin nor pyrin activate the NF-_κ_B-signaling pathway when expressed in 293T cells containing physiological levels of ASC.
The cryopyrin disease-associated mutants exhibit increased ASC-dependent caspase-1 activation ability
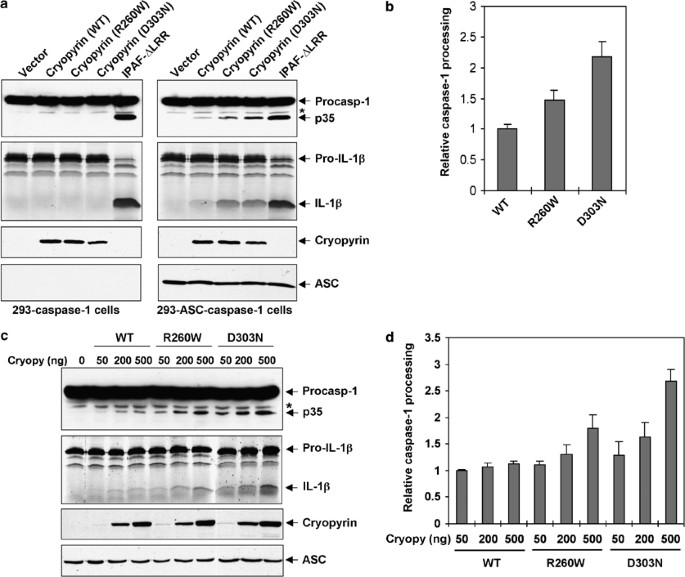
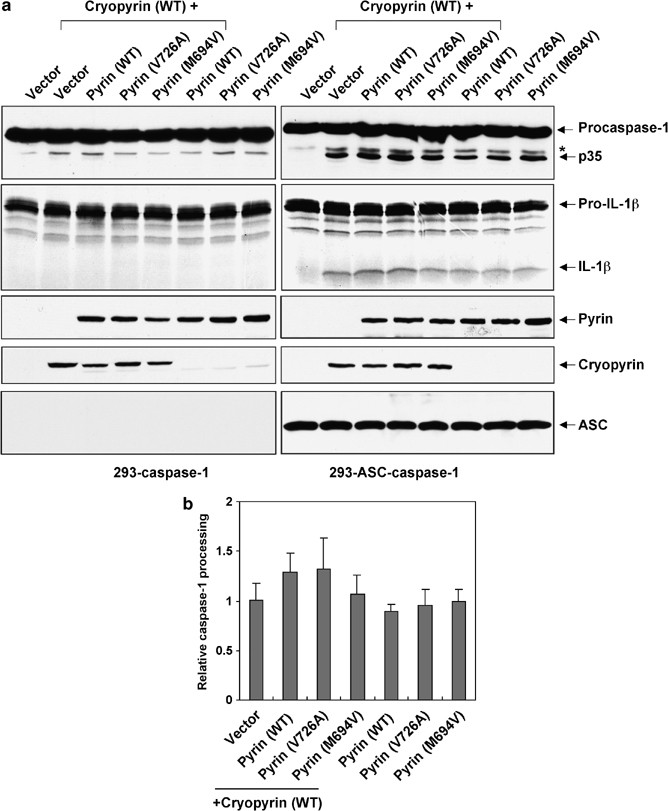
Constitutive IL-1_β_ secretion was observed in macrophages from patients with cryopyrin R260W mutation.35 Since processing of pro-IL-1_β_ is mediated by caspase-1 we investigated whether the cryopyrin disease-associated mutants have an enhanced ability to activate procaspase-1 in the absence of ligand stimulation. To do that, we generated stable HEK293T cell clones expressing either procaspase-1 alone (293-caspase-1) or procaspase-1 and ASC together (293-ASC-caspase-1). These cells were transfected with a constitutively active Ipaf as a positive control,37 or with WT cryopyrin or the cryopyrin mutants R260W and D303N. Expression of Ipaf, which binds directly to procaspase-1 via CARD–CARD interactions independent of ASC,37 induced autoprocessing of procaspase-1 in both 293-caspase-1 and 293-ASC-caspase-1 cells (Figure 2a). However, expression of WT cryopyrin or the cryopyrin mutants R260W and D303N resulted in autoprocessing of procaspase-1 in the 293-ASC-caspase-1 cells, but not in the 293-caspase-1 cells (Figure 2a, right panel), indicating that cryopyrin requires ASC to induce autoprocessing of procaspase-1. The processed caspase-1 was active as evidenced by its ability to cleave pro-IL-1_β_ to generate mature IL-1_β_ (Figure 2a, 2nd panel from top). Quantitative analysis of the activity of the WT compared to the activity of the cryopyrin mutants revealed that the activity of the cryopyrin mutants were significantly higher than that of the WT cryopyrin (Figure 2b). This was confirmed by expression of increasing amounts of the WT cryopyrin or the cryopyrin mutants in 293-ASC-caspase-1 cells (Figure 2c and d). This later experiment clearly showed that the cryopyrin mutants induced more, dose-dependent, caspase-1 activation compared to the WT cryopyrin, when the WT and the mutant cryopyrin proteins were expressed at equivalent levels. Among the two cryopyrin mutants, the D303N was the most active in inducing caspase-1 activation and pro-IL-1_β_ processing (Figure 2c and d). Combined, these studies demonstrate that the disease-associated cryopyrin mutants have an enhanced ASC-dependent ability to activate more caspase-1 than the WT cryopyrin, a result consistent with the reported increased secretion of mature IL-1_β_ in monocytes from patients harboring the cryopyrin R260W mutation.35
Figure 2
Activation of caspase-1 by cryopyrin and the cryopyrin disease-associated mutants. (a) 293-caspase-1 cells (left panels) or 293-ASC-caspase-1 cells (right panels) were transfected with equal amounts (250 ng) of empty vector (vector), or the indicated cryopyrin proteins (2nd–4th lanes), or Ipaf-ΔLRR (5th lanes). After transfection cells were lysed, and the total cell lysates were Western blotted with anti-Flag antibody to detect caspase-1 (1st panels from top) and cryopyrin (3nd panels from top) or anti-ASC monoclonal antibody (4th panels from top). Cell lysates from the transfected cells were also incubated with 35S-labeled pro-IL-1_β_ and the reaction products were fractionated by SDS-PAGE and detected by autoradiography (2nd panels from top). The processed caspase-1 p35 subunit and the mature IL-1_β_ are indicated. The asterisk indicates a nonspecific band. (b) Quantitative analysis of cryopyrin-induced caspase-1 processing. 293-ASC-caspase-1 cells were transfected with equal amounts (250 ng) of the indicated cryopyrin plasmids. After transfection the cell lysates were analyzed by Western blotting as in (a) and the amounts of caspase-1 p35 fragment were determined by densitometry. The relative caspase-1 processing was calculated as the amount of p35 fragment produced by each cryopyrin protein divided by the amount of p35 produced by WT cryopyrin. The values were normalized relative to the expression level of each cryopyrin protein. The values were calculated from at least three independent experiments. (c) 293-ASC-caspase-1 cells were transfected with increasing amounts of the indicated cryopyrin proteins. After transfection the cells were lysed and assayed as in (a). The asterisk indicates a nonspecific band. (d) Quantitative analysis of cryopyrin-induced caspase-1 processing. 293-ASC-caspase-1 cells were transfected with increasing amounts of the indicated cryopyrin proteins and analyzed as in (c). The relative caspase-1 processing was calculated as the amount of p35 fragment produced by each cryopyrin transfection divided by the amount of p35 produced by the lowest concentration of WT cryopyrin. The values were normalized relative to the expression level of each cryopyrin protein in each transfection. The values were calculated from at least three independent experiments
As with the 293-ASC cells (see Figure 1c and e), the stable expression of procaspase-1 alone (293-caspase-1 cells) or together with ASC (293-ASC-caspase-1) did not induce NF-_κ_B activation in response to expression of WT or mutant cryopyrin, or pyrin proteins (Figure 1f and data not shown), confirming that cryopyrin and pyrin proteins do not induce NF-_κ_B activation even in the presence of ASC and procaspase-1. Furthermore, additional experiments using an endogenous downstream NF-_κ_B target gene (IL-8), confirmed the above results (Figure 1g). The results demonstrate clearly that expression of TRAF6 in the 293-ASC-caspase-1 cells leads to induction of the IL-8 gene (Figure 1g, 4th lane), whereas expression of cryopyrin or the cryopyrin disease-associated D303N mutant in the same cells does not induce IL-8 (Figure 1g, 2nd and 3rd lanes). Similar results were obtained with the 293-ASC or 293-caspase-1 cells (data not shown). As with the NF-_κ_B reporter gene assay (Figure 1b and c), coexpression of cryopyrin with TRAF6 in these cells also suppressed TRAF6-induced IL-8 gene expression (Figure 1g, 5th lane).
