Completing the map of human genetic variation (original) (raw)
A plan to identify and integrate normal structural variation into the human genome sequence.
Large-scale studies of human genetic variation have focused largely on understanding the pattern and nature of single-nucleotide differences within the human genome. Recent studies that have identified larger polymorphisms, such as insertions, deletions and inversions, emphasize the value of investing in more comprehensive and systematic studies of human structural genetic variation. We describe a community resource project recently launched by the National Human Genome Research Institute (NHGRI) to sequence large-insert clones from many individuals, systematically discovering and resolving these complex variants at the DNA sequence level. The project includes the discovery of variants through development of clone resources, sequence resolution of variants, and accurate typing of variants in individuals of African, European or Asian ancestry. Sequence resolution of both single-nucleotide and larger-scale genomic variants will improve our picture of natural variation in human populations and will enhance our ability to link genetics and human health.
Background
The information gained from the sequencing of the human genome1,2 has begun to revolutionize human biology and genetic medicine. Advances in genomic technologies and bioinformatics, combined with an enormous reduction in cost, have led to genome sequencing projects for dozens of species. It is anticipated that the sequencing of individual human genomes will ultimately be required for a comprehensive genetic understanding of disease3, although at present the cost of such efforts is prohibitive. The discovery of functionally important genetic variation lies at the core of these endeavours, and there has been considerable progress in understanding the common patterns of single-nucleotide polymorphism (SNP) in humans. Indeed, of the estimated 10–15 million common human SNPs, a significant fraction have now been identified and genotyped among population samples (HapMap release 21)4,5.
By contrast, our understanding of structural variation in the human genome is more recent and rudimentary. In its broadest sense, structural variation can be defined as all genomic changes that are not single base-pair substitutions6,7,8. Such variation includes insertions, deletions, inversions, duplications and translocations of DNA sequences, and encompasses copy-number differences (also known as copy-number variants, CNVs)9,10,11. During the past two years, several genome-wide surveys8,12,13,14,15,16,17,18,19 have described large-scale (>100 kb), intermediate-scale (500 bp–100 kb) and fine-scale (1–500 bp) structural variations in the human genome. These studies have revealed that structural changes are ubiquitous and common, and frequently involve the rearrangement of genes. Along with SNPs, it is important that we establish a baseline for normal structural variation in order to facilitate the future discovery and characterization of disease-causing mutations in patients.
Previous efforts to find such variants have relied on array-based methods, comparing patterns of fluorescence intensity across the genome and between individuals. This approach has been the focus of the Copy Number Variation Project, an international consortium effort initiated in 2004 to comprehensively identify copy-number variants in the 269 samples analysed by the International HapMap Project10. Remarkably, the project has revealed considerable variation between normal human genomes, with more than 1,447 copy-number variant regions spanning 12% of the reference DNA sequence18. Although these array-based studies are very important, most are not able to identify which specific DNA sequences have been altered, nor the molecular events that have given rise to these structural genomic variants. Moreover, array-based technologies dependent on the detection of copy-number differences are unable to detect structural variation events that have arisen as a result of balanced chromosomal rearrangements (such as inversions or reciprocal translocations of chromosomal segments). In most cases the frequency of such balanced events is unknown, although analyses of genomic sequence14,19 suggest that 1–20% of all structural variation may in fact be balanced and does not involve copy-number changes14,19.
Biomedical relevance
Some of the earliest human genetic traits to be mapped — such as colour blindness, rhesus blood group sensitivity, classical haemophilia and forms of beta- and alpha-thalassaemia20,21,22 — result from complex structural alterations in genes and gene families23,24,25,26,27. At the other end of the spectrum are large, structural rearrangements of chromosomes known to cause genomic disorders that typically involve millions of base pairs of sequence (for example, Prader–Willi syndrome and velocardiofacial syndrome)27. Structural genetic variation can confer phenotypes through several mechanisms28. These include gene dosage (copy-number variation); gene disruption; gene fusions at the junction; position effects in which the rearrangement alters the regulation of a nearby gene; and unmasking of recessive mutations or functional SNPs on the remaining allele. Another possible mechanism could occur through perturbations of gene expression that normally result from the pairing of homologous alleles, as has been observed in Drosophila29.
