Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe (original) (raw)
- Short Report
- Published: 21 January 2009
- Valerio Onofri2,
- Francesca Brisighelli3,4,
- Ilaria Boschi4,
- Francesca Scarnicci4,
- Mara Masullo4,
- Gianmarco Ferri5,
- Sergio Tofanelli6,
- Adriano Tagliabracci2,
- Leonor Gusmao7,
- Antonio Amorim7,8,
- Francesco Gatto9,
- Mirna Kirin10,
- Davide Merlitti11,
- Maria Brion3,
- Alejandro Blanco Verea3,
- Valentino Romano12,
- Francesco Cali13 &
- …
- Vincenzo Pascali4
European Journal of Human Genetics volume 17, pages 848–852 (2009)Cite this article
- 6877 Accesses
- 36 Citations
- 67 Altmetric
- Metrics details
Abstract
To investigate the male genetic legacy of the Arab rule in southern Europe during medieval times, we focused on specific Northwest African haplogroups and identified evolutionary close STR-defined haplotypes in Iberia, Sicily and the Italian peninsula. Our results point to a higher recent Northwest African contribution in Iberia and Sicily in agreement with historical data. southern Italian regions known to have experienced long-term Arab presence also show an enrichment of Northwest African types. The forensic and genomic implications of these findings are discussed.
Similar content being viewed by others
Introduction
After the collapse of the Roman Empire in Europe, the Arab dominance across the Mediterranean was one of the most impressive historical events that occurred in this region. Arabs appeared on the southern shores of the Mediterranean in the early seventh century and quickly conquered North Africa. They spread their language and religion to the native Northwest (NW) African Berber populations, which represented the bulk of the Muslim army that later conquered southern Europe.1, 2 Referred to either as Moors (in Iberia) or Saracens (in South Italy and Sicily), their arrival in Europe dates to 711 AD, rapidly subduing most of Iberia and Sicily (831 AD). Among European kingdoms their presence was seen as a constant danger, and only by the fifteenth century was the Iberian reconquest completed.3 In the thirteenth century Frederick II destroyed Arab rule in Sicily and between 1221 and 1226 he moved all the Arabs of Sicily to the city of Lucera, north of Apulia.3 Lucera was later destroyed by Charles II (1301) but an Arab community was recorded in Apulia in 1336.3 Guerrilla warfare was still conducted by Arabs in Sicily even after Frederick II's actions.3
So far, Y chromosome studies attempting to estimate the medieval North African (MNA) contribution to southern Europe have focused almost exclusively on the North African haplogroup E3b1b1b-M81, and have only partially taken into consideration the evolutionary relationships among haplotypes.4, 5, 6, 7 To generate a more comprehensive view of the genetic legacy of the MNA dominance in Europe, we systematically screened for Y chromosome haplotypes within three NW African specific haplogroups, across multiple southern European populations, and performed additional genotyping to refine the available genetic data. Our results confirm a general correlation between historical and genetic data: Iberia and Sicily are the regions with the highest MNA male legacy.
Materials and methods
Identification of recently introgressed NW African haplotypes
Given the historical indication of a prevalently Berber origin for the Arab groups invading southern Europe,2, 3 we focused on NW African specific haplogroups as markers of MNA contribution to this region. Haplogroups E1b1b1b (M81 derived), E1b1b1a-β (M78 derived chromosomes showing the rare DYS439 allele 10) and a subset of J1 (M267 derived) were identified in the literature as being NW Africa specific, together accounting for between 58 and 90% of males in populations from this area, but never above 13% in Europe.8, 9, 10, 11 We note that the other lineages present in these populations would also have been brought over to Europe, and any account of the total MNA contribution to present day Europe should take these into consideration.
Given a number of investigated loci n, and a mutation rate μ (estimated using locus specific data as in reference12), it is possible to obtain the posterior distribution of the Time to the Most Recent Common Ancestor for any pair of haplotypes differing at k loci, using the approach implemented in reference.13 The selected method is based on the infinite alleles model, a reasonable approximation when few mutations are expected to occur, as in the temporal framework evaluated here. So, considering 9 loci and 40 generations (approximately 1200 years ago with a 31-year generation length14), either 0 or 1 mutational difference is the most likely consequence. Two mutations are only slightly less likely, but overlap with other much more ancient events, for example 80 generations or 2400 years ago. Posterior distributions for more ancient events have probability peaks centred on a higher number of differences, with 0–1 mutations being extremely unlikely (data not shown). Therefore, following this, European Y chromosomes within the three haplogroups identical to, or with one mutational difference from, NW African STR haplotypes were considered compatible with an MNA ancestry. In Iberia and peninsular Italy, they account for 90, 78 and 42% of the E1b1b1b, E1b1b1a-β and J1 chromosomes respectively.