The cryopyrin disease-associated mutants exhibit increased oligomerization with ASC compared to WT cryopyrin
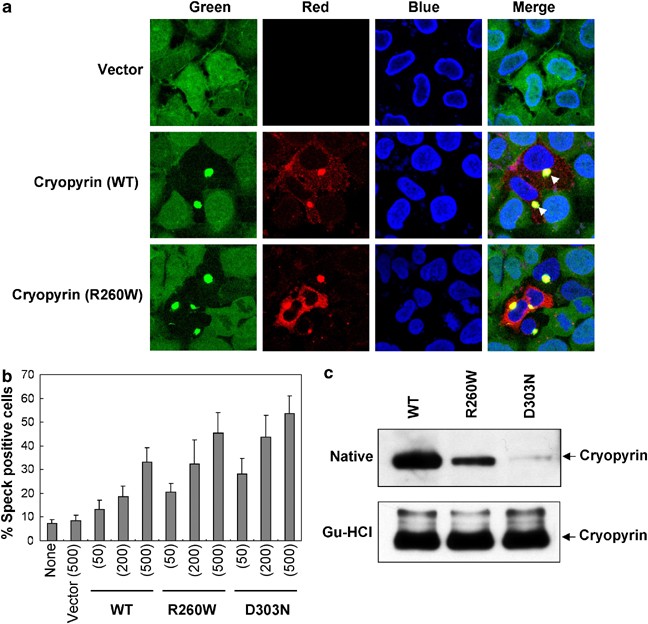
The molecular mechanism responsible for the constitutive activity of the disease-associated cryopyrin mutants is unclear. Since the majority of the disease-associated cryopyrin mutations are missense mutations located within the nucleotide and oligomerization domain (NOD), these mutations could cause conformational changes in this domain leading to increased oligomerization or aggregation of the mutant cryopyrin with ASC. Consequently, the oligomerized cryopyrin–ASC complex could serve as a platform to recruit and oligomerize procaspase-1, leading to its autoactivation. To test this hypothesis, we generated a stable 293T cell line expressing a GFP-tagged ASC to visualize the oligomerization of ASC in response to expression of the cryopyrin proteins by confocal microscopy. The ASC–GFP-expressing 293T cells were transfected with an empty plasmid or a plasmid encoding Flag-tagged WT cryopyrin or the cryopyrin mutants R260W and D303N. The transfected cells were stained with monoclonal anti-Nalp3 and rhodamine-red-conjugated anti-mouse IgG to visualize cryopyrin. In almost all the cells, the empty plasmid-transfected cells showed a homogeneous diffused ASC–GFP fluorescence over the entire cytoplasm and nucleus (Figure 3a, top panel, green channel). However, when the ASC–GFP cells were transfected with the cryopyrin disease-associated mutants (R260W or D303N) the entire ASC–GFP fluorescence colocalized with the cryopyrin mutants in bright perinuclear specks or aggregates in all the cells expressing the mutant cryopyrin proteins (Figure 3a, bottom panels). A similar but less dramatic aggregation of ASC–GFP and cryopyrin was seen in cells expressing the WT cryopyrin (Figure 3a, middle panel). A quantitative analysis of the percentage of cryopyrin-induced ASC–GFP aggregates in a large population of transfected cells revealed that the disease-associated mutant cryopyrin proteins induce significantly more specks/aggregates than the WT cryopyrin (Figure 3b). Interestingly, when the two mutants were compared with each other we found that the D303N mutant could induce significantly more ASC oligomerization than the R260W mutant (Figure 3b). These results suggest that the disease-associated mutations increase the tendency of cryopyrin to form oligomerized complexes with ASC, with the D303N mutant exhibiting the highest activity.
Figure 3
Cryopyrin induces oligomerization of ASC. (a) 293-ASC–GFP cells were transfected with empty vector (1st panels from top), or with the WT cryopyrin (2nd panels from top) or the R260W cryopyrin mutant (3rd panels from top). The cells were stained with anti-Nalp3/cryopyrin and rhodamine-red-conjugated secondary antibody (red channel) and nuclei were counterstained with DAPI (blue channel). The green channel represents the ASC–GFP fluorescence. The arrowheads indicate the perinuclear cryopyrin-ASC–GFP specks. Note that only cells expressing exogenous cryopyrin (red channel, cells in the center of the field) show cryopyrin–ASC specks. In the cells that do not express exogenous cryopyrin, ASC is seen in the entire cytoplasm (green channel). (b) 293-ASC–GFP cells were transfected with empty vector (vector) or the indicated amounts (ng) of cryopyrin expression plasmids. After transfection the percentages of cells containing cryopyrin–ASC–GFP specks were calculated as described under Materials and Methods. (c) The indicated His6-tagged WT and mutant cryopyrin proteins were expressed in Sf-9 cells and then bound to Ni2+-affinity beads in a native (upper panel) or guanidine-HCl (lower panel) buffer as described under Materials and Methods. The bead-bound proteins were then fractionated by SDS-PAGE and immunoblotted with anticryopyrin/Nalp-3 antibody
The cryopyrin disease-associated mutants are less soluble than the WT cryopyrin
To understand why the mutant cryopyrin proteins induce more ASC oligomerization than the WT cryopyrin, we expressed His6-tagged WT and cryopyrin mutants in Sf-9 cells with the baculovirus system. Cells were lysed in a native or a 6 M guandine-HCl-containing buffer and the soluble proteins were bound to Ni2+-affinity resin. Western blot analysis of the Ni2+-bound proteins revealed dramatically less mutant cryopyrins than WT cryopyrin bound to the Ni2+-affinity resin when the cells were lysed in a native buffer (Figure 3c, upper panel). The least binding was observed with the D303N mutant (Figure 3c, upper panel, third lane). However, there was no significant difference in the amounts of the mutant and WT cryopyrins bound to the Ni2+-affinity resin when the cells were lysed in a 6 M guandine-HCl-containing buffer (Figure 3c, lower panel). These results demonstrate clearly that the mutant proteins are less soluble under native conditions than the WT protein due to increased self-oligomerization/aggregation, which could explain their increased ability to induce more ASC oligomerization (Figure 3a and b). This is likely responsible for their higher caspase-1-activating ability in our cell-based reconstitution system (Figure 2a and b), and for the increased secretion of mature IL-1_β_ in monocytes from patients harboring the cryopyrin R260W mutation.35 Remarkably, the least soluble cryopyrin mutant, D303N, was also the most effective in inducing caspase-1 activation (compare Figure 3c with Figure 2b and d). This suggests that mutations that induce more self-oligomerization of cryopyrin may cause more caspase-1 activation, leading to a more severe disease phenotype in patients. Indeed, patients carrying the D303N mutation display a more severe disease phenotype compared to patients with the R260W mutation.38 It remains to be determined whether this correlation applies to other cryopyrin mutations.