In addition to their roles in rare mendelian diseases and genomic disorders, several common structural genetic variants (>1% minor allele frequency) have been shown to be important in both normal phenotypic variability and disease susceptibility (Table 1). For example, deletions of the UGT2B17 gene contribute to ethnic and interindividual differences in testosterone metabolism and risk of prostate cancer30,31. Increased copy number of the CCL3L1 gene is associated with reduced susceptibility to HIV infection and progression to AIDS32. Similarly, individuals with fewer copies of the DEFB4 gene have a higher risk of developing colonic Crohn's disease33, and reduced FCGR3 copy number predisposes people to glomerulonephritis34.
Table 1 Common structural polymorphisms and disease

These examples highlight the importance of structural variation to disease and disease susceptibility, and suggest several concepts of potentially broad relevance. First, the number of copies of a given gene or family of genes can be a direct risk factor for specific diseases. Second, in some cases copy number alone is not sufficient to explain phenotypic differences caused by structural genetic variation. In the examples of rhesus blood group sensitivity, colour blindness and the alpha- and beta-thalassaemias, it is the precise DNA sequence structure (that is, the formation of fusion genes or the position of a gene with respect to functional promoters) that provides the most meaningful associations between genotype and disease23,24,25. Third, normal structural genetic variation can increase the risk of secondary, pathogenic rearrangement. For example, there is increasing evidence to support the suggestion that normal inversion polymorphisms can be predisposing factors for common microdeletion syndromes11. This is reminiscent of the 'premutation' class of allele associated with triplet-repeat diseases. Finally, structural genetic variants may be associated with genes related to immune response, host defence, drug response and environmental interaction, leading to different phenotypic effects35.
Although whole-genome SNP-based association studies hold great promise for the discovery of variants and genes influencing common diseases, the genetic complexity of structural genetic variants adds another level of information that needs to be incorporated into this approach. Specifically, the presence of structurally variant sequences can result in the misinterpretation of marker genotypes and their segregation patterns36 or in a reduction of reliable SNP genotyping assays using various commercial genotyping platforms37,38, as well as in the Single Nucleotide Polymorphism database (dbSNP) and the HapMap database4,5. This, in turn, limits the utility of linkage disequilibrium with reliably assayed SNPs to 'tag' structural variants in disease-association studies. Although there is a growing number of examples of linkage disequilibrium between structural genetic variants and nearby polymorphic markers16,17,37, our ability to type all structurally variant regions (and SNPs within them) using current genome-wide technologies is limited.
The initiative
The fact that segments of the human genome vary substantially in copy number indicates that any single human genome carries only a subset of the full complement of human DNA sequences. Given that the public human genome reference sequence assembly represents what is essentially one version of that structure at any given site, it is incomplete. Like the initial requirement for a high-quality human genome reference sequence, there is now a need to generate a quality reference set of sequenced structural variants from many normal individuals and to discover new sequences that may be common, but are missing from the reference genome. Association studies of disease are likewise dependent on the 'completeness' of the reference sequence.
In 2005, the NHGRI Large-Scale Genome Sequencing Program (http://www.genome.gov/10001691) identified structural variation as an area of interest. Two NHGRI working groups put forward a proposal to characterize structural variation in the human genome of phenotypically normal individuals to achieve several goals. First, to systematically discover structural variations of as little as 5 kb in length. Second, to capture forms of natural genetic variation, such as inversions, that result from balanced chromosomal rearrangements and that cannot currently be detected by array-based technologies. Third, to provide sequence-based resolution of normal human structural genetic variation. The proposed aim was to bring knowledge of structural variation in the human genome to the level that has now been achieved for SNPs. Such information would complement SNP-based data as a valuable resource for genetic association studies of human disease.
The proposal was reviewed and approved by the National Advisory Council for Human Genome Research. It was recognized that this initiative would be large and complex in scope, and potentially competitive with other applications of large-scale sequencing efforts of medical interest. Sequencing costs associated with each additional human genome are estimated at US$800,000 per individual, with an additional $150,000 per individual assigned to targeted finishing and infrastructure costs. A two-to-three-year timeline was projected for completion of all sequencing aspects of this proposal. These costs and timelines are regarded as preliminary, and are subject to change owing to technological improvements. The plan for implementation includes regular assessment of the data as they emerge to ensure that the initiative is yielding the expected information and warrants the continued use of sequencing capacity to generate additional data.