Samples
A NW African database was constructed for haplotype comparisons including more than 400 samples genotyped at nine STR loci (DYS19, DYS389 I–II, DYS390, DYS391, DYS392, DSY393, and the bi-allelic DYS385). The database included 127 Berbers from Tunisia;15, 16 102 South Tunisians;17 109 Moroccan Arab and Berber speakers;18 50 Moroccan and 52 Tunisians (unpublished data). NW African specific haplogroups were identified by further genotyping of samples that were previously described elsewhere.5, 6, 7, 19, 20, 21 We also included a Basque dataset22, 23 and two novel Italian samples (Lucera and Veneto; Table 1). Within these populations, all E1b1b1a chromosomes were scored for the DYS439 locus to identify the E1b1b1a-β cluster9 and the M267 marker was investigated in those chromosomes previously identified as J*(xJ2). Alternatively, the DYS458 .2 allele was used to identify the J1 types within J*(xJ2) chromosomes.24 All the individuals within E1b1b1b, E1b1b1a-β and J1 were also genotyped for the same nine STRs as the NW Africans (DYS19, DYS389 I–II, DYS390, DYS391, DYS392, DYS393 and DYS385). The DYS385 bilocal locus was considered as two different loci, the smaller allele assigned to locus DYS385a and the larger to DYS385b. A previous investigation25 showed that misassignment would influence only a minimal fraction of the haplotypes and so this can be assumed to have a negligible effect on our estimates. A Sicilian population was also included (samples overlapping in references26, 27). Sicilian genotypes were screened for E1b1b1* and J*(xJ2) lineages, and did not include DYS439. Within the E1b1b1* and J*(xJ2) haplogroups, 8 and 3 chromosomes, respectively, were found close to NW African types. These samples were then made available for further genotyping, to include DYS439, M78, M81 and M267. We note that because of partial sampling across NW Africa, a subset of the European chromosomes with true MNA ancestry could potentially fail to be identified. However, given the general homogeneity observed across NW Africa, the number of populations included, and the large dataset used, we believe that this is unlikely to influence our results.
Table 1 Historically introduced NW African types in Italy and Iberia
Results and discussion
To address the degree of historical NW African contribution, we used a combined SNP-STR approach. The coalescent times for the three NW African specific haplogroups ranges between 5000 and 24 000 years, spanning a number of historical scenarios each potentially explaining their presence on the Northern Mediterranean shores.9, 10 It follows that estimating MNA genetic legacy on the basis of haplogroups' occurrence only would be misleading. To avoid this limitation, we have extended our analysis to include STR data whose high mutation rate allows one to focus on more recent events. We screened more than 2300 South European samples (Figure 1; Table 1) to identify those haplotypes which are evolutionary close to NW African chromosomes. Total frequencies for these chromosomes range between 0 and 19% across southern Europe, the highest being in Cantabria and comprising a sample from the Pas Valley, previously shown to have an extremely high frequency of the North African haplogroup E1b1b1b.9 Our estimates of NW African chromosome frequencies were highest in Iberia and Sicily, in accordance with the long-term Arab rule in these two areas.3 The chromosome frequencies in the two samples were not significantly different from each other (Fisher's exact test _P_=0.83) but were both significantly different from the peninsular Italy sample (P<0.01). An inspection of Table 1 reveals a non-random distribution of MNA types in the Italian peninsula, with at least a twofold increase over the Italian average estimate in three geographically close samples across the southern Apennine mountains (East Campania, Northwest Apulia, Lucera). When pooled together, these three Italian samples displayed a local frequency of 4.7%, significantly different from the North and the rest of South Italy (P<0.01), but not from Iberia and Sicily (_P_=0.12 and _P_=0.33, respectively). Arab presence is historically recorded in these areas following Frederick II's relocation of Sicilian Arabs.3 In Iberia, a non-random distribution might also potentially be present, as suggested by our lower estimates in the northeast (Basque region and Catalans), but more samples across the peninsula will be required to properly address this issue. Assuming that a large population in regions such as Iberia, Sicily and Italy was present in the past, the ratio between Y chromosomes with a MNA ancestry and other types will have stayed approximately constant across time. Smaller areas, however, would have been influenced by drift, in the Pas Valley for example. Consistent with historical data,3 no population in Central Europe or the Balkans shows the presence of recently introgressed NW African types9, 10, 28 besides a few chromosomes in Albania and Romania.29
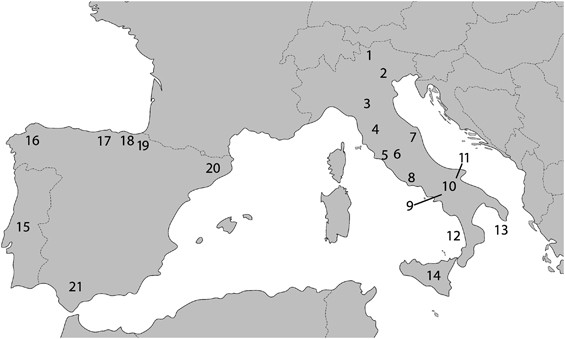
Figure 1
Geographical location of the investigated southern European samples. Numbers are same as in Table 1.