Pyrin induces ASC-dependent caspase-1 activation
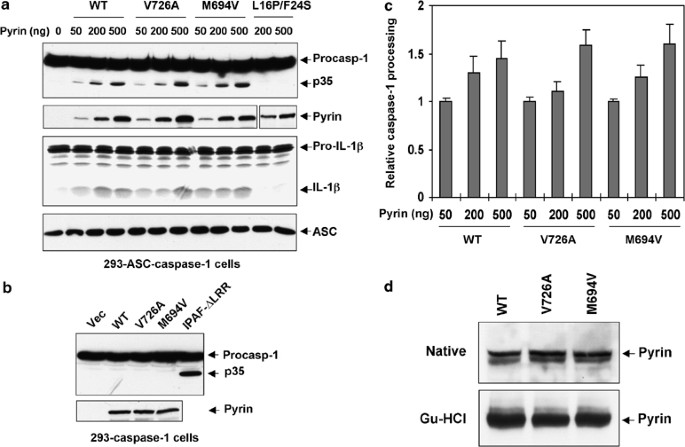
Mutations in pyrin are associated with the autoinflammatory disease familial mediteranian fever (FMF), suggesting that pyrin is an important regulator of the inflammatory process. Pyrin has been shown to interact with ASC,18 but whether this interaction leads to potentiation or inhibition of caspase-1 activation is still unclear. Based on evidence that a C-terminal truncation of pyrin in mice increased IL-1_β_ processing while ectopic expression of WT pyrin in mouse RAW cells impaired IL-1_β_ processing, a recent study proposed that the physiological function of pyrin is to inhibit the proteolytic activation of caspase-1 by sequestering ASC and preventing its interactions with caspase-1.11 However, the findings that many pyrin mutations in the C-terminal B30.2/rfp/SPRY domain are associated with FMF,16 and that human pyrin is induced as an immediate-early gene by proinflammatory molecules10 argue that the activity of pyrin could be proinflammatory. To test whether pyrin has an anti- or proinflammatory activity, we assayed the activity of WT pyrin, and two FMF-associated pyrin mutants, M694V and V726A, in our cell-based 293-ASC-caspase-1 reconstitution system. As shown in Figure 4a, expression of increasing amounts of WT pyrin or the FMF-associated pyrin mutants in 293-ASC-caspase-1 cells resulted in a dose-dependent autoprocessing of procaspase-1. In contrast, expression of these proteins in the 293–caspase-1 cells, which lacks ASC, did not induce autoprocessing of procaspase-1 (Figures 4b and 5a left panels), indicating that ASC is required for pyrin to recruit and induce autoprocessing of procaspase-1. Consistent with this result, expression of a pyrin mutant with two point mutations in the PYD (L16P/F24S), which are expected to prevent the interaction of pyrin with ASC (see Figure 6, bottom panels), also failed to induce autoprocessing of procaspase-1 (Figure 4a, 11th and 12th lanes). Surprisingly, unlike the disease-associated cryopyrin mutants, the two FMF-associated pyrin mutants did not show a significant increase in activity above that of the WT pyrin with regard to caspase-1 activation (Figure 4c). In addition, there was no significant difference in the solubility between the mutant pyrin proteins and the WT pyrin in a native buffer (Figure 4d), indicating that these pyrin mutations do not increase pyrin oligomerization. One explanation for these results is that these FMF-associated pyrin mutations might have been acquired during evolution to confer enhanced responsiveness to certain pathogen-associated molecular patterns (PAMP).16 In the absence of these PAMPs the difference between the activity of the WT and the mutant pyrin proteins might not be detectable.
Figure 4
Activation of caspase-1 by pyrin. (a) 293-ASC-caspase-1 cells were transfected with increasing amounts of the indicated pyrin proteins. After transfection cells were lysed, and total cell lysates were Western blotted with anti-Flag antibody to detect caspase-1 (1st panel from top) and the pyrin mutant (2nd panel from top, 11th and 12th lanes), or anti-Myc antibody to detect WT, V726A and M694V pyrins (2nd panel from top, 2nd–10th lanes), or anti-ASC monoclonal antibody (4th panel from top). Cell lysates from the transfected cells were also incubated with 35S-labeled pro-IL-1_β_ and the reaction products were fractionated by SDS-PAGE and detected by autoradiography (3rd panel from top). The processed caspase-1 p35 subunit and the mature IL-1_β_ are indicated. (b) 293-caspase-1 cells were transfected with equal amounts (500 ng) of empty vector (vec, 1st lane), or the indicated WT and mutant pyrin proteins (2nd–4th lanes), or Ipaf-ΔLRR (5th lane). After transfection cells were lysed, and the total cell lysates were fractionated by SDS-PAGE and detected by autoradiography as in (a). (c) Quantitative analysis of pyrin-induced caspase-1 processing. 293-ASC-caspase-1 cells were transfected with increasing amounts of the indicated pyrin proteins and analyzed as in (a). The relative caspase-1 processing was calculated as in Figure 2d legend. The values were calculated from at least three independent experiments. (d) The indicated His6-tagged WT and mutant pyrin proteins were expressed in Sf-9 cells and then bound to Ni2+-affinity beads in a native (upper panel) or guanidine-HCl (lower panel) buffer as described under Materials and Methods. The bead-bound proteins were then fractionated by SDS-PAGE and immunoblotted with anti-His6 antibody
Figure 5
Pyrin does not interfere with cryopyrin induced caspase-1 activation. (a) 293-caspase-1 cells (left panels) or 293-ASC-caspase-1 cells (right panels) were transfected with the indicated pyrin constructs alone (750 ng each) (6th to 8th lanes), or together with WT cryopyrin (750 ng) (3rd–5th lanes). Cells transfected with empty vector alone (1st lane) or together with cryopyrin (2nd lane) were used as controls. After transfection the cells were lysed and assayed as in (a). The asterisk indicates a nonspecific band. (b) Quantitative analysis of pyrin plus cryopyrin-induced caspase-1 processing. 293-ASC-caspase-1 cells were transfected with the indicated pyrin constructs alone (5th–7th columns) or together with WT cryopyrin (2nd–4th columns) and analyzed as in (a). First column represents cells transfected with empty vector together with cryopyrin. The relative caspase-1 processing was calculated as the amount of p35 fragment produced by each transfection divided by the amount of p35 produced by WT cryopyrin plus vector (1st column). The values were calculated from at least three independent experiments