An overview
The objective of this initiative is to characterize the pattern of human genetic structural variation at the nucleotide level from a collection of phenotypically normal individuals. In principle, the discovery and analysis of human structural genetic variation involves three straightforward steps: identifying variants, sequencing to resolve each variant's structure, and genotyping in larger samples to establish frequency and linkage disequilibrium characteristics. Identifying structural genetic variants has been challenging, especially doing so in a manner that allows for follow-up sequencing to define the variant at the nucleotide level.
The initiative will expand on a recently published strategy that exploits clusters of discordant end-sequence pairs from large-insert genomic clones with a known distribution of insert sizes14 (Fig. 1). The strategy maps the end-sequence pairs from a 10–12-fold redundant, whole-genome clone library from each individual to the human genome reference sequence assembly. This creates a clone tiling path of the second human genome compared with the reference and identifies discordant regions in which multiple clones show statistically significant discrepancies by length and/or orientation. These regions contain putative sites of insertion, deletion or inversion (Fig. 2).
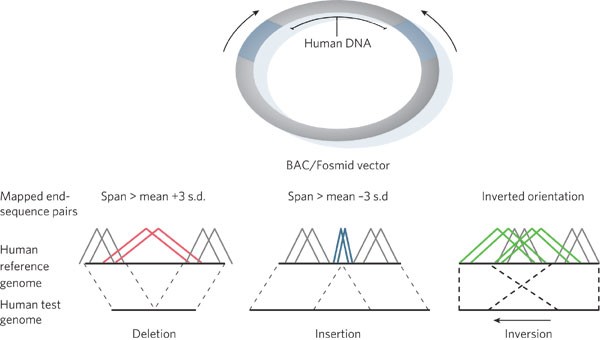
Figure 1: Paired-end sequence approach.
Genomic libraries are constructed from fragmented DNA and subcloned into circular vectors such as BACs or fosmids. The ends of these fragment inserts are directly sequenced from universal vector primers near the subcloning site (arrows) and are termed end-sequence pairs or paired-end sequences. End-sequence pairs are mapped to their best location in the human reference genome sequence assembly. End-sequence pairs that are discordant in terms of length (> 3 s.d. from the mean insert length) and/or orientation when mapped against the reference genome assembly may be indicative of deletions, insertions or inversion, as indicated (red, blue and green, respectively). End-sequence pairs consistent in terms of length and orientation are shown as grey.
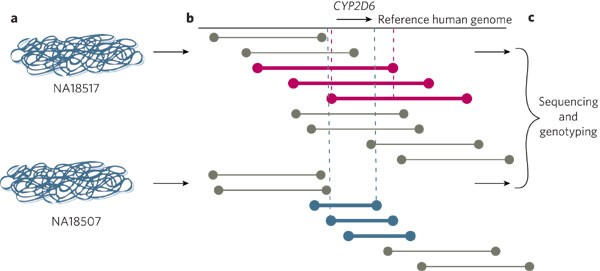
Figure 2: Sequencing structural variation.
a, Genomic clone libraries are constructed from different human DNA samples (Yoruban Nigerian samples NA18517 and NA18507). b, The inserts of ∼1 million fosmid clones are end-sequenced for each individual and aligned against the reference human genome. This provides a tiling path for each individual's genome against the reference sequence. The amount of DNA sequence between the ends of a clone (between end-sequence pairs) is known approximately, even before the clones are sequenced. The end sequences of each clone are mapped to the reference sequence. If they map to sites that are farther apart in the reference sequence than in the test sequence clone, there is a deletion in the test sequence, relative to the reference sequence. Conversely, if the end sequences map to sites that are closer in the reference sequence than in the test sequence, there is an insertion in the test sequence. Overlapping clones refine the location of the insertion or deletion (dashed lines), in this case, near the CYP2D6 gene. c, Sequencing of the corresponding clone provides sequence-based resolution of the insertion or deletion and allows genotyping assays to be developed to type a large number of individuals.