The increasing use of highly structured distributions of Y chromosome types to investigate the ethnic/geographic origin of unknown samples30 gives the identification of regions in Italy enriched with recently introgressed NW African types forensic relevance. We found that more than 56% of the Italian individuals identified here as having a recent NW African do not have a match in a large Italian Y chromosome dataset comprising almost 1200 individuals.31 Of these, 31% instead perfectly overlap with types from NW African populations, potentially providing misleading advice to investigators. Such results are also of interest in the light of the expanding business of genealogical services offering Y chromosome analysis to identify an individual's ethnic ancestry. Our results clearly confirm that conclusions based on single chromosomes should be taken very cautiously.32 What are the expected genomic consequences of this historically recent admixture event? Suppose that 40 generations ago there was a 5% male introgression of African DNA into the European gene pool, corresponding to a total contribution of 2.5% of genetic material. Immediately after the admixture event, a fraction of chromosomes within Europe would have African ancestry. Recombination since this event will have substantially reduced the size of the fragments of African ancestry within European haplotypes, and with these parameters we would today expect to see an approximately exponential distribution (measuring size using genetic distance) of fragment sizes, with a mean value of roughly 2.6 cM. Assuming a genome-wide average recombination rate of 1.3 cM/Mb,33 2.5% of a typical present day southern European genome would consist on average of 2 Mb regions of African DNA. We therefore believe that signatures of this event would be correctly identified using modern dense genotype data.34 By using northern Italian and Mozabite samples recently genotyped for a large SNP autosomal dataset35 as the best available proxy of Italian and northern African populations, we estimated that about 41.5% of more than 640 000 genotyped SNPs showed an absolute allele frequency difference of at least 10% between the two groups. Such frequency differences (and sometimes even smaller) between cases and controls characterized the vast majority of the inferred disease-causing SNPs in a recent genome-wide investigation.36 In general then, it is critical to take population structure into account so as to avoid false positives in case–control association studies.37 Thus, an understanding of similar historical admixture events is likely to aid researchers conducting such studies.
References
- Davies RHC : A History of Medieval Europe. London, UK: Longmann Group Limited, 1988, pp 83–101.
Google Scholar - Hitti P : The Arabs: A Short History. Washington DC: Gateway, 1990.
Google Scholar - Norman D : The Arabs and Medieval Europe. London, UK: Longmann Group Limited, 1975.
Google Scholar - Rosser ZH, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Article CAS PubMed PubMed Central Google Scholar - Bosch E, Calafell F, Comas D et al: High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 2001; 68: 1019–1029.
Article CAS PubMed PubMed Central Google Scholar - Brion M, Quintans B, Zarrabeitia M et al: Micro-geographical differentiation in Northern Iberia revealed by Y-chromosomal DNA analysis. Gene 2004; 329: 17–25.
Article CAS PubMed Google Scholar - Beleza S, Gusmão L, Lopes A et al: Micro-phylogeographic and demographic history of Portuguese male lineages. Ann Hum Genet 2006; 70: 181–194.
Article CAS PubMed Google Scholar - Arredi B, Poloni ES, Paracchini S et al: A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet 2004; 75: 338–345.
Article CAS PubMed PubMed Central Google Scholar - Cruciani F, La Fratta R, Santolamazza P et al: Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 2004; 74: 1014–1022.
Article CAS PubMed PubMed Central Google Scholar - Semino O, Magri C, Benuzzi G et al: Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
Article CAS PubMed PubMed Central Google Scholar - History and geography of human Y-chromosome in Europe: a SNP perspective Paolo Francalacci & Daria Sanna. Journal of Anthropological Sciences (J Anthropol Sci) 2008; 86: 59–89.
