Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain (original) (raw)
Introduction
During the last three decades, the resistance to antibiotics has increased and disseminated all over the world. Bacteria have developed several ways to resist against almost all antibiotics used and few new effective antibiotics have been discovered so far1,2. The return in the pre-antibiotic era3 seems to be a reality for some infections with multidrug-resistant (MDR) or extremely-drug resistant (XDR) bacteria4. Pseudomonas aeruginosa is one of these “superbugs”; and infections associated with multidrug-resistant P. aeruginosa are having a substantial impact on hospital costs and mortality rates. P. aeruginosa is an invasive, Gram negative opportunistic pathogen that causes a wide range of severe infections including bacteraemia, pneumonia, meningitis, urinary tract and wound infections5. Moreover_, P. aeruginosa_ is naturally resistant to multiple antibiotics; this is due to its natural low outer membrane permeability and to many adaptive resistance mechanisms (loss of porins, surexpression of efflux pumps, presence of many beta-lactamases or carbapenemases…)5,6,7,8,9. Most frequently pandrug-resistant P. aeruginosa are isolated from wound or respiratory tract infections: resistance including third-generation cephalosporin, carbapenems, fluoroquinolones and aminosides. The last effective antibiotic was often colistin, an old and highly toxic molecule10,11. There is therefore, an urgent need for new antibacterials, with an innovative mechanism of action.
Among various approaches to develop new antibacterial agents is one dedicated to cationic compounds12. In this work we focused on a polycationic calixarene-based guanidinium compound. Calixarenes are rigid oligomeric phenol macrocycles spatially organized, purely synthetic, with a structure completely different from antibiotics currently used in therapy13. Pioneer works demonstrated that our lead compound, the tetra _para_-guanidinoethylcalix[4]arene13 (named CX1) has a real antibacterial activity with a broad spectrum, including MDR bacteria14. The main interest of this new drug is that because of an innovative structure, it will take bacteria some time to find a mechanism of resistance. Moreover we have demonstrated in vitro that this compound is not able to select resistant mutant15. However the mechanism of action of this new cationic antibacterial drug has not yet been extensively studied. The initial hypothesis is that the introduction of positive charges on the calixarene core (i.e. guanidinium functions) leads to a constrained tetra cation able to disorganize the bacterial cell wall. P. aeruginosa possesses a highly negatively charged outer membrane and so is a good candidate to study the interaction with CX1.
Since it's invention in 1986, Atomic Force Microscopy (AFM)16,17 has created new paradigms in life nanoscience. It gives access to the ultrastructural (imaging, Single Molecule Force Spectroscopy (SMFS)) and nanomechanical (force spectroscopy) properties of single living cells18,19,20,21,22. For the study of live bacteria, AFM provides the opportunity to investigate the surface nanostructure under controlled aqueous conditions23,24. Therefore it is ideal to study the nanoscale effects of anti-infective drugs on bacteria25,26.
Results
The approach that we have developed includes several technical aspects of the AFM (imaging living cells, supported bilayers, SMFS, nanomechanical measurements). The experimental components and principle of our approach are described in Fig. 1. P. aeruginosa cells were immobilized27 by taking advantage of the electrostatic interaction between the bacteria's negative charges and a positively charged surface. To this end, glass slides were coated with PolyEthylenImine (PEI), a polycation. Bacteria were then incubated on the PEI coated glass slides for an hour at room temperature28.
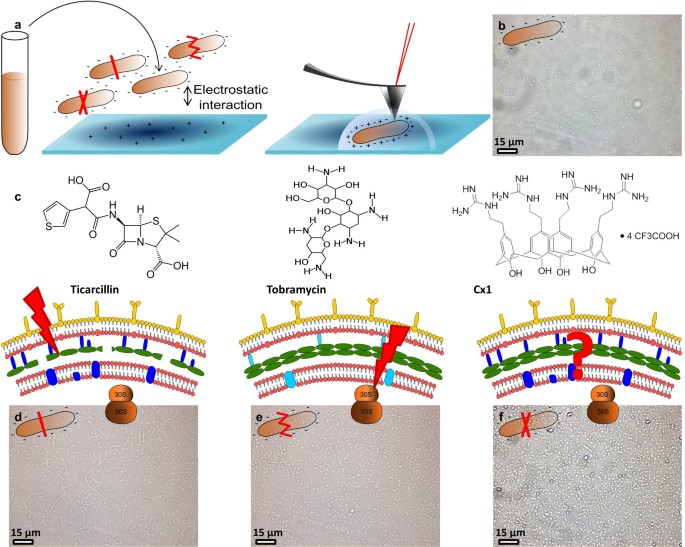
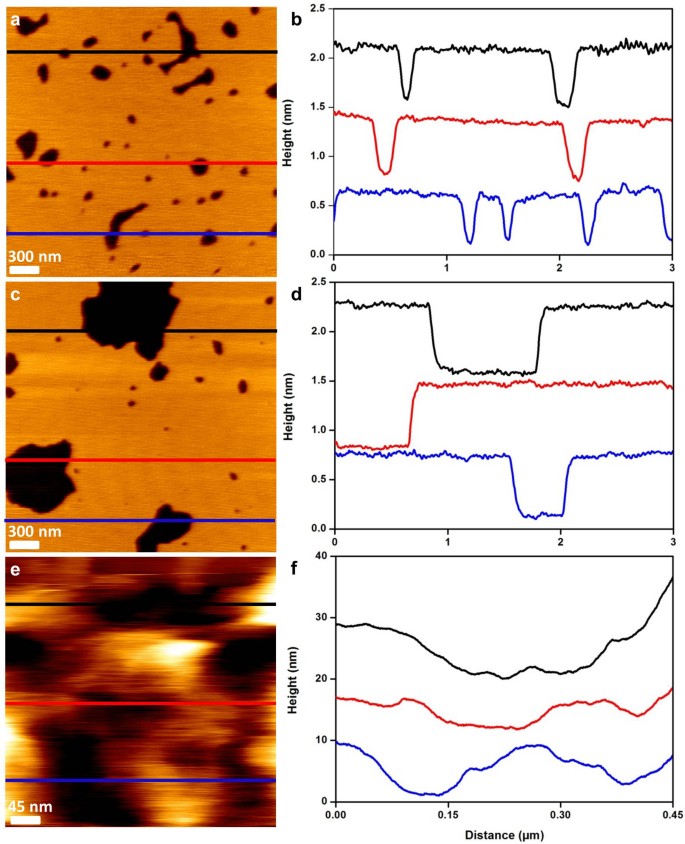
Figure 1
Schematic representation of the strategy used.