Specifics of the plan
To obtain 95% of the common variation (minor allele frequency >5%), the plan is to make fosmid clone libraries (∼40 kb inserts) from the genomic DNA of 48 unrelated females already genotyped in the HapMap, and BAC clone libraries and from 14 unrelated HapMap males with the concomitant production of ∼50 Gb of human sequence in the form of end-sequence pairs (see white paper at http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/ StructuralVariationProject.pdf for sample size rationale). The large insert (∼150 kb) BAC clone libraries will provide a mechanism by which to obtain sequence information on structural variants18 that are too large to be encompassed in the fosmid inserts, such as those associated with segmental duplications39 and the highly repetitive palindromic sequences of the Y chromosome40,41. Individuals studied in the International HapMap Project are ideal for this research because they are being characterized for structural variation by other means16,17,18,37 may be used for genome-wide variation discovery with full data release, and have already been genotyped for 3.4 million SNPs, making it possible to correlate structural variation with what is currently known about the genetic architecture of the region in question. Hence, genome libraries will be constructed from representative individuals with European, Asian and African ancestry.
Each human genome library will be constructed to tenfold physical coverage per individual and inserts will be end-sequenced. This should capture >98% of each parental haplotype in clones, even after allowing for cloning biases, sequence failure and failure of the end sequence to map to the genome14. The most important parameter for detecting structural variation in this plan is the insert size variance in both the fosmid and BAC libraries. With standard deviations of 1.5 kb for fosmid libraries, for example, it is possible to detect several hundred sites of structural variation as small as 5 kb per individual. The wider insert size distribution of BAC clones will require putative structural variant clones to be validated by fingerprinting before complete insert sequencing. A further benefit of this initiative is that it is expected to yield ∼15-fold greater coverage of human genomic sequence, providing ample substrate for the recovery of previously unknown rare SNPs and smaller insertion/deletion polymorphisms7,8.
A key aspect of the plan is to sequence all genomic clones that are discordant with the reference sequence in terms of length or orientation. On the basis of preliminary studies, we expect to identify several thousand sites of structural variation. These will be sequenced to a high degree, allowing base-pair resolution of the structural variants16. This amount of sequencing is well within the capacity of the genome centres. It is important to note that although some variants will be the result of simple insertions or deletions, others will be embedded in complex regions of the genome, and will have many rearrangements with respect to the reference sequence14,42. Clones from the library resource may also be useful to various research groups for other reasons. They could be used to close gaps in the human genome sequence and for follow-up investigation of positive 'hits' in whole-genome or candidate-region association studies by providing rapid and fairly complete characterization of all SNPs and structural variation on one or more associated haplotypes. In addition, they could be used to compare the ability of platforms to accurately detect different types of variation.
Another goal of the initiative is to genotype the discovered variants in the full set of HapMap samples, thus contributing to an integrated map of SNPs and structural variants. This is especially important because of the many genome-wide association studies currently in progress or planned for the near future. Investigators interpreting these data will encounter the structural variants only through their SNP genotype data. Recognizing that no single technology can adequately genotype all forms of structural variation9,11,43, this effort, among others, would stimulate technological improvements that would allow rapid, inexpensive and comprehensive assessment of all forms of structural variation. The immediate plan is to use the sequence-validated structural genetic variants from the 62 individuals (48 HapMap females and 14 HapMap males) to evaluate new technologies and to perform cross-platform comparisons of existing technologies, providing a better understanding of false positives and false negatives. The integration of the resulting structural genetic variant map with SNPs will offer clues to their evolutionary history in the genome. Structural variants that arose only once would be expected to show linkage disequilibrium with SNPs on their original haplotype, whereas structural variants that arise repeatedly would be expected to show little linkage disequilibrium with nearby SNPs. Identifying the structural genetic variants in linkage disequilibrium with nearby SNPs would also allow these variants to be tagged by SNPs, facilitating efficient identification of this subset of variants in subsequent association studies.
All sequence data from this initiative, including the corresponding end-sequence pairs and assembled clone insert sequences, will be deposited in NIH-sponsored public databases — the Trace Archive and GenBank, respectively — according to standards already established for large-scale sequencing efforts (http://www.wellcome.ac.uk/doc_wtd003208.html). Incorporating information from larger and more complex rearrangements presents new challenges to the bioinformatics community. The NIH SNP database (dbSNP) is designed to accept several classes of smaller variant, including SNPs, microsatellite repeats and small insertion or deletion events but not larger variants. It will be necessary, for example, to integrate alternative views of the human genome organization which are linked to the sequenced clones, provide sequence alignments of the structural genetic variants to the reference sequence, and flag regions in which mRNAs or genes could potentially be affected.
