Molecular architecture of native HIV-1 gp120 trimers (original) (raw)
Main
It is estimated that over 33 million individuals are at present infected with HIV (http://www.unaids.org). The development of an effective vaccine is therefore a challenge of fundamental medical interest. It has been widely recognized that a better understanding of the structure of trimeric Env in its various conformational states is likely to be an important element in the overall strategy for vaccine development4. Although X-ray crystallographic methods have led to atomic models for HIV-1 gp120 monomers complexed to antibodies in the presence and absence of CD4 (refs 2, 3, 5), determination of the structures of intact trimers on native viruses has nevertheless remained elusive. Theoretical models for the structure of the trimer that take into account constraints determined from biochemical and mutagenesis studies of monomeric gp120 (refs 6, 7) have been advanced, but the advent of electron tomographic methods8 provides a unique opportunity for direct experimental determination of the structure of the intact trimer on the virus in its native state. Here we report structural analysis of native HIV-1 Env using alignment and classification procedures that take into account the missing wedge that arises from the limited angular range used for data collection in electron tomography. Our approach takes advantage of complexes containing monomeric gp120 for which there are known X-ray structures, allowing us to derive models for trimeric gp120 in unliganded and CD4-bound states.
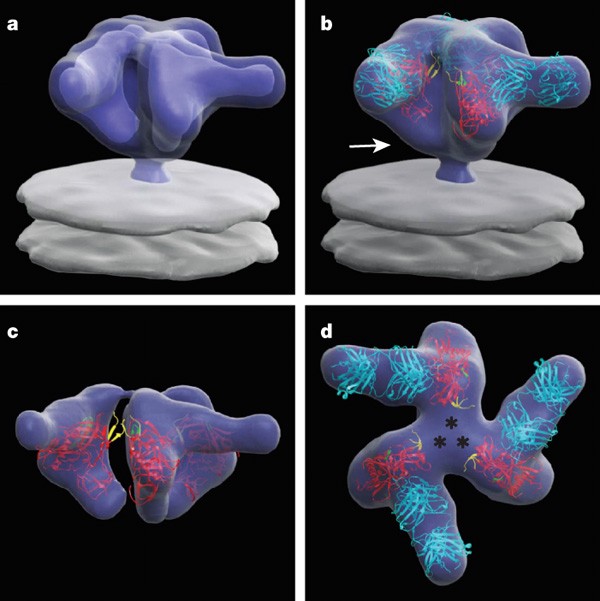
We first analysed tomograms obtained with viruses complexed with Fab fragments from the potent, broadly cross-reactive, neutralizing antibody b12, as an atomic model of the complex of a disulphide-bond stabilized version of monomeric gp120 core with the Fab fragment of b12 is available3 (see Supplementary Methods and Supplementary Figs 1–3 for a detailed description of methods). The contributions of the Fab fragment to the experimentally derived density map can be easily spotted (Fig. 1). The X-ray coordinates for the gp120–Fab complex were docked as a rigid body into the map using automated fitting procedures (Supplementary Fig. 4), resulting in a description of the molecular structure of the b12-complexed trimer. The X-ray structure of monomeric gp120 in complex with b12-Fab includes only ∼58% of the gp120 polypeptide sequence, and lacks most of the residues in the V1/V2 loops (residues 121–203), V3 loop (residues 300–328) and portions of the amino and carboxy termini (residues 1–82 and 493–511, respectively). Inspection of the extra densities in the density map that are not occupied by the coordinates reveal the likely locations of these regions, as well as the probable location of gp41 in the native trimer (Fig. 1). In particular, the unassigned densities adjacent to the V1/V2 stem have a size consistent with that expected from the ∼80 residues missing in the V1/V2 loop, implying that the three V1/V2 loop regions come together to form the apex of the mushroom-shaped Env trimer.
Figure 1: Averaged 3D structure of the HIV-1 spike in complex with b12-Fab.
a, Perspective view of the surface of the density map shown at two thresholds; one to include the entire spike (outer), and another to highlight the Fab and gp120 components (inner). b–d, Front (b, c) and top (d) views of the map fitted with X-ray coordinates of the complex of the Fab fragment of b12 (cyan) with gp120 (red, PDB ID, 2NY7); only gp120 coordinates are shown in c, which is at the inner threshold. Likely locations of the V1/V2 loop and gp41 regions are indicated by asterisks in d and the white arrow in b, respectively. The stumps of the V1/V2 and V3 loop regions are shown in yellow and green, respectively.