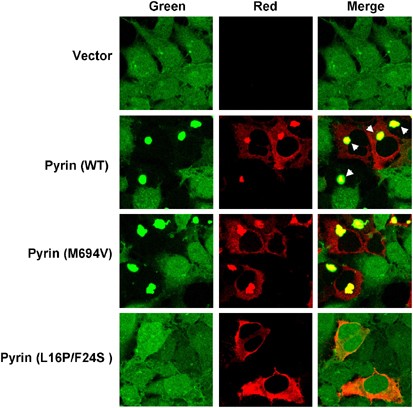
Figure 6
Pyrin induces ASC oligomerization. 293-ASC–GFP cells were transfected with empty vector (1st panel from top), or with the WT pyrin (2nd panel from top), the pyrin mutant M694V (3rd panel from top), or the pyrin mutant L16P/F24S (4th panel from top). The cells in the second and third panels were stained with anti-Myc and rhodamine-red-conjugated secondary antibody to visualize WT and M694V pyrin proteins (red channel). The cells in the fourth panels were stained with anti-Flag and rhodamine-red-conjugated anti-mouse IgG to visualize the pyrin L16P/F24S mutant (red channel). The green channel represents the ASC–GFP fluorescence. The arrowheads indicate the perinuclear pyrin-ASC–GFP specks. Note that the pyrin mutant L16P/F24S does not induce pyrin-ASC–GFP specks
Pyrin does not interfere with cryopyrin-induced caspase-1 activation
Recent observations suggested that pyrin negatively regulates the cryopyrin-signaling pathway by disrupting the cryopyrin–ASC interactions.30 To determine the effect of pyrin on cryopyrin-induced caspase-1 activation, we cotransfected pyrin or the FMF-associated pyrin mutants together with cryopyrin into 293-caspase-1 or 293-ASC-caspase-1 cells. As shown in Figure 5a right panel, coexpression of pyrin or the FMF-associated pyrin mutants together with cryopyrin in 293-ASC-caspase-1 cells did not inhibit processing of procaspase-1. On the contrary, there was a slight increase in procaspase-1 processing in the cells that express the two proteins (Figure 5b). These results indicate that the expression of the two proteins together in the same cell does not interfere with caspase-1 activation by either of the two proteins. No activation of caspase-1 by expression of the two proteins was observed in 293-caspase-1 cells, which lacks ASC (Figure 5a, left panels). Collectively, our results indicate that pyrin, like cryopyrin, is a proinflammatory molecule that is capable of activating procaspase-1 in an ASC-dependent manner.
Pyrin induces ASC oligomerization
The ability of pyrin to induce ASC-dependent procaspase-1 activation suggests that pyrin, like cryopyrin, could induce ASC oligomerization. To visualize ASC oligomerization by pyrin, we expressed WT pyrin or the pyrin mutants M694V and L16P/F24S in the 293-ASC-GFP cells. Expression of the WT pyrin or the pyrin mutant M694V in these cells resulted in complete oligomerization and colocalization of ASC-GFP with the WT pyrin in bright perinuclear specks (Figure 6). No ASC-GFP specks were observed when the cells were transfected with the PYD mutant L16P/F24S, indicating that this mutant does not interact with ASC. The lack of interaction with ASC is thus responsible for the inability of this mutant to induce procaspase-1 activation (see Figure 4a, 11th and 12th lanes). Together, these results indicate that the interaction of pyrin with ASC induces ASC oligomerization, a step that is essential to promote activation of procaspase-1.
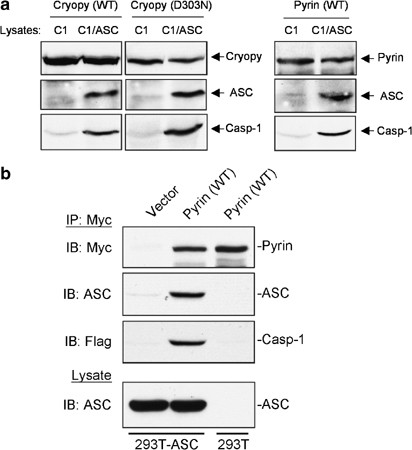
Pyrin forms a ternary inflammasome complex with ASC and procaspase-1
The ability of pyrin to oligomerize ASC (Figure 6) and to induce ASC-dependent caspase-1 activation (Figures 4 and 5) suggests that pyrin is capable of assembling a functional inflammasome complex with ASC and procaspase-1. To test this hypothesis we generated C-terminal His6-tagged pyrin and cryopyrin proteins in the baculovirus system and immobilized them on Ni2+-affinity beads. The immobilized proteins were incubated with extracts from the 293-caspase-1 or 293-ASC-caspase-1 cells. As shown in Figure 7a, pyrin, like cryopyrin proteins, was able to recruit procaspase-1 only in the presence of ASC (2nd lane in each panel) but not in its absence (1st lane in each panel), indicating that the formation of a pyrin–ASC or cryopyrin–ASC complex is a prerequisite for recruitment of procaspase-1. We also noticed that the D303N cryopyrin mutant recruits more procaspase-1 and ASC compared with the WT cryopyrin, consistent with its increased caspase-1-activating ability.
Figure 7
Pyrin recruits ASC and procaspase-1 to form an inflammasome complex. (a) His6-tagged WT pyrin (right panel) or cryopyrin proteins (WT and D303N mutant, left panels) were expressed in Sf-9 cells and bound to Ni2+-affinity beads. The bead-bound proteins were then incubated with extracts prepared from 293-caspase-1 (C1) or 293-ASC-caspase-1 (C1/ASC) cells. After incubation the complexes were fractionated by SDS-PAGE and immunoblotted with the appropriate antibodies to detect cryopyrin, pyrin, ASC and procaspase-1. (b) 293-ASC or 293T cells were transfected with C-terminal Myc-tagged pyrin plasmid. 293-ASC cells were also transfected with an empty vector control. After transfection the cell lysates were immunoprecipitated with an anti-Myc antibody and protein G-sepharose beads. The beads were then incubated with lysates containing procaspase-1 prepared from 293-caspase-1 cells. The bead-bound proteins were then fractionated by SDS-PAGE and immunoblotted with the appropriate antibodies to detect pyrin, ASC and procaspase-1
To provide additional evidence that the pyrin–ASC complex could recruit procaspase-1, we immunoprecipitated a pyrin–ASC complex from pyrin-transfected 293-ASC cells, and incubated it with lysates from 293-caspase-1 cells (Figure 7b). As expected the pyrin–ASC complex (2nd lane), but not pyrin alone (3rd lane), was able to recruit procaspase-1. Taken together, our data indicate that pyrin, like cryopyrin, can assemble a ternary inflammasome complex with ASC and procaspase-1.