We propose the integration of sequence-defined structural genetic variation with the reference sequence and other genetic variation as part of dbSNP. The integrated information should include mapping data, size, structural properties, individual source and linkage disequilibrium with nearby variants, and could be treated as STS-like features (intervals defined by flanking sequence) when annotated against the reference genome assembly. As breakpoints are localized by sequencing and validation, the record can be expanded into sequenced haplotype alternatives. Similarly, public dissemination will benefit from integration with data on common genome browsers (such as that of the University of California, Santa Cruz, and ENSEMBL) as well as other public databases (for example, http://projects.tcag.ca/variation and http://humanparalogy.gs.washington.edu/structuralvariation).
Concluding remarks
Although there is no single approach that can adequately catalogue all human structural genetic variation, the plan outlined here is based on the successful bottom-up strategy that was essential to the Human Genome Project and, later, the HapMap project. This strategy will dovetail with top-down approaches such as that used by the Copy Number Variation Project10, which used array-based technology to discover the landscape of larger events in the same HapMap samples. The clone-based approach has a number of advantages. First, it couples discovery to sequence resolution at the nucleotide level. Second, it is genome-wide. Third, it is not biased by frequency. And, finally, it allows the detection and characterization of structural variants that result from balanced chromosomal rearrangements (such as inversions) as well as insertions that are not represented in the human genome reference sequence. The limitations of this approach include cost, the limited number of samples that can currently be analysed and the logistics associated with the generation and management of such a large-scale clone resource.
The data and clone resources generated by this initiative will provide insight into the composition and evolution of the human genome, including sequence information on thousands of larger structural variants. Such information cannot be obtained by simply reducing the costs of sequencing and generating more sequence data of lower quality and shorter read length44. The complexity of these regions demands high-quality sequence data, which can only be provided, at present, by strategic sequencing of large-insert clones. Data collected from a large number of phenotypically normal individuals will provide an important resource to assess the significance of newly discovered structural genetic variants and of those found to be enriched in patients with disease.
Although the primary goal of this initiative is to sequence most of the common structural genetic variants, this approach should enable the identification, characterization and genotyping of both common and rare variants. Therefore, these studies will provide a unique perspective by comprehensively comparing individual genomes against the current human reference sequence (Fig. 2), foreshadowing the development of rapid and complete individual genome sequencing44. Ultimately, approaches that couple high-throughput genome sequencing and paired-end sequence detection of structural variation may make it possible (and economically feasible) to analyse both SNPs and structural variants simultaneously in clinical samples. Meaningful interpretation of common and rare structural variants among patients will benefit from the most complete characterization of all forms of natural DNA sequence variation in the human genome.
References
- IHGSC. Initial sequencing and analysis of the human genome. Nature 409, 860– 921 (2001).
- Venter, J. C. et al. The sequence of the human genome. Science 291, 1304– 1351 (2001).
Article ADS CAS Google Scholar - Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. A vision for the future of genomics research. Nature 422, 835– 847 (2003).
Article ADS CAS Google Scholar - IHMC. A haplotype map of the human genome. Nature 437, 1299– 1320 (2005).
- Hinds, D. A. et al. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072– 1079 (2005).
Article ADS CAS Google Scholar - Weber, J. L. et al. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 71, 854– 862 (2002).
Article Google Scholar - Bhangale, T. R., Rieder, M. J., Livingston, R. J. & Nickerson, D. A. Comprehensive identification and characterization of diallelic insertion–deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 14, 59– 69 (2005).
Article CAS Google Scholar - Mills, R. E. et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16, 1182– 1190 (2006).
Article CAS Google Scholar - Feuk, L., Carson, A. R. & Scherer, S. W. Structural variation in the human genome. Nature Rev. Genet. 7, 85– 97 (2006).
Article CAS Google Scholar - Freeman, J. L. et al. Copy number variation: new insights in genome diversity. Genome Res. 16, 949– 961 (2006).