(a) cells cultivated in Mueller Hinton broth for 20 hours at 35°C are immobilized on a polyethylenimine coated glass slide for AFM experiments. (b) optical image of the surface covered with immobilized untreated P. aeruginosa ATCC 27853. (c) molecules used in the study and their targets. (d) optical images of P. aeruginosa ATCC 27853 treated by ticarcillin (4 µg/mL), (e) by tobramycin (0.25 µg/mL) and (f) by CX1 (32 µg/mL).
In the first part of this study, we characterized the effects of ticarcillin and tobramycin on the structure and the nanomechanical properties of P. aeruginosa ATCC 27853 (reference strain) and PaR3 (clinical strain resistant to almost all antibiotics, antibiogram in Supplemantary data 1). In a second part, by comparing the nano-effects (cell shape alteration, elasticity modifications, cell wall disorganization) caused by them with the ones caused by CX1, we get a better understanding of the mechanism of action of CX1 (Fig.1).
Morphology and surface roughness
The morphological effects, of ticarcillin and tobramycin on the reference strain are presented in Table 1. Supplementary data 2 and 3 present the raw data of respectively morphology and surface roughness. The results show only one analysis, these features were observed on at least 5 bacteria coming from 3 independent cultures. Bacteria in native conditions (without treatment) show a smooth surface. They are 2.2 µm long, 1.1 µm large and 453.5 nm high. We confirmed at the nanoscale, that bacteria growing in the presence of ticarcillin formed filaments of 6 to 18 µm long. For tobramycin, we showed that treated bacteria have a deformed cell wall. These effects of the antibiotics are not observed on the multidrug resistant strain PaR3, demonstrating that these two molecules have no effect on the morphology of the bacteria. Concerning our lead compound, CX1, we showed that it causes an alteration of the bacterial cell wall on the two different strains, without size modifications. We also focused on the surface roughness as it is a feature that characterizes a bacterial species25,26,29 (Table 1 and Supplementary Data 3) and showed that the surface of the reference strain was modified by the three molecules: the roughness is increased from 0.2 nm to 0.6 nm in presence of ticarcillin or tobramycin and to 1.0 nm in presence of CX1. PaR3 presented a smooth surface with no modification of the roughness when grown with antibiotics. When treated by CX1, the surface aspect is modified, showing perforations and the roughness is increased from 0.6 to 1.5 nm.
Table 1 Recapitulative table of the morphology and roughness results obtained on P. aeruginosa ATCC 27853 and PaR3. L stands for length, W stands for width and H stands for height. The analyses were performed on at least 5 different bacteria coming from 3 independent cultures
Nanomechanical properties
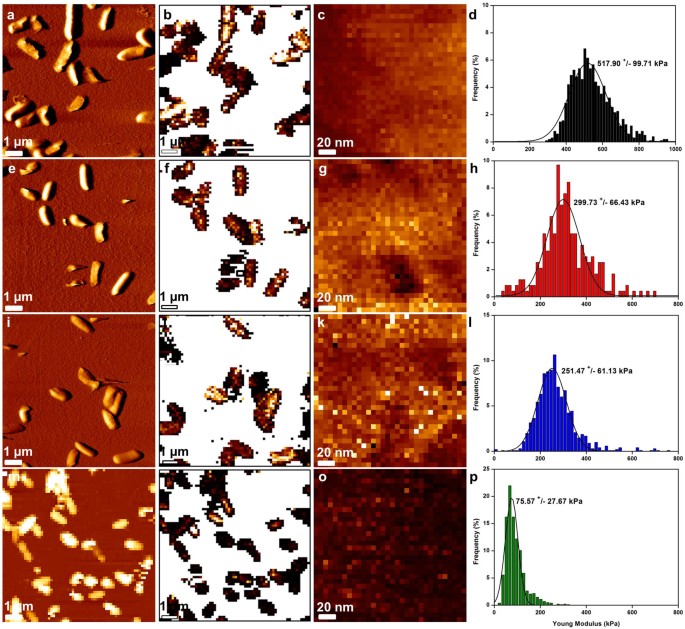
In view of the role of the bacterial cell wall conferring rigidity and protection, we then addressed the pertinent question as to whether the observed structural changes were correlated with differences in the cell wall mechanical properties. To this end, PaR3 treated with ticarcillin, tobramycin or CX1 were probed using nanoindentation measurements (Fig. 2). Several bacteria were probed (global effect) and then local measurements (surface elasticity) were performed on each bacterium present on the global force map (n = 5). These experiments were also conducted on P. aeruginosa ATCC 27853 (Supplementary data 4). To this end, arrays of 32 by 32 force curves were recorded on each bacterium. All the force curves were then converted into indentation curves and fitted with the Hertz model F = ((2.E.tanα)/(π.(1-ν2)).