- Walsh B : Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics 2001; 158: 897–912.
CAS PubMed PubMed Central Google Scholar - Gusmão L, Sánchez-Diz P, Calafell F et al: Mutation rates at Y chromosome specific microsatellites. Hum Mutat 2005; 26: 520–528.
Article PubMed Google Scholar - Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, Stefansson K : A population wide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet 2003; 72: 1370–1389.
Article CAS PubMed PubMed Central Google Scholar - Frigi S, Pereira F, Pereira L et al: Data for Y-chromosome haplotypes defined by 17 STRs (AmpFLSTR® Yfiler™) in two Tunisian Berber communities. Forensic Sci Int 2006; 160: 80–83.
Article CAS PubMed Google Scholar - Cherni L, Pereira L, Goios A et al: Y-chromosomal STR haplotypes in three ethnic groups and one cosmopolitan population from Tunisia. Forensic Sci Int 2005; 152: 95–99.
Article CAS PubMed Google Scholar - Ayadi I, Ammar-Keskes L, Rebai A : Haplotypes for 13 Y-chromosomal STR loci in South Tunisian population (Sfax region). Forensic Sci Int 2006; 164: 249–253.
Article CAS PubMed Google Scholar - Quintana-Murci L, Bigham A, Rouba H et al: Y-chromosomal STR haplotypes in Berber and Arabic-speaking populations from Morocco. Forensic Sci Int 2004; 140: 113–115.
Article CAS PubMed Google Scholar - Onofri V, Alessandrini F, Turchi C et al: Y-chromosome genetic structure in sub-Apennine populations of Central Italy by SNP and STR analysis. Int J Legal Med 2007; 121: 234–237.
Article PubMed Google Scholar - Capelli C, Brisighelli F, Scarnicci F et al: Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter. Mol Phylogenet Evol 2007; 44: 228–239.
Article PubMed Google Scholar - Ferri G, Alù M, Corradini B et al: Slow and fast evolving markers typing in Modena males (North Italy). Forensic Sci Int Genet, (in press).
- García O, Martín P, Gusmão L et al: A Basque Country autochthonous population study of 11 Y-chromosome STR loci. Forensic Sci Int 2004; 145: 65–68.
Article PubMed Google Scholar - Alonso S, Flores C, Cabrera V et al: The place of the Basques in the European Y-chromosome diversity landscape. Eur J Hum Genet 2005; 13: 1293–1302.
Article CAS PubMed Google Scholar - Myres NM, Ekins JE, Lin AA et al: Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405. Croat Med J 2007; 48: 450–459.
CAS PubMed PubMed Central Google Scholar - Niederstätter H, Berger B, Oberacher H, Brandstätter A, Huber CG, Parson W : Separate analysis of DYS385a and b versus conventional DYS385 typing: is there forensic relevance? Int J Legal Med 2005; 119: 1–9.
Article PubMed Google Scholar - Capelli C, Redhead N, Romano V et al: Population structure in the Mediterranean basin: a Y chromosome perspective. Ann Hum Genet 2006; 70: 207–225.
Article CAS PubMed Google Scholar - Robino C, Inturri S, Gino S et al: Y-chromosomal STR haplotypes in Sicily. Forensic Sci Int 2006; 159: 235–240.
Article CAS PubMed Google Scholar - Berger B, Lindinger A, Niederstätter H et al: Y-STR typing of an Austrian population sample using a 17-loci multiplex PCR assay. Int J Legal Med 2005; 119: 241–246. Erratum in: Int J Legal Med 2006; 120: 255.
Article PubMed Google Scholar - Bosch E, Calafell F, González-Neira A et al: Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet 2006; 70: 459–487.
Article CAS PubMed Google Scholar - Wetton JH, Tsang KW, Khan H : Inferring the population of origin of DNA evidence within the UK by allele-specific hybridization of Y-SNPs. Forensic Sci Int 2005; 152: 45–53.
Article CAS PubMed Google Scholar - Presciuttini S, Caglià A, Alù M et al: Y-chromosome haplotypes in Italy: the GEFI collaborative database. Forensic Sci Int 2001; 122: 184–188.
Article CAS PubMed Google Scholar - King TE, Parkin EJ, Swinfield G et al: Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur J Hum Genet 2007; 15: 288–293.
Article CAS PubMed Google Scholar - Yu A, Zhao C, Fan Y et al: Comparison of human genetic and sequence-based physical maps. Nature 2001; 409: 951–953.