Discussion
Many cells of lymphoid origin express ASC, procaspase-1 and a large number of proinflammatory NLR proteins. The majority of NLR proteins contain an N-terminal PYD followed by a NOD/NACHT domain and a regulatory C-terminal LRR domain, which is believed to function as a ligand-binding domain.5, 26, 36 Many NLR proteins interact with ASC via PYD–PYD interactions, but their specific ligands and their exact role in inflammation is largely unknown.5, 26 One way to gain insights into the function of the individual NLRs is to reconstitute the NLR complexes in a cell line, such as 293T, that is deficient in ASC, procaspase-1 and proinflammatory NLRs. Here we have described a 293T cell-based reconstitution system that allows the determination of the pro- or anti-inflammatory potential of molecules that interact with ASC and procaspase-1. Unlike the previously described systems in which ASC and procaspase-1 are transiently expressed with different regulatory molecules, the new system utilizes 293T cells that stably express ASC and/or procaspase-1 at physiological levels thus eliminating any artificial results due to variations in ASC and procaspase-1 expression levels. Using this system we demonstrated that cryopyrin and pyrin or their corresponding disease-associated mutants do not induce ASC-dependent activation of NF-_κ_B, thus making it less likely that these proteins regulate inflammation through NF-_κ_B signaling. Rather, these proteins appear to exclusively induce ASC-dependent procaspase-1 activation through ASC oligomerization.
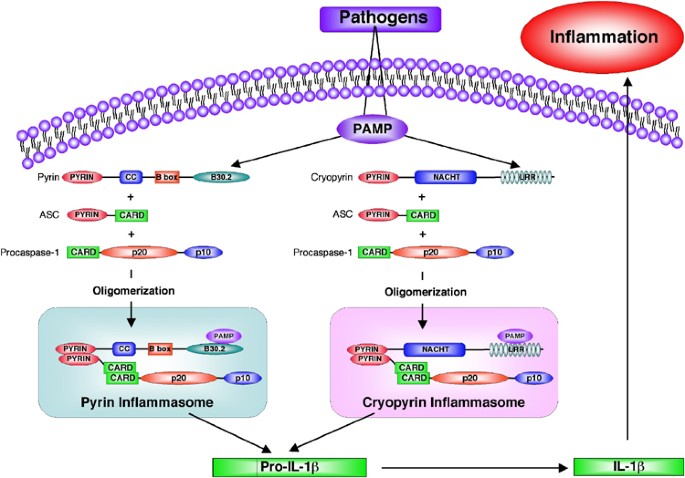
Our data support a model in which pyrin and cryopyrin regulate inflammation through assembly of two distinct and independent inflammasome complexes with ASC and procaspase-1 (Figure 8). What triggers the assembly of these inflammasome complexes is currently unknown. Recent findings that cryopyrin activity is regulated by bacterial peptidoglycans (PGN) independent of the LPS receptor TLR-4, raises the possibility that PGNs or its metabolized product muramyl dipeptide (MDP) interacts directly with the LRR of cryopyrin in the cytoplasm to regulate its activity.36 These interactions could induce oligomerization of cryopyrin, leading to recruitment of ASC and procaspase-1 to this complex. As a result, procaspase-1 is activated by the induced proximity mechanism, and is then capable of processing pro-IL-1_β_ into the mature IL-1_β_ cytokine, which mounts the inflammatory response. Supporting this model crude LPS, which contains PGN could induce procaspase-1 activation independent of the TLR adaptors MyD88 and Trif,39 suggesting that activation of caspase-1 could be mediated by direct interaction of LPS or PGN with cryopyrin or other related NLR family members in the cytoplasm.
Figure 8
A hypothetical model illustrating the formation of the cryopyrin and pyrin inflammasomes in response to pathogen infection. Recognition of PAMPS by the LRR or B30.2 domains of cryopyrin or pyrin proteins leads to recruitment and oligomerization of ASC and procaspase-1 into two distinct inflammasome complexes. These complexes are then capable of processing pro-IL-1_β_ into the mature IL-1_β_, which induces inflammation after its release from the infected cell
Several missense cryopyrin disease-associated mutations have been described in patients with FCAS, MWS, and NOMID/CINCA.8, 9 Among these diseases NOMID/CINCA syndrome display the most severe phenotype with neonatal onset, chronic polymorphonuclear meningitis leading to progressive neurologic impairment, and recurrent flare-ups of joint inflammation.38 The molecular mechanisms for the development of these autoinflammatory diseases are not clear, but recent co-immunoprecipitation data showed that the cryopyrin disease-associated mutant proteins exhibit increased interaction with ASC.31 However, we were unable to reproduce these results by co-immunoprecipitation because coexpression of the cryopyrin disease-associated mutants and ASC resulted in the formation of insoluble intracellular ASC–cryopyrin complexes/specks (Figure 3). Nevertheless, in vitro pull-down assays revealed that the D303N mutant does indeed interact more with ASC than with the WT protein (Figure 7a). In addition, using a 293T cell line that stably expresses a GFP-tagged ASC we were able to observe that the cryopyrin disease-associated mutants can interact with ASC and can also induce more ASC oligomerization/aggregation than the WT cryopyrin (Figure 3b). The reason for the increased ASC oligomerization by the mutant proteins is perhaps due to mutation-induced conformational changes in the oligomerization domain. A computer-generated model of the cryopyrin structure has suggested that the R260W and D303N mutations are located on one side of the protein, near the nucleotide-binding cleft, within a region possibly involved in oligomeric interactions, based on the known oligomeric structures of AAA+ ATPases.38 Therefore, these mutations may cause conformational changes leading to enhanced/increased oligomerization of the mutant cryopyrins. Indeed, analysis of the solubility of the mutant and the WT cryopyrins in a native buffer revealed that the mutant proteins are less soluble than the WT protein, which could explain their tendency to form more oligomers than the WT protein. Collectively, our data suggest that the cryopyrin disease-associated mutations may cause conformational changes in the oligomerization domain leading to self–self interaction and oligomerization. Thus, the cryopyrin disease-associated mutations may mimic activation induced by LPS or PGN (Figure 8).
Contrary to the current model in which it was suggested that pyrin inhibits caspase-1 activation by binding and sequestering ASC,11 our data clearly demonstrate that pyrin forms a ternary complex with ASC and procaspase-1 and that formation of this complex leads to caspase-1 autoactivation. Indeed, pyrin was as effective as cryopyrin in inducing ASC-dependent caspase-1 activation, and coexpression of the two proteins in the same cell did not interfere with caspase-1 activation, thus ruling out the possibility that association of pyrin with ASC prevents ASC from binding to caspase-1. Considering that most FMF-mutations are located in the C-terminal B30.2/rfp/SPRY domain of pyrin,16 which has been speculated to function in ligand binding,40 or signal transduction,41 it is likely that this domain regulates the activity of pyrin. In analogy with the C-terminal LRR domain of NLR family members, one could speculate that this domain represents a ligand-binding domain that senses PAMPs. Interaction of PAMPs with this domain induces oligomerization and binding of pyrin to ASC, which results in caspase-1 activation and IL-1_β_ processing (Figure 8). The FMF-associated mutations might represent gain of function mutations that were acquired during evolution to sense a wider variety of PAMPs than the WT protein does. This would increase the immune response to a broad class of pathogens and confer a selective survival advantage to FMF carriers. Consistent with this hypothesis human population studies revealed extremely high allele frequencies for several different pyrin mutations, leading to the conclusion that the mutant alleles confer a selective advantage.42, 43, 44 It was also reported that some of the very same amino acids mutated in FMF carriers are often present as WT in primates and other species.16
The recently reported data that mice expressing a C-terminal truncated version of pyrin have a heightened sensitivity to endotoxic shock and an increased caspase-1 activation and IL-1_β_ production in macrophages from these mice after stimulation,11 is not inconsistent with our model. Several reports have shown that deleting the regulatory LRR domains of several members of the NLR family such as Ipaf, cryopyrin and Nalp1 results in generation of constitutively active molecules.19, 36, 37 Considering that the C-terminal SPRY domain of pyrin is a regulatory domain then deleting it could result in a constitutively active pyrin. Alternatively, since macrophages from pyrin-truncation mice exhibit a defect in apoptosis compared to macrophages from WT mice in response to IL-4 and LPS treatment,11 it is possible that accumulation of activated macrophages in the pyrin-truncation mice could be responsible for their reported heightened sensitivity to endotoxic shock.