δ2, where F is the force (experimentally measured), E the Young modulus, α the opening angle of the tip (measured using MEB 35°, data not shown), ν the Poisson ratio (arbitrarily assumed to be 0.5) and δ the indentation (experimentally measured). This procedure gives access to the Young Modulus values that are represented on the histograms on Fig. 2. For each experiment, AFM tips were calibrated using the thermal noise method30, the spring constant values ranged from 0.012 to 0.019 N/m. The global force maps give information about the multidrug resistant bacterial population behavior towards the different molecules; it seems that only CX1 treated cells have an affected cell wall with a global decreased elasticity compared to antibiotics treated cells. These information were then confirmed with the local nanoindentation measurements that show that untreated PaR3 cells have a Young Modulus of 520 ± 100 kPa, whereas ticarcillin or tobramycin-treated bacteria had a Young Modulus respectively of 300 ± 66 kPa and 252 ± 61 kPa. After treatment by CX1, bacteria presented a Young modulus that drops to 76 ± 28 kPa. These results showed that interestingly, treatment by ticarcillin and tobramycin decreases the cell wall elasticity, but in a reasonable range. CX1, however, dramatically decreases the cell wall elasticity.
Figure 2
Mapping of P. aeruginosa R3 cell surface elasticity.
(a) vertical deflection image of native cells, (e) ticarcillin-treated cells (4 µg/mL), (i) tobramycin-treated cells (0.25 µg/mL). (m) height image (z-range = 800 nm) of CX1-treated cells (32 µg/mL). (b), (f), (j) and (n), elasticity maps (z-range = 1.5 MPa) corresponding to the vertical deflection images. (c), (g), (k) and (o), local elasticity maps (z-range = 800 kPa) recorded on one bacterium from the corresponding vertical deflection images. (d), (h), (l) and (p), distributions of Young Modulus values corresponding to the local elasticity maps.
Single molecule force spectroscopy
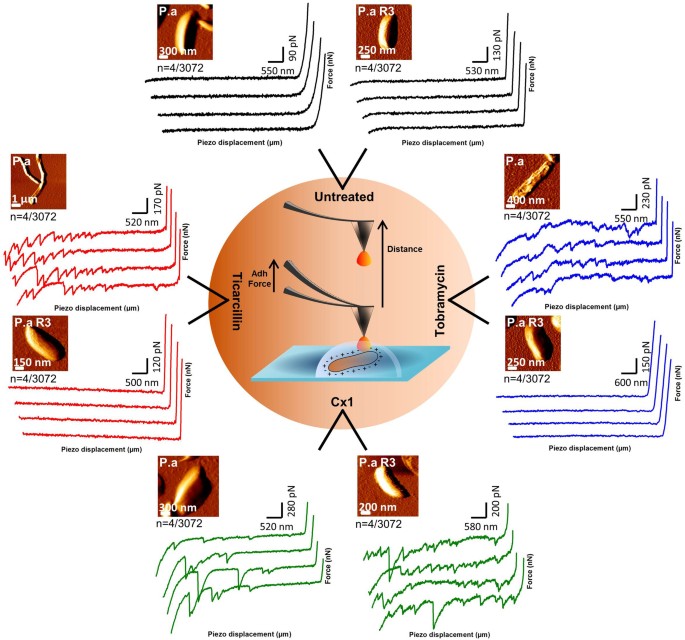
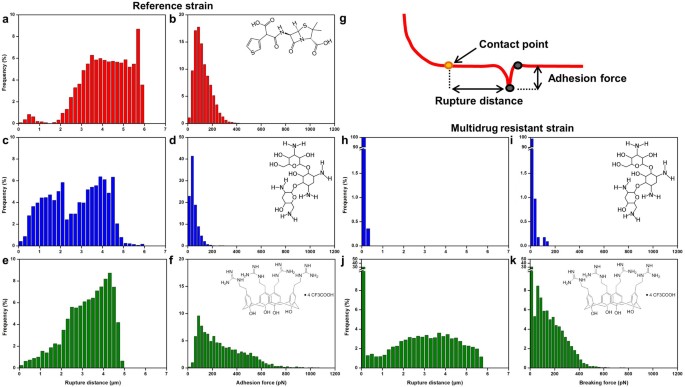
At this stage of the work, we know that CX1 disorganizes the cell wall of P. aeruginosa, but we still do not know how. To go further, dendritips31,32 were functionalized with lectin ConcanavalinA (ConA). ConA binding structure and specificity have been well determined for mannose-containing structures33,34,35, including recognition of biantennary, complex N-glycans36 and for terminal glucose35. These lectin tips were then used to perform adhesion force maps on bacteria in their native environment, or after treatment by the three molecules. Fig. 3 shows the force curves recorded on the two strains in the different conditions. We can see that force curves recorded on both untreated strains showed no adhesions (Fig. 3b). After ticarcillin or tobramycin treatment, the reference strain showed force curves presenting many adhesions and PaR3 showed nothing but flat curves (Fig. 3c and 3e). In Fig. 4, the distributions of the breaking forces and the ruptures distances (as sketched in Fig. 4g) were represented in histograms, for the conditions for which force curves showed adhesions. First, for the reference strain, with ticarcillin (Fig. 4a and 4b), the adhesion forces reached 98 ± 56 pN. Only a small fraction of these adhesions happened between 0 and 1 µm (3%), the major part ranging from 2 to 6 µm. With tobramycin (Fig. 4c) the force curves showed multiple adhesions. The adhesion force histogram (Fig. 4d) shows that forces only reach 37 ± 1 pN which is more or less three times less than with ticarcillin. Similar results were obtained with CX1; adhesions were ranging from 0 to 5 µm (Fig. 4e) and forces reached 186 ± 206 pN (Fig. 4f). For PaR3, we have previously showed that only CX1 conducted to a decrease in the elasticity of the cell wall. On the force curves recorded on PaR3 with a lectin probe, we observed adhesions ranging from 0 to 6 µm (Fig. 4j) and that reached 135 ±127 pN (Fig. 4k). The pulled out molecules are surprisingly long (up to 6 µm), much longer than the bacteria itself.