Article CAS PubMed Google Scholar - Frazer KA, Ballinger DG, Cox DR et al: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Article CAS PubMed Google Scholar - Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Article CAS PubMed Google Scholar - Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Article Google Scholar - Marchini J, Cardon LR, Phillips MS et al: The effects of human population structure on large genetic association studies. Nat Genet 2004; 36: 512–517.
Article CAS PubMed Google Scholar
Acknowledgements
We thank Elena Bosch and Walther Parson for kindly providing unpublished data; Giovanni Destro-Bisol for commenting a preliminary version of the article; Dr Trincucci for support in the sampling of the Lucera inhabitants; Marcello Menegatti, Cristian Sossai and the Associazione Culturale ‘Borghi dell'Ovest’ for the Veneto samples. CC thanks Simon Myers and Garrett Hellenthal for comments and suggestions on the genomic structure implication following recent admixture events, Jim Wilson for support and Prof Francesco Sabatini for discussion on the history of Lucera. CC is a RCUK Academic Fellow.
Author information
Authors and Affiliations
- Department of Zoology, University of Oxford, Oxford, UK
Cristian Capelli - Institute of Legal Medicine, Universita' Politecnica delle Marche, Policlinico Torrette, Ancona, Italy
Valerio Onofri & Adriano Tagliabracci - Medicine Genomic Group, Hospital-University complex of Santiago (CHUS), University of Santiago de Compostela, Spain
Francesca Brisighelli, Maria Brion & Alejandro Blanco Verea - Instituto di Medicina Legale, Universita' Cattolica del S. Cuore, Rome, Italy
Francesca Brisighelli, Ilaria Boschi, Francesca Scarnicci, Mara Masullo & Vincenzo Pascali - Department of Diagnostic and Laboratory Service and Legal Medicine, Section of Legal Medicine, University of Modena and Reggio Emilia, Italy
Gianmarco Ferri - Department of Biology, Anthropology Unit, University of Pisa, Italy
Sergio Tofanelli - IPATIMUP – Institute of Molecular Pathology and Immunology of the University of Porto, Portugal
Leonor Gusmao & Antonio Amorim - Faculty of Sciences, University of Porto, Portugal
Antonio Amorim - Biotechnology Unit, Istituto Zooprofilattico Sperimentale Lazio e Toscana, Rome, Italy
Francesco Gatto - Public Health Sciences, University of Edinburgh, Edinburgh, Scotland
Mirna Kirin - Scuola Normale Superiore di Pisa, Pisa, Italy
Davide Merlitti - Dipartimento di Oncologia Sperimentale e Applicazioni Cliniche Università di Palermo, Italy
Valentino Romano - Oasi Institute for Research on mental Retardation and Brain Aging (IRCCS), Troina, Italy
Francesco Cali
Authors
- Cristian Capelli
You can also search for this author inPubMed Google Scholar - Valerio Onofri
You can also search for this author inPubMed Google Scholar - Francesca Brisighelli
You can also search for this author inPubMed Google Scholar - Ilaria Boschi
You can also search for this author inPubMed Google Scholar - Francesca Scarnicci
You can also search for this author inPubMed Google Scholar - Mara Masullo
You can also search for this author inPubMed Google Scholar - Gianmarco Ferri
You can also search for this author inPubMed Google Scholar - Sergio Tofanelli
You can also search for this author inPubMed Google Scholar - Adriano Tagliabracci
You can also search for this author inPubMed Google Scholar - Leonor Gusmao
You can also search for this author inPubMed Google Scholar - Antonio Amorim
You can also search for this author inPubMed Google Scholar - Francesco Gatto
You can also search for this author inPubMed Google Scholar - Mirna Kirin
You can also search for this author inPubMed Google Scholar - Davide Merlitti
You can also search for this author inPubMed Google Scholar - Maria Brion
You can also search for this author inPubMed Google Scholar - Alejandro Blanco Verea
You can also search for this author inPubMed Google Scholar - Valentino Romano
You can also search for this author inPubMed Google Scholar - Francesco Cali
You can also search for this author inPubMed Google Scholar - Vincenzo Pascali
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toCristian Capelli.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Capelli, C., Onofri, V., Brisighelli, F. et al. Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe.Eur J Hum Genet 17, 848–852 (2009). https://doi.org/10.1038/ejhg.2008.258
- Received: 27 May 2008
- Revised: 27 November 2008
- Accepted: 02 December 2008
- Published: 21 January 2009
- Issue Date: June 2009
- DOI: https://doi.org/10.1038/ejhg.2008.258