Article CAS Google Scholar - Sharp, A. J., Cheng, Z. & Eichler, E. E. Structural variation of the human genome. Annu. Rev. Genom. Hum. Genet. 7, 407– 442 (2006).
Article CAS Google Scholar - Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nature Genet. 36, 949– 951 (2004).
Article CAS Google Scholar - Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525– 528 (2004).
Article ADS CAS Google Scholar - Tuzun, E. et al. Fine-scale structural variation of the human genome. Nature Genet. 37, 727– 732 (2005).
Article CAS Google Scholar - Hinds, D. A., Kloek, A. P., Jen, M., Chen, X. & Frazer, K. A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nature Genet. 38, 82– 85 (2006).
Article CAS Google Scholar - Conrad, D. F., Andrews, T. D., Carter, N. P., Hurles, M. E. & Pritchard, J. K. A high-resolution survey of deletion polymorphisms in the human genome. Nature Genet. 38, 75– 81 (2006).
Article CAS Google Scholar - McCarroll, S. A. et al. Common deletion polymorphisms in the human genome. Nature Genet. 38, 86– 92 (2006).
Article CAS Google Scholar - Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444– 454 (2006).
Article ADS CAS Google Scholar - Khaja, R. et al. Genome assembly comparison identifies structural variants in the human genome. Nature Genet. 38, 1413– 1418 (2006).
Article CAS Google Scholar - Wilson, E. B. The sex chromosomes. Arch. Mikrosk. Anat. Entwicklungsmech 77, 249– 271 (1911).
Article Google Scholar - Cooley, T. B. & Lee, P. A series of cases of splenomegaly in children with anemia and peculiar bone changes. Trans. Am. Pediatr. Soc. 37, 29 (1925).
Google Scholar - Levine, P., Katzin, E. M. & Burnham, L. Isoimmunization in pregnancy: its possible bearing on the etiology of erythroblastosis foetalis. J. Am. Med. Assoc. 116, 825– 827 (1941).
Article Google Scholar - Deeb, S. S. The molecular basis of variation in human color vision. Clin. Genet. 67, 369– 377 (2005).
Article CAS Google Scholar - Wagner, F. F. & Flegel, W. A. The molecular basis of the Rh blood group phenotypes. Immunohematol. 20, 23– 36 (2004).
CAS Google Scholar - Fucharoen, S. & Winichagoon, P. Thalassemia and abnormal hemoglobin. Int. J. Hematol. 76 (Suppl. 2), 83– 89 (2002).
Article Google Scholar - Lupski, J. R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417– 422 (1998).
Article CAS Google Scholar - Stankiewicz, P. & Lupski, J. R. Genomic architecture, rearrangements and genomic disorders. Trends Genet. 18, 74– 82 (2002).
Article CAS Google Scholar - Lupski, J. R. & Stankiewicz, P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 1, e49 (2005).
Article Google Scholar - Duncan, I. W. Transvection effects in Drosophila. Annu. Rev. Genet. 36, 521– 556 (2002).
Article CAS Google Scholar - Jakobsson, J. et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J. Clin. Endocrinol. Metab. 91, 687– 693 (2006).
Article CAS Google Scholar - Park, J. et al. Deletion polymorphism of UDP-glucuronosyltransferase 2B17 and risk of prostate cancer in African American and Caucasian men. Cancer Epidemiol. Biomarkers Prev. 15, 1473– 1478 (2006).
Article CAS Google Scholar - Gonzalez, E. et al. The Influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307, 1434– 1440 (2005).
Article ADS CAS Google Scholar - Fellermann, K. et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 79, 439– 448 (2006).
Article CAS Google Scholar - Aitman, T. J. et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439, 851– 855 (2006).
Article ADS CAS Google Scholar - Buckland, P. R. Polymorphically duplicated genes: their relevance to phenotypic variation in humans. Ann. Med. 35, 308– 315 (2003).
Article CAS Google Scholar - Lupski, J. R. et al. DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell 66, 219– 232 (1991).
Article CAS Google Scholar - Locke, D. P. et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275– 290 (2006).