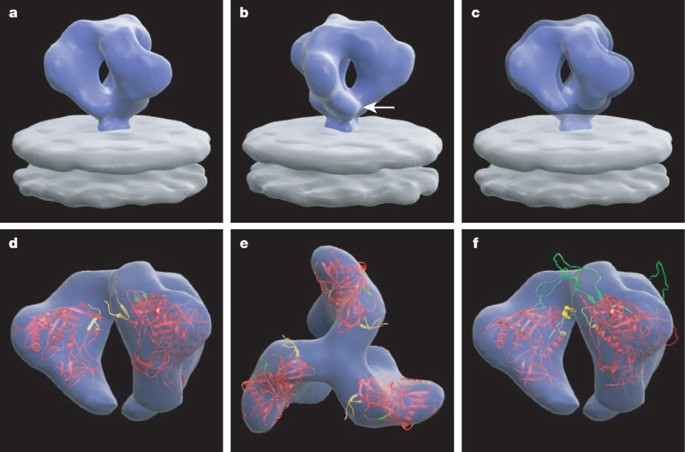
Analysis of the native, undecorated Env trimer (Fig. 2a–c) shows the general shape and arrangement of gp120 monomers in the native spike, which are comparable to those obtained for the Env–b12 complex (also see Supplementary Video). Overall, the spike has a height of ∼120 Å, and a maximal width of ∼150 Å, which tapers from ∼80 Å at the base of the gp120 regions to ∼35 Å at the junction with the membrane. The best fit of gp120 into the density map is shown in Fig. 2d, e with the conformation of gp120 derived in complex with b12 (identical to gp120 coordinates shown in Fig. 1), and from the conformation obtained in a complex with the X5 antibody5 (Fig. 2f; see Supplementary Fig. 4 for a detailed description of the fitting procedures used). The latter complex includes the V3 loop region (in the CD4-bound conformation, which is likely to be very different from the conformation in the unliganded state), but the gp120 constructs used to obtain both crystal structures lack significant portions of the V1/V2 loop and the N and C termini of gp120. The regions of the V1/V2 stem that are included in the structure display high temperature factors indicating potential flexibility2, and also may not reflect their actual positions in the intact loop region because of the truncation. Nevertheless, inspection of the fits confirms and extends the general conclusion drawn from Fig. 1—that the V1/V2 and V3 regions on each monomer are near the apex of the trimer, and that the three gp41 components form a mushroom-shaped structure at the base of the gp120 trimer. The residues likely to be glycosylated, as well as the variable loops (V1–V4), are all generally located in solvent accessible regions, as suggested in a previously proposed theoretical model for trimer architecture6.
Figure 2: Averaged 3D structure of the trimeric glycoprotein spike on native HIV-1.
a, b, Perspective/front views of the surface of the density map; the white arrow in b points to the likely location of gp41 in the map. c, Same view as in a but shown using two thresholds to illustrate both the overall shape (outer), and the contribution of the gp120 containing regions (inner). d, e, Front and top views of the map with coordinates for the gp120 core derived from the complex with b12 (PDB ID, 2NY7). f, Front view of the map fitted with coordinates for the gp120 core derived from the complex with X5 (PDB ID, 2B4C). The gp120 core is shown in red, and the regions of the V1/V2 loop and V3 loop included in the coordinates are shown in yellow and green, respectively.
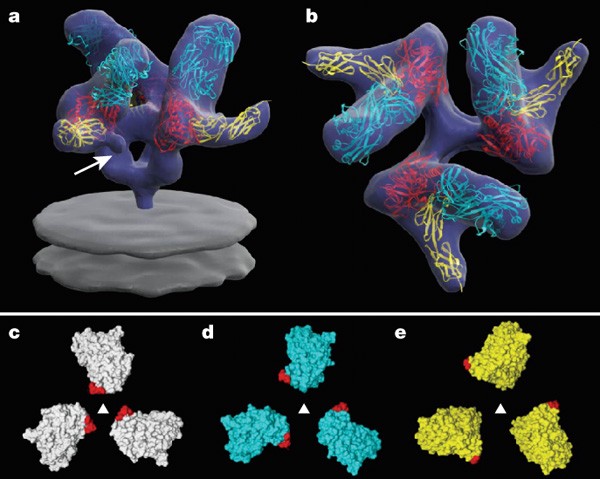
Understanding the nature of conformational changes in the Env trimer induced by CD4 binding9 is at the heart of defining the molecular mechanisms underlying HIV entry into cells. We therefore carried out electron tomographic analysis of HIV-1 complexed to CD4 and the Fab fragment of the 17b antibody, both because the 17b antibody has been shown10 to stabilize and lock gp120 in the CD4-bound conformation, and because the crystallographic structure of the ternary complex of gp120 with CD4 and the 17b Fab fragment has been determined2. Inspection of the averaged structure of the Env trimer bound to CD4 and 17b (Fig. 3a, b) shows a dramatic conformational change in comparison with that of the unliganded trimer (Fig. 2). As in the case of the previous maps, the constraints provided by the presence of CD4 and 17b ensure that there is a single, unambiguous fit of the coordinates into the density map (see Supplementary Fig. 4), and provide the basis to understand the conformational change induced by CD4 binding. There is clear additional density in the averaged map for the V1/V2 loop (∼70 residues) that is missing in the crystal structure of the complex (Fig. 3a). Relative to gp120 in the unliganded trimer, each gp120 monomer in the CD4 complex displays a rotation of about 45° around an axis parallel to the central three-fold axis, an out of plane rotation of about 15°, and an upward displacement of the overall centre of mass by ∼15 Å. There are also discernible changes in the gp41 region adjacent to the viral membrane, and a new feature is observed at the centre of the spike that is not present in the unliganded spike or in the complex with b12. The most likely interpretation of this feature is that it arises from rearrangements of gp41 that eventually lead to formation of the six-helix bundle structure, and towards fusion between viral and target cell membranes11.
Figure 3: 3D structure of the HIV-1 spike in complex with CD4 and 17b-Fab.
a, b, Front and top views showing the X-ray coordinates of the ternary complex (PDB ID, 1GC1) of the gp120 core (red) with CD4 (yellow) and the Fab fragment of 17b (cyan) fitted into the map as a rigid body using automated fitting procedures27. The arrow in a points to the likely location of the V1/V2 loop region, which was partially deleted in the construct used to obtain crystals of the ternary complex2. c–e, Top views showing superposition of the X-ray coordinates for the gp120 trimer derived from the maps for the unliganded (white), b12-bound (cyan) and CD4/17b-bound (yellow) states of the trimeric spike, with locations of the V1/V2 stem regions indicated in red.