Materials and Methods
Expression constructs
Mammalian expression constructs for WT and disease-associated mutant of cryopyrin (pcDNA-cryopyrin-Flag, pcDNA-cryopyrin-R260W-Flag, pcDNA-cryopyrin-D303N-Flag) and pyrin (pcDNA-pyrin-His/Myc, pcDNA-pyrin-M694V-His/Myc, pcDNA-pyrin-V726A-His/Myc) and expression constructs for Ipaf-ΔLRR, and Flag-procaspase-1 were described previously.31, 37, 45 The expression plasmid for the pyrin mutant L16P/F24S was constructed in pcDNA3 with a C-terminal Flag tag. The expression plasmid for pro-IL-1_β_ was generated by inserting a PCR generated pro-IL-1_β_ cDNA in pcDNA3 (Invitrogen). pMSCV-ASC and pEGFP-N1-ASC were generated by inserting the cDNA of full-length human ASC in the pMSCV or pEGFP-N1 expression plasmids, respectively. The nucleotide sequences of all constructs were confirmed by automated sequencing.
Cell culture and stable cell lines
293T cells were cultured in DMEM/F-12 supplemented with 10% fetal bovine serum, 200 _μ_g/ml penicillin and 100 _μ_g/ml streptomycin sulfate. Sf9 cells were grown in Sf-900 II SFM supplemented with 10% heat-inactivated fetal bovine serum and 10 _μ_g/ml gentamycin. The N-terminal Flag-tagged human procaspase-1-expressing 293T cells (293-caspase-1 cells) were generated by stable transfection of 293T cells with a pcDNA3 expression vector containing the full-length human procaspase-1 cDNA (pcDNA-Flag-procaspase-1). Stable cell clones were selected using G418. The 293-ASC-caspase-1 cells or 293-ASC cells were generated from the 293-caspase-1 cells or 293T cells, respectively, by stable transfection with pMSCV–ASC expression plasmid. Stable cell clones were selected using puromycin. 293T cells expressing C-terminal GFP-tagged full-length ASC were generated by stable transfection with the expression plasmid pEGFP-N1-ASC and the empty pMSCV vector. Stable cell clones were selected using puromycin.
Expression of cryopyrin and pyrin proteins in the baculovirus system
The recombinant baculovirus transfer vectors for the WT and mutant cryopyrin and pyrin proteins were constructed in pVL-1393 with C-terminal His6 tags. The cryopyrin and pyrin transfer vectors were cotransfected with Sapphire™ baculovirus DNA (Orbigen) into Sf-9 cells to generate the recombinant baculoviruses according to the manufacturer's recommendation. To express cryopyrin or pyrin proteins in Sf-9 cells, 1 × 107 cells were infected with the respective recombinant baculoviruses at multiplicity of infection of 10. Cells were then seeded into T175 flasks and incubated for 48 h at 27°C. The expression of cryopyrin and pyrin proteins in Sf-9 cells was determined by Western blotting with anti-cryopyrin/Nalp3 antibody (Alexis) or anti-His6 antibody, respectively.
Cryopyrin and pyrin solubility assay
Sf-9 cells were infected with recombinant cryopyrin or pyrin baculoviruses for 48 h as described above. The infected cells were lysed by syringing (20 × ) in a native buffer (20 mM Hepes, pH 7.5, 1.5 mM MgCl2, 100 mM NaCl, 1% Triton X100, 0.1 mM PMSF) or a denaturation buffer (50 mM Tris pH 7.5, 8.0 M guanidine-HCl) and centrifuged at 16 000 × g for 10 min. The cell lysates were then bound to Ni2+-affinity beads and washed several times. The bead-bound proteins were then fractionated by SDS-PAGE and immunoblotted with the appropriate antibodies to detect cryopyrin and pyrin proteins.
Caspase-1 processing and IL-1_β_ cleavage assay
Cells (2 × 105) were plated in six-well plates, and 16 h later, cells were transfected with the indicated plasmid using LipofectAMINE (Invitrogen). Cells were harvested 24 h after transfection, and lysed in a buffer containing 20 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 0.2 mM PMSF, 2 μ_g/ml leupeptin, 2 mM DTT, 1 mM Na3VO4, 10 mM NaF by passing the cells several times through a 21 G needle. Lysates were then clarified by centrifugation at 16 000 × g for 10 min, and the supernatants were normalized for protein concentration. The normalized supernatants were then subjected to SDS-PAGE followed by immunoblotting with anti-Flag antibody to detect caspase-1 processing. The expression of the transfected pyrin-Myc or cryopyrin-Flag proteins was detected by Western blotting with the anti-Myc and anti-Flag antibodies, respectively. The expression of ASC was detected by Western blotting with the ASC monoclonal antibody.17 For IL-1_β cleavage assay, pro-IL-1_β_ was in vitro translated in the presence of 35S-methione using a TNT reticulocyte system (Promega), and incubated with the supernatant (50 _μ_g) at 30°C for 90 min. The reactants were then analyzed by SDS-PAGE followed by autoradiography.
NF-_κ_B reporter gene assays
Cells in 6-well plates were transfected with 5 × _κ_B-luciferase reporter plasmid (30 ng), p_β_-gal (60 ng), cryopyrin or pyrin (0.4 _μ_g). and TRAF6 (0.4 _μ_g) as indicated. The total amount of DNA (0.8 _μ_g) was kept constant by inclusion of empty vector DNAs. The luciferase activity was determined using a Dual-Luciferase Assay System (Promega). A LacZ-expressing plasmid was used for normalizing transfection efficiencies.
Induction of the endogenous NF-_κ_B target gene IL-8 was determined 20 h after transfection of cells in six-well plates with the indicated constructs (500 ng) by semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) using the IL-8 primers; 5′-primer, ATGACTTCCAAGCTGGCC and 3′-primer CTATGAATTCTCAGCCCTC. The amplification products were separated on a 2% agarose gel and visualized by ethidium bromide staining.