Article CAS Google Scholar - Wirtenberger, M., Hemminki, K. & Burwinkel, B. Identification of frequent chromosome copy-number polymorphisms by use of high-resolution single-nucleotide-polymorphism arrays. Am. J. Hum. Genet. 78, 520– 522 (2006).
Article CAS Google Scholar - Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78– 88 (2005).
Article CAS Google Scholar - Rozen, S. et al. Abundant gene conversion between arms of massive palindromes in human and ape Y chromosomes. Nature 423, 873– 876 (2003).
Article ADS CAS Google Scholar - Repping, S. et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nature Genet. 38, 463– 467 (2006).
Article CAS Google Scholar - Schmutz, J. et al. The DNA sequence and comparative analysis of human chromosome 5. Nature 431, 268– 274 (2004).
Article ADS CAS Google Scholar - Eichler, E. E. Widening the spectrum of human genetic variation. Nature Genet. 38, 9– 11 (2006).
Article CAS Google Scholar - Bentley, D. R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16, 545– 552 (2006).
Article CAS Google Scholar - Lackner, C., Cohen, J. C. & Hobbs, H. H. Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2, 933– 940 (1993).
Article CAS Google Scholar - Rao, Y. et al. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol. Pharmacol. 58, 747– 755 (2000).
Article CAS Google Scholar
Acknowledgements
We thank R. Spielman and three anonymous reviewers for helpful comments.
Author information
Authors and Affiliations
- Department of Genome Sciences, University of Washington, Seattle, 98195, Washington, USA
Evan E. Eichler, Deborah A. Nickerson & Robert H. Waterston - Department of Howard Hughes Medical Institute, University of Washington, Seattle, 98195, Washington, USA
Evan E. Eichler - Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, 02142, Massachusetts, USA
David Altshuler & Charles Lee - Department of Genetics, Washington University School of Medicine, St. Louis, 63110, Missouri, USA
Anne M. Bowcock - National Human Genome Research Institute, National Institutes of Health, Bethesda, 20892, Maryland, USA
Lisa D. Brooks, Adam Felsenfeld & Mark Guyer - Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, CB4 5RW, UK
Nigel P. Carter - National Center for Biotechnology Information, National Library of Medicine, Bethesda, 20894, Maryland, USA
Deanna M. Church & Stephen T. Sherry - Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, 02115, Massachusetts, USA
Charles Lee - Department of Molecular and Human Genetics, Department of Pediatrics, and Texas Children's Hospital, Baylor College of Medicine, Houston, 77030, Texas, USA
James R. Lupski - Genome Technology Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, 20892, Maryland, USA
James C. Mullikin - Department of Human Genetics, University of Chicago, Chicago, 60637, Illinois, USA
Jonathan K. Pritchard - Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 11724, USA
Jonathan Sebat - Agencourt Bioscience Corporation, Beverly, 01915, Massachusetts, USA
Douglas Smith - Johns Hopkins University School of Medicine, Baltimore, 21025, Maryland, USA
David Valle
Consortia
The Human Genome Structural Variation Working Group
- Evan E. Eichler
- , Deborah A. Nickerson
- , David Altshuler
- , Anne M. Bowcock
- , Lisa D. Brooks
- , Nigel P. Carter
- , Deanna M. Church
- , Adam Felsenfeld
- , Mark Guyer
- , Charles Lee
- , James R. Lupski
- , James C. Mullikin
- , Jonathan K. Pritchard
- , Jonathan Sebat
- , Stephen T. Sherry
- , Douglas Smith
- , David Valle
- & Robert H. Waterston
Ethics declarations
Competing interests
The author declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to E.E.E. (eee@gs.washington.edu).
Author Contributions E.E.E., D.A.N., D.A., A.F., J.R.L. and S.T.S. wrote the manuscript. A.M.B, L.D.B., N.P.C., D.M.C., M.G., C.L., J.C.M., J.K.P., J.S., D.S., D.V. and R.H.W. contributed to the plan design and provided comments and suggestions during preparation of the manuscript.
Rights and permissions
About this article
Cite this article
The Human Genome Structural Variation Working Group. Completing the map of human genetic variation.Nature 447, 161–165 (2007). https://doi.org/10.1038/447161a
- Published: 09 May 2007
- Issue Date: 10 May 2007
- DOI: https://doi.org/10.1038/447161a