Figure 3
Force spectroscopy of the ConA-tip interactions.
Schematic representation of the force curves (retract segment) obtained with ConcanavalinA functionalized tips on P. aeruginosa ATCC 27853 and PaR3 in native conditions, treated by ticarcillin (4 µg/mL), tobramycin (0.25 µg/mL) and CX1 (32 µg/mL). The 4 force curves (n) presented by conditions were chosen out of 3072 curves recorded on 3 different bacteria coming from 3 independent cultures.
Figure 4
Force spectroscopy of the ConA-tips interactions.
(a), adhesion force histograms (n = 1024 force curves) obtained on P. aeruginosa ATCC 27853 ticarcillin-treated cells (4 µg/mL), (c), tobramycin-treated cells (0.25 µg/mL) and (e), CX1-treated cells (32 µg/mL). (b), (d) and (f), corresponding rupture distance histograms. (g), schematic representation of how the retract segment of the force curves were analyzed. (h), adhesion force histograms (n = 1024 force curves) obtained on P. aeruginosa R3 tobramycin-treated cells (0.25 µg/mL) and (j), CX1-treated cells (32 µg/mL). (i) and (k), corresponding rupture distance histograms.
Effects of CX1 on supported bilayers
In order to mimic the effect of CX1 on the outer membrane of P. aeruginosa, we created phospholipidic bilayers supported on mica leaves with lipids widely found in bacterial outer membranes; POPE (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosophoethanolamine) and POPG (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol) (2:1)37. The method was previously described and has proven useful for the understanding of surfactin effect on lipid bilayer38. The results showed in Fig. 5 present these bilayers treated by CX1 at a concentration of 10 µg/mL. We have seen that in 1 hour, holes were created in the synthetic membrane. Those holes could be compared to the one observed in the cell wall of PaR3 treated by CX1 (Fig. 5e and 5f), which allowed us to think that CX1, with its spatial organization and its charges, is able to create perforations in the bacterial outer membrane.
Figure 5
POPE:POPG (2:1) supported bilayers.
(a), height images (z-range = 1.5 nm) of POPE:POPG (2:1) supported bilayers at t = 0 minutes after treatment by CX1 (0.01 mg/mL) and (c), 1 hour after treatment. (e), height images (z-range = 600 nm) of PaR3 treated by CX1 (32 µg/mL). (b), (d) and (f), cross sections taken along the colored lines on the images.
Discussion
We choose to work with two strains of P. aeruginosa; a reference strain susceptible to antibiotics (ATCC 27853) and a clinical isolate (PaR3, ABC Platform® Bugs Bank) collected from a respiratory sample, resistant to almost all antibiotics (antibiotic susceptibility profile is given in Supplementary data 1). This isolate is resistant to ticarcillin and tobramycin, two antibiotics widely used in P. aeruginosa infections. The first one belongs to the β-lactams family and inhibits the peptidoglycan synthesis by interacting with the Penicillin Binding Proteins (PBP). After treatment by ticarcillin, filamentous forms of P. aeruginosa are described28,39. This is caused by the fact that β-lactams like ticarcillin activate the SOS system of bacteria, therefore inhibiting the cell division39. Tobramycin belongs to the aminoglycosides family and interacts with the 30S ribosomal sub-unit. This leads to the synthesis of abnormal proteins which are then incorporated to the cell wall, which loses its integrity.
The results of surface roughness, consistent with the morphology analysis, showed that only CX1 is able to alter the cell wall of PaR3 and this is our first clue; thus, we have originally emphasized the fact that PaR3 is resistant to ticarcillin and tobramycin. However, we can hypothesize that the resistance mechanism must either have an energy cost, or result in a cell wall modification since the elasticity is a little decreased by classical antibiotics. So after 24 hours of growing in the presence of the antibiotics, PaR3 cell wall seems to be affected in an insignificant way. With CX1, the elasticity decreased, indicating that the integrity of the wall is compromised. PaR3 is unable to resist to the disorganization of the cell wall induced by CX1, whereas it resists to the one induced by ticarcillin and tobramycin. So, we showed that an innovative molecule, like CX1, is able to disorganize the cell wall of a MDR P. aeruginosa. The bacterial cell wall plays several roles, it is a barrier that withstands the osmotic pressure, it gives shape to the cells, ensures communication with the environment. Thus the cell wall disorganization is another proof showing that CX1 is an efficient antimicrobial molecule against a resistant strain of P. aeruginosa.