Confocal microscopy
ASC–GFP expressing 293T cells grown on cover slips were transfected with the pyrin or cryopyrin expression plasmids. The transfections were carried out using Lipofectamine PLUS-reagent (Invitrogen) according to the manufacturer's instructions. All the plasmids for transfection were prepared by using Qiagen columns. After 24–48 h of transfection cells were stained with anti-Nalp3 antibody (Alexis) (for WT, R260W and D303N cryopyrins), anti-Flag antibody (for pyrin mutant L16P/F24S), or anti-Myc (for WT, M694V and V726A pyrins), and rhodamine-red-conjugated secondary antibodies, and nuclei were counterstained with DAPI. Cells were observed using a Zeiss LSM 510 Meta confocal microscope.
Speck quantitation assay
ASC–GFP expressing 293T cells grown in six-well plates were transfected with different amounts of the pyrin or cryopyrin expression plasmids as above. At 48 h after transfection the number of cells containing ASC–GFP specks were counted in randomly selected multiple fields. The percentage of ASC–GFP speck-positive cells was calculated as the number of specks divided by the total number of transfected cells. At least three independently transfected wells were analyzed per experiment.
Immunoprecipitation and pull-down assays
293-ASC or 293T cells were transfected with a C-terminal Myc-tagged pyrin plasmid (2 _μ_g) in 100 mm dishes and 24 h after transfection the transfected cells were lysed by syringing (20 × ) in an IP buffer (20 mM Hepes, pH 7.4,. 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 0.1 mM PMSF, 2 _μ_g/ml leupeptin, 1 mM Na3VO4, 5 mM NaF) and centrifuged at 16 000 × g for 10 min. The pyrin–ASC complex and pyrin alone were immunoprecipitated from the lysates with an anti-Myc antibody (Abcam) and immobilized on protein G-sepharose beads. The beads were then incubated with lysates containing procaspase-1 prepared from 293-caspase-1 cells in IP buffer for 3 h at 4°C. The bead-bound proteins were then fractionated by SDS-PAGE and immunoblotted with the appropriate antibodies to detect pyrin, ASC and procaspase-1.
In vitro pull-down assays were performed with recombinant C-terminal His6-tagged cryopyrin and pyrin proteins expressed in the baculovirus system. Cryopyrin and pyrin proteins were isolated from Sf-9 cell lysates by Ni2+-affinity purification on Ni2+-affinity beads. The bead-bound proteins were then incubated for 2 h at 4°C with extracts prepared from 293-caspase-1 or 293-ASC-caspase-1 cells. After incubation the complexes were fractionated by SDS-PAGE and immunoblotted with the appropriate antibodies to detect cryopyrin, pyrin, ASC and procaspase-1.
Abbreviations
CARD:
caspase-associated recruitment domain
PYD:
pyrin domain
DD:
death domain
DED:
death effector domain
NOD:
nucleotide-binding oligomerization domain
LRR:
leucine-rich repeat
NLR:
NACHT-LRR
IL:
interleukin
LPS:
lipopolysaccharide
TNF:
tumor necrosis factor
ASC:
apoptosis-associated speck-like protein containing a CARD
PFS:
periodic fever syndromes
FMF:
familial Mediterranean fever
FCAS:
familial cold autoinflammatory syndrome
FCU:
familial cold urticaria
MWS:
Muckle-Wells syndrome
NOMID:
neonatal-onset multisystem inflammatory disease
CINCA:
chronic infantile neurological cutaneous and articular syndrome
PSTPIP1:
proline serine threonine phosphatase-interacting protein 1
PAPA:
pyogenic arthritis, pyoderma gangrenosum, and acne syndrome
IKK:
I_κ_B kinase
PAMP:
pathogen-associated molecular patterns
PGN:
peptidoglycans
MDP:
muramyl dipeptide
WT:
wildtype
RT:
reverse transcription
PCR:
polymerase chain reaction
NACHT:
domain present in NAIP, CIITA, HET-E and TP1
References
- Gumucio DL, Diaz A, Schaner P, Richards N, Babcock C, Schaller M and Cesena T (2002) Fire and ICE: the role of pyrin domain-containing proteins in inflammation and apoptosis. Clin. Exp. Rheumatol. 20: S45–S53
CAS PubMed Google Scholar - Hull KM, Shoham N, Chae JJ, Aksentijevich I and Kastner DL (2003) The expanding spectrum of systemic autoinflammatory disorders and their rheumatic manifestations. Curr. Opin. Rheumatol. 15: 61–69
Article CAS PubMed Google Scholar - McDermott MF and Aksentijevich I (2002) The autoinflammatory syndromes. Curr. Opin. Allergy Clin. Immunol. 2: 511–516
Article PubMed Google Scholar - Stehlik C and Reed JC (2004) The PYRIN connection: novel players in innate immunity and inflammation. J. Exp. Med. 200: 551–558
Article CAS PubMed PubMed Central Google Scholar - Tschopp J, Martinon F and Burns K (2003) NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4: 95–104
Article CAS PubMed Google Scholar - Consortium TFF (1997) A candidate gene for familial Mediterranean fever. The French FMF Consortium. Nat. Genet. 17: 25–31
Article Google Scholar - Consortium TIF (1997) Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium. Cell 90: 797–807
Article Google Scholar - Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A and de Saint Basile G (2002) Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71: 198–203
Article CAS PubMed PubMed Central Google Scholar - Hoffman HM, Mueller JL, Broide DH, Wanderer AA and Kolodner RD (2001) Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle–Wells syndrome. Nat. Genet. 29: 301–305
Article CAS PubMed PubMed Central Google Scholar - Centola M, Wood G, Frucht DM, Galon J, Aringer M, Farrell C, Kingma DW, Horwitz ME, Mansfield E, Holland SM, O'Shea JJ, Rosenberg HF, Malech HL and Kastner DL (2000) The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood 95: 3223–3231
CAS PubMed Google Scholar - Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP and Kastner DL (2003) Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol. Cell 11: 591–604
Article CAS PubMed Google Scholar - Bertin J and DiStefano PS (2000) The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 7: 1273–1274
Article CAS PubMed Google Scholar - Fairbrother WJ, Gordon NC, Humke EW, O'Rourke KM, Starovasnik MA, Yin JP and Dixit VM (2001) The PYRIN domain: a member of the death domain-fold superfamily. Protein Sci. 10: 1911–1918
Article CAS PubMed PubMed Central Google Scholar - Martinon F, Hofmann K and Tschopp J (2001) The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 11: R118–R120
Article CAS PubMed Google Scholar - Pawlowski K, Pio F, Chu Z, Reed JC and Godzik A (2001) PAAD – a new protein domain associated with apoptosis, cancer and autoimmune diseases. Trends Biochem. Sci. 26: 85–87
Article CAS PubMed Google Scholar - Schaner P, Richards N, Wadhwa A, Aksentijevich I, Kastner D, Tucker P and Gumucio D (2001) Episodic evolution of pyrin in primates: human mutations recapitulate ancestral amino acid states. Nat. Genet. 27: 318–321
Article CAS PubMed Google Scholar - Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T and Sagara J (1999) ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 274: 33835–33838
Article CAS PubMed Google Scholar - Richards N, Schaner P, Diaz A, Stuckey J, Shelden E, Wadhwa A and Gumucio DL (2001) Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J. Biol. Chem. 276: 39320–39329
Article CAS PubMed Google Scholar - Martinon F, Burns K and Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10: 417–426