The next step in this investigation has been to explore at the molecular level, the effects of the antibiotics and of CX1. To this end we used lectin functionalized AFM probes and looked at what could be pulled out from the surface (fig. 3 and 4). PaR3 showed nothing but flat curves with ticarcillin and tobramycin. This can be explained by the fact that PaR3 cell wall was not disorganized and no molecules could be pulled out from the surface by the functionalized AFM tip. But interestingly, when CX1 was used to treat the different bacterial strains, multiple adhesions could be seen on the force curves (Fig. 3d). These new results are consistent with the nanomechanical evidence. CX1 disorganizes the bacterial cell at the molecular level. On the reference strain, antibiotics and CX1 disorganized the cell wall leading to a dramatic decrease of the elasticity (Supplementary data 4) and to the stretching of surprisingly long glycans (lectin recognition) molecules. We must, therefore, have unfolded a super coiled molecule. Recently, Andre et al studied the architecture of peptidoglycan in Bacillus subtilis40 and showed that glycan strands could be polymerized and crosslinked to form a peptidoglycan rope, which would then be coiled into a helical cable. Hayhurst et al41 worked on the same bacteria and showed that glycan strands were up to 5 µm, so way longer than the cell itself. The authors also proposed a coiled-coil model for peptidoglycan architecture. Following these ideas, our AFM tips could pull the peptidoglycan and uncoil it on large distances, consistently with the force curves observed in Fig. 3. In Gram-negative bacteria, a thin peptidoglycan layer is overlaid by a bilayers of phospholipids and lipopolysaccharids containing membrane proteins (for example porins, Braun lipoproteins)40. When bacteria are in native conditions, the peptidoglycan is very well organized and covered by the outer membrane, thus inaccessible for the lectin probe, resulting in no adhesive behavior when probed with a lectin tip. But when the ATCC strain is treated by ticarcillin, the landscape is different. Ticarcillin binds to transmembrane enzymes: carboxypeptidase and transpeptidase respectively responsible for the cleavage of the DAla-DAla motif and the assembly of the cleaved pentapeptides. The bacteria are thus unable to grow normally which leads to the activation of their SOS system. This results in extraordinary long bacteria (sometimes longer than 10 µm). Nevertheless, at the extremity of these “spaghetti”-like bacteria, a 2 µm long part of normal peptidoglycan remains. So when pulling with the lectin probe on the first 2 µm at the extremity of the bacteria, nothing happens. Then, new abnormal peptidoglycan is pulled out, which results in the force curves presented in Fig. 3.
With tobramycin, it is the protein synthesis of the ATCC 27853 strain that is altered. Braun lipoproteins and porins are essential components of the outer membrane structure. It is therefore not surprising that we can access to the peptidoglycan through the altered membrane. As the Young modulus is highly affected by tobramycin, it is also straight that the peptidoglycan is affected, but this is not yet described. However tobramycin inhibits the synthesis of all proteins among which the enzymes involved in the peptidoglycan synthesis. It is therefore not surprising that the peptidoglycan of tobramycin treated cells is abnormal and could lead to the force curves profile presented in Fig. 3c.
Finally, with CX1, it is even clearer that the peptidoglycan is affected for both the ATCC and the multidrug resistant strain PaR3; the Young modulus drops, indeed, dramatically. However, it must be noticed that for the ATCC strain; the force curves in Fig. 3d show less adhesive events with CX1 treatment than with ticarcillin or tobramycin treatment. This is not true with PaR3 which seems very affected by CX1.
A keen analyze conducted on the force curves presented on Fig. 3 shows that the distance between each adhesive event, when occurring, is of 280 ± 155 nm (277> n > 290 according to the condition). As Vollmer et al.42 proposed in a recent work, the glycan strands of P. aeruginosa are 17 nm long. In line with this data and consistently with the architectural model of the peptidoglycan proposed by Dufrêne and Foster's teams, it seems like our functionalized AFM tip pulls out from the damaged bacteria the peptidoglycan by “packs” of glycans strands. In Gram-negative bacteria, the peptidoglycan layer is encored to the outer membrane by the Braun lipoproteins. These “packs” of glycan strands could then correspond to the distances between the Braun lipoproteins all along the bacteria. Ticarcillin, tobramycin and CX1 treatments induce, indeed, the same distance rupture between the adhesive events. This means that although the 3 molecules have completely different mechanism of action, they induce somehow the same disorder. Therefore the hypothesis of the distance between the Braun lipoproteins is consolidated. Braun lipoproteins synthesis is inhibited by tobramycin and they make the link between the peptidoglycan (ticarcillin inhibits its synthesis) and the outer membrane (CX1 deeply alter phospholipid bilayers as it will be demonstrated in the next paragraph). If these new data gives light on the architecture of Gram negative bacteria cell wall, the fundamental mechanism of action of CX1 still remains unclear. We suppose that, when the positively charged calixarene, interacts with the negatively charged ultrastructures of the bacterial surface (i.e. phospholipids and lipopolysaccharides), its particular three-dimensional organization causes disruptions of the outer membrane of the cell wall as suggested by the increased surface roughness (Supplementary data 4) which is in line with the mechanism of action described for cationic antimicrobial peptides43,44,45,46.
These AFM experiments, conducted on two P. aeruginosa strains, a susceptible one (ATCC 27853) and a resistant one (PaR3), has allowed us to evaluate the effects of three different antibacterial molecules. The in depth analysis of AFM raw data collected on treated bacteria is an original way to get fundamental knowledge on the bacterial cell wall organization. These results confirm that CX1 is efficient on resistant bacteria and has a different mechanism of action as tobramycin and or ticarcillin. Also, these very new results allowed us to make an hypothesis on the potential mechanism of action of the CX1; it interacts with the surface of the Gram-negative bacteria and creates holes in the outer membrane. This hypothesis was then confirmed by the experiments conducted on the supported biomembranes that showed destruction by creation of holes. Interestingly it has been demonstrated that CX1 has no side effects on eukaryotic HaCaT cells14 or on membranes made of zwiterrionic phospholipids (DMPC, DMPS)47.
It is also obvious that CX1 damage the peptidoglycan as we saw on the nanoindentations results and therefore there is no reason why CX1 could not reach and damage the inner membrane. The next step is now to determine if CX1 has also an intracellular target, which has not been explored yet.
Methods
Bacteria growth conditions
The bacteria (ATCC 27853) (reference strain for the Comité de l'Antibiogramme de la Société Française de Microbiologie, CA-SFM, the European Committee on Antimicrobial Susceptibility Testing, EUCAST and the Clinical and Laboratory Standards Institute, CLSI) and Pa R3 (isolated from a respiratory sample, ABC platform® Bugs Bank ) were stocked at −80 °C, revivified on Mueller Hinton Agar (Difco, 225250-500 g) and grown in Mueller Hinton Broth (Difco, 275730-500 g) for 24 hours at 35°C under static conditions.
Antibiotic treatments
The antibiotics were added during the 18 to 24 hours before the experiments.
Before AFM measurements were conducted, bacteria were grown in Mueller Hinton broth containing the antibiotics at a concentration of 4 µg/mL for ticarcillin (Sigma, T5639-1 g), 0.25 µg/mL for tobramycin (Sigma, T4014-100 mg) and 32 µg/mL for CX1 for 24 hours at 35°C.
Sample preparation for AFM experiments
Cells were concentrated by centrifugation, washed 2 times in Milli-Q water, re-suspended in PBS 1X (Sigma, P2194-10PAK) to a concentration of ~ 108 cells/mL and immobilized on PEI (Fluka P3142-100 mL) coated glass slides (prepared as described elsewhere e.g.48). Briefly, freshly oxygen activated glass slides were covered by a 0.2% PEI solution in deionized water and left for incubation overnight. Then the glass slides were rinsed with 20 mL of Milli-Q water and nitrogen dried. A total of 1 mL of the bacterial suspension was then applied to the PEI coated glass slide, allowed to stand for one hour and rinsed with PBS 1X. Images were recorded in PBS 1x in contact mode with MLCT AUHW cantilever (nominal spring constant 0.01 N/m). The applied force was kept as low as possible around 200 pN. For imaging and force spectroscopy we used an AFM Nanowizard II and III (JPK instruments, Berlin, Germany). The cantilevers spring constant were measured by the thermal noise methods30,49 ranging 14.56 to 15.20 mN/m. The functionalized tips were produced according to a french patent of the authors31 described later in sensors and actuators32. Briefly, AFM tips are functionalized with dendrimers presenting CHO functions able to covalently link with NH2 functions of proteins. Those dendritips are then incubated with the lectin concanavalin A (Sigma, L7647-100MG, 100µg/mL) for 1 hour, before being used for force spectroscopy experiments.
Phospholipid bilayers
POPE (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosophoethanolamine) and POPG (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol) (Avanti Polar Lipids) were dissolved in CHCl3 (2:1) and mixed in glass tubes to obtain the desired concentration. The solvent was evaporated with nitrogen and dried in a dessicator. Dried films were maintained under reduced pressure overnight and thereafter rehydrated using PBS 10 mM, 1 mM CaCl2, 1 mM MnCl2, pH 7.4. To obtain small unilamellar vesicles (SUVs), the suspension was sonicated to clarity (3 cycles of 3 min) using a 500W probe sonicator (Fisher Bioblock Scientific, France; 35% of the maximal power) while keeping the suspension in an ice bath. The suspension was finally centrifuged (5 min, 15000 g). The SUV solution was then put into contact with freshly cleaved mica substrates for 45 min at room temperature. Then samples were imaged using hyperdrive mode from Nanowizard III JPK instrument and PPP-NCHAuD-10 probes provided by Nanosensors.
References
- Taubes, G. The Bacteria fight back. Science 321, 356–361 (2008).
CAS PubMed Google Scholar - Payne, D. J. Desperately seeking new antibiotics. Science 321, 1644–1645 (2008).
CAS PubMed Google Scholar - Paterson, D. L. & Lipman, J. Returning to the pre-antibiotic era in the critically ill: The XDR problem. Crit. Care Med. 35, 1789–1791 (2007).
PubMed Google Scholar - Lipsitch, M., Bergstrom, C. T. & Levin, B. R. The epidemiology of antibiotic resistance in hospitals: Paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 97, 1938–1943 (2000).