Article CAS PubMed Google Scholar - Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z and Alnemri ES (2002) The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 277: 21119–21122
Article CAS PubMed Google Scholar - Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS and Bertin J (2002) PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J. Biol. Chem. 277: 29874–29880
Article CAS PubMed Google Scholar - Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S and Dixit VM (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430: 213–218
Article CAS PubMed Google Scholar - Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA and Kastner DL (2003) Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc. Natl. Acad. Sci. USA 100: 13501–13506
Article CAS PubMed PubMed Central Google Scholar - Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S and Lovett M (2002) Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum. Mol. Genet. 11: 961–969
Article CAS PubMed Google Scholar - McDermott MF (2004) A common pathway in periodic fever syndromes. Trends Immunol. 25: 457–460
Article CAS PubMed Google Scholar - Ting JP and Davis BK (2005) Caterpiller: a novel gene family important in immunity, cell death, and diseases. Annu. Rev. Immunol. 23: 387–414
Article CAS PubMed Google Scholar - McDermott MF (2002) Genetic clues to understanding periodic fevers, and possible therapies. Trends Mol. Med. 8: 550–554
Article CAS PubMed Google Scholar - Anderson JP, Mueller JL, Rosengren S, Boyle DL, Schaner P, Cannon SB, Goodyear CS and Hoffman HM (2004) Structural, expression, and evolutionary analysis of mouse CIAS1. Gene 338: 25–34
Article CAS PubMed PubMed Central Google Scholar - Manji GA, Wang L, Geddes BJ, Brown M, Merriam S, Al-Garawi A, Mak S, Lora JM, Briskin M, Jurman M, Cao J, DiStefano PS and Bertin J (2002) PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. J. Biol. Chem. 277: 11570–11575
Article CAS PubMed Google Scholar - Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N and Nunez G (2003) Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem. Biophys. Res. Commun. 302: 575–580
Article CAS PubMed Google Scholar - Dowds TA, Masumoto J, Zhu L, Inohara N and Nunez G (2004) Cryopyrin-induced interleukin 1beta secretion in monocytic cells: enhanced activity of disease-associated mutants and requirement for ASC. J. Biol. Chem. 279: 21924–21928
Article CAS PubMed Google Scholar - O'Connor Jr W, Harton JA, Zhu X, Linhoff MW and Ting JP (2003) Cutting edge: CIAS1/cryopyrin/PYPAF1/NALP3/CATERPILLER 1.1 is an inducible inflammatory mediator with NF-kappa B suppressive properties. J. Immunol. 171: 6329–6333
Article CAS PubMed Google Scholar - Stehlik C, Fiorentino L, Dorfleutner A, Bruey JM, Ariza EM, Sagara J and Reed JC (2002) The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor kappaB activation pathways. J. Exp. Med. 196: 1605–1615
Article CAS PubMed PubMed Central Google Scholar - Karin M and Delhase M (2000) The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin. Immunol. 12: 85–98
Article CAS PubMed Google Scholar - Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN and Tschopp J (2004) NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle–Wells autoinflammatory disorder. Immunity 20: 319–325
Article CAS PubMed Google Scholar - Martinon F, Agostini L, Meylan E and Tschopp J (2004) Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14: 1929–1934
Article CAS PubMed Google Scholar - Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T and Alnemri ES (2001) Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276: 28309–28313
Article CAS PubMed Google Scholar - Neven B, Callebaut I, Prieur AM, Feldmann J, Bodemer C, Lepore L, Derfalvi B, Benjaponpitak S, Vesely R, Sauvain MJ, Oertle S, Allen R, Morgan G, Borkhardt A, Hill C, Gardner-Medwin J, Fischer A and de Saint Basile G (2004) Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood 103: 2809–2815
Article CAS PubMed Google Scholar - Yamamoto M, Yaginuma K, Tsutsui H, Sagara J, Guan X, Seki E, Yasuda K, Akira S, Nakanishi K, Noda T and Taniguchi S (2004) ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells 9: 1055–1067
Article CAS PubMed Google Scholar - Henry J, Mather IH, McDermott MF and Pontarotti P (1998) B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol. Biol. Evol. 15: 1696–1705
Article CAS PubMed Google Scholar - Schultz J, Milpetz F, Bork P and Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864
Article CAS PubMed PubMed Central Google Scholar - Aksentijevich I, Torosyan Y, Samuels J, Centola M, Pras E, Chae JJ, Oddoux C, Wood G, Azzaro MP, Palumbo G, Giustolisi R, Pras M, Ostrer H and Kastner DL (1999) Mutation and haplotype studies of familial Mediterranean fever reveal new ancestral relationships and evidence for a high carrier frequency with reduced penetrance in the Ashkenazi Jewish population. Am. J. Hum. Genet. 64: 949–962
Article CAS PubMed PubMed Central Google Scholar - Stoffman N, Magal N, Shohat T, Lotan R, Koman S, Oron A, Danon Y, Halpern GJ, Lifshitz Y and Shohat M (2000) Higher than expected carrier rates for familial Mediterranean fever in various Jewish ethnic groups. Eur. J. Hum. Genet. 8: 307–310
Article CAS PubMed Google Scholar - Tunca M, Kirkali G, Soyturk M, Akar S, Pepys MB and Hawkins PN (1999) Acute phase response and evolution of familial Mediterranean fever. Lancet 353: 1415
Article CAS PubMed Google Scholar - Mansfield E, Chae JJ, Komarow HD, Brotz TM, Frucht DM, Aksentijevich I and Kastner DL (2001) The familial Mediterranean fever protein, pyrin, associates with microtubules and colocalizes with actin filaments. Blood 98: 851–859
Article CAS PubMed Google Scholar
Acknowledgements
We thank Dr. Gabriel Nunez for the WT and mutant cryopyrin expression plasmids and Dr. Daniel Kastner for the WT and mutant pyrin expression plasmids. I Ibrahimi is on sabbatical leave from the University of Jordan. He is partially supported by the distinguished scholar award from the Arab fund for social and economic development. This work was supported by NIH grants AG14357 and AG13487 (to ESA).
Author information
Author notes
- J-W Yu and J Wu: These authors contributed equally to this work
Authors and Affiliations
- Department of Microbiology and Immunology, Center for Apoptosis Research, Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia, 19107, PA, USA
J-W Yu, J Wu, Z Zhang, P Datta, I Ibrahimi, T Fernandes-Alnemri & E S Alnemri - Molecular Oncology, Institute on Aging and Adaptation, Shinshu University Graduate School of Medicine, 3-1-1 Asahi, Asahi, 390-8621, Japan
S Taniguchi & J Sagara
Authors
- J-W Yu
- J Wu
- Z Zhang
- P Datta
- I Ibrahimi
- S Taniguchi
- J Sagara
- T Fernandes-Alnemri
- E S Alnemri
Corresponding author
Correspondence toE S Alnemri.
Additional information
Edited by DR Green
Supplementary information accompanies the paper on the Cell Death and Differentiation web site(http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, JW., Wu, J., Zhang, Z. et al. Cryopyrin and pyrin activate caspase-1, but not NF-_κ_B, via ASC oligomerization.Cell Death Differ 13, 236–249 (2006). https://doi.org/10.1038/sj.cdd.4401734
- Received: 10 February 2005
- Revised: 06 June 2005
- Accepted: 22 June 2005
- Published: 22 July 2005
- Issue Date: 01 February 2006
- DOI: https://doi.org/10.1038/sj.cdd.4401734