ADS CAS PubMed Google Scholar - Hauser, A. R. & Ozer, E. A. Pseudomonas aeruginosa. Nat. rev. Microbiol. 9, Poster (2011).
- Skiada, A., Markogiannakis, A., Plachouras, D. & Daikos, G. L. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 37, 187–193 (2011).
CAS PubMed Google Scholar - Lister, P. D., Wolter, D. J. & Hanson, N. D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 22, 582–610 (2009).
CAS PubMed PubMed Central Google Scholar - Strateva, T. & Yordanov, D. Pseudomonas aeruginosa: a phenomenon of bacterial resistance. J. Med. Microbiol. 59 (2009).
- Breidenstein, E. B. M., de la Fuente-Núñez, C. & Hancock, R. E. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426 (2011).
CAS PubMed Google Scholar - Mortensen, N. P. et al. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25, 3728-3733 (2009).
CAS PubMed Google Scholar - Soon, R. L. et al. Effect of colistin exposure and growth phase on the surface properties of live Acinetobacter baumannii cells examined by atomic force microscopy. Int. J. Antimicrob. Agents 38, 493–501 (2011).
CAS PubMed PubMed Central Google Scholar - Yin, L. M., Edwards, M. A., Li, J., Yip, C. M. & Deber, C. M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 287, 7738–7745 (2012).
CAS PubMed PubMed Central Google Scholar - Mourer, M., Duval, R. E., Finance, C. & Regnouf-de-Vains, J.-B. Functional organisation and gain of activity: The case of the antibacterial tetra-para-guanidinoethyl-calix[4]arene. Bioorg. Med. Chem. Lett. 16, 2960–2963 (2006).
CAS PubMed Google Scholar - Grare, M. et al. In vitro activity of para-guanidinoethylcalix[4]arene against susceptible and antibiotic-resistant Gram-negative and Gram-positive bacteria. J. Antimicrob. Chemother. 60, 575–581 (2007).
CAS PubMed Google Scholar - Grare, M. et al. In vitro mutation studies with a new promising compound, the para-guanidinoethylcalix[4]arene. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3951 (2008).
- Binnig, G., Rohrer, H., Gerber, C. & Weibel, E. Tunneling through a controllable vacuum gap. Appl. Phys. Lett. 40, 178–180 (1982).
ADS CAS Google Scholar - Binnig, G. & Quate, C. F. Atomic force microscope. Phys. rev. lett. 56, 930–933 (1986).
ADS CAS PubMed Google Scholar - Muller, D. J., Helenius, J., Alsteens, D. & Dufrene, Y. F. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 5, 383–390 (2009).
PubMed Google Scholar - Dufrêne, Y. F. & Müller, D. J. Atomic Force Microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 3, 261–269 (2008).
ADS PubMed Google Scholar - Cross, S. E., Jin, Y.-S., Rao, J. & Gimzewski, J. K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2, 780–783 (2007).
ADS CAS Google Scholar - Buzhynskyy, N., Girmens, J.-F., Faigle, W. & Scheuring, S. Human cataract lens membrane at subnanometer resolution. J. Mol. Biol. 374, 162–169 (2007).
CAS PubMed Google Scholar - Stolz, M. et al. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nat. Nanotechnol. 4, 186–192 (2009).
ADS CAS PubMed Google Scholar - Liu, S. & Wang, Y. Application of AFM in microbiology: a review. Scanning 32, 61–73 (2010).
CAS PubMed Google Scholar - Dorobantu, L. S. & Gray, M. R. Application of atomic force microscopy in bacterial research. Scanning 32, 74–96 (2010).
CAS PubMed Google Scholar - Alsteens, D. et al. Organization of the mycobacterial cell wall: a nanoscale view. Pflugers Arch. 456, 117–125 (2008).
CAS PubMed Google Scholar - Francius, G., Domenech, O., Mingeot-Leclercq, M. P. & Dufrêne, Y. F. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin. J. Bact. 190, 7904–7909 (2008).
CAS PubMed Google Scholar - El Kirat, K., Burton, I., Dupres, V. & Dufrene, Y. F. Sample preparation procedures for biological atomic force microscopy. J. Microsc. 218, 199–207 (2005).
MathSciNet CAS PubMed Google Scholar - Formosa, C., Grare, M., Duval, R. E. & Dague, E. Nanoscale effects of antibiotics on P. aeruginosa. Nanomed. Nanotechnol. Biol. Med. 8, 12–16 (2012).
CAS Google Scholar - Francius, G. et al. Conformational analysis of single polysaccharide molecule on live bacteria. ACS Nano 2, 1921–1929 (2008).
CAS PubMed Google Scholar - Emerson Iv, R. J. & Camesano, T. A. On the importance of precise calibration techniques for an atomic force microscope. Ultramicroscopy 106, 413–422 (2006).
Google Scholar - Dague, E., Jauvert, E. & Trevisiol, E. France Pointe de microscope à force atomique modifiée et biomodifiée, 2010 n° dépôt 10 57932, 30/09/2010
- Jauvert, E. et al. Probing single molecule interactions by AFM using bio-functionalized dendritips. Sens. Actuators B Chem. 168, 436–441 (2012).
CAS Google Scholar - Hardman, K. D. & Ainsworth, C. F. Structure of concanavalin A at 2.4-A resolution. Biochem. 11, 4910–4919 (1972 ).
CAS Google Scholar - Naismith, J. H. & Field, R. A. Structural Basis of Trimannoside Recognition by Concanavalin A. J. Biol. Chem. 271, 972–976 (1996).
CAS PubMed Google Scholar - Gupta, D., Dam, T. K., Oscarson, S. & Brewer, C. F. Thermodynamics of lectin-carbohydrate interactions. J. Biol. Chem. 272, 6388–6392 (1997).
CAS PubMed Google Scholar - Moothoo, D. N. & Naismith, J. H. Concanavalin A distorts the beta-GlcNAc-(1-2)-Man linkage of beta-GlcNAc-(1-2)-alpha-Man-(1-3)-[beta-GlcNAc-(1-2)-alpha-Man-(1-6)]-Man upon binding. Glycobiol. 8, 173–181 (1998).
CAS Google Scholar - Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. (In Press).
- Francius, G. et al. Nanoscale membrane activity of surfactins: Influence of geometry, charge and hydrophobicity. Biochim. Biophys. Acta 1778, 2058–2068 (2008).
CAS PubMed Google Scholar - Prior, R. B. & Warner, J. F. Morphological alterations of Pseudomonas aeruginosa by ticarcillin: a scanning electron microscope study. Antimicrob. Agents Chemother. 6, 853–855 (1974).
CAS PubMed PubMed Central Google Scholar - Andre, G. et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 1, 27 (2010).
ADS PubMed Google Scholar - Hayhurst, E. J., Kailas, L., Hobbs, J. K. & Foster, S. J. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. 105, 14603–14608 (2008).
ADS CAS PubMed Google Scholar - Vollmer, W. & Seligman, S. J. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18, 59–66 (2010).
CAS PubMed Google Scholar - Jenssen, H., Hamill, P. & Hancock, R. E. W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 (2006).
CAS PubMed PubMed Central Google Scholar - Finlay, B. B. & Hancock, R. E. W. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2, 497–504 (2004).
CAS PubMed Google Scholar - McPhee, J. B., Lewenza, S. & Hancock, R. E. W. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50, 205–217 (2003).
CAS PubMed Google Scholar - Hancock, R. E. W. & Patrzykat, A. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disord. 2, 79–83 (2002).
CAS PubMed Google Scholar - Sautrey, G. et al. Membrane activity of tetra-_p_-guanidinoethylcalix[4]arene as a possible reason for its antibacterial properties. J. Phys. Chem. B 115, 15002–15012 (2011).
CAS PubMed Google Scholar - Francius, G., Tesson, B., Dague, E., Martin-Jézéquel, V. & Dufrêne, Y. F. Nanostructure and nanomechanics of live Phaeodactylum tricornutum morphotypes. Environ. Microbiol. 10, 1344–1356 (2008).
CAS PubMed Google Scholar - Hutter, J. L. & Bechhoefer, J. Calibration of Atomic Force Microscope tips. Rev. Sci. Instrum. 64, 1868 (1993).
ADS CAS Google Scholar
Acknowledgements
ED is a researcher of Centre National de la Recherche Scientifique (CNRS). CF received funding from Direction Générale de l'Armement (DGA) and Agence Nationale pour la Recherche (ANR). We thank the program Young Scientist from ANR and especially the AFMYST project. The authors thank UMS 3039 ITAV for the use of the AFM facilities.
Author information
Authors and Affiliations
- Centre National de la Recherche Scientifique, Laboratoire d'Analyse et d'Architecture des systèmes (LAAS), Toulouse, France
C. Formosa, E. Jauvert, A. Coutable & E. Dague - Centre National de la Recherche Scientifique, Toulouse, France
C. Formosa, E. Jauvert, A. Coutable & E. Dague - Université de Toulouse, Toulouse, France
C. Formosa, E. Jauvert, A. Coutable & E. Dague - SRSMC, Université de Lorraine - CNRS, Nancy, France
C. Formosa, J. B. Regnouf-de-Vains, M. Mourer & R. E. Duval - ABC Platform®, Nancy, France
R. E. Duval - Laboratoire de Bactériologie Hygiène, Institut Fédératif de Biologie, Toulouse, France
M. Grare
Authors
- C. Formosa
You can also search for this author inPubMed Google Scholar - M. Grare
You can also search for this author inPubMed Google Scholar - E. Jauvert
You can also search for this author inPubMed Google Scholar - A. Coutable
You can also search for this author inPubMed Google Scholar - J. B. Regnouf-de-Vains
You can also search for this author inPubMed Google Scholar - M. Mourer
You can also search for this author inPubMed Google Scholar - R. E. Duval
You can also search for this author inPubMed Google Scholar - E. Dague
You can also search for this author inPubMed Google Scholar
Contributions
ED and RED are the leads author of the paper. ED, RED, MG and CF conceived-designed the experiments and wrote the article. CF made the experimental work and the data analysis work. EJ and AC worked on the experimental protocols and prepared respectively dendritips and phospholipid bilayers. MM and JBRdV conceived CX1 as a possible antibacterial agent and furnished batch used in this study. All the authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Formosa, C., Grare, M., Jauvert, E. et al. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain.Sci Rep 2, 575 (2012). https://doi.org/10.1038/srep00575
- Received: 08 June 2012
- Accepted: 30 July 2012
- Published: 14 August 2012
- DOI: https://doi.org/10.1038/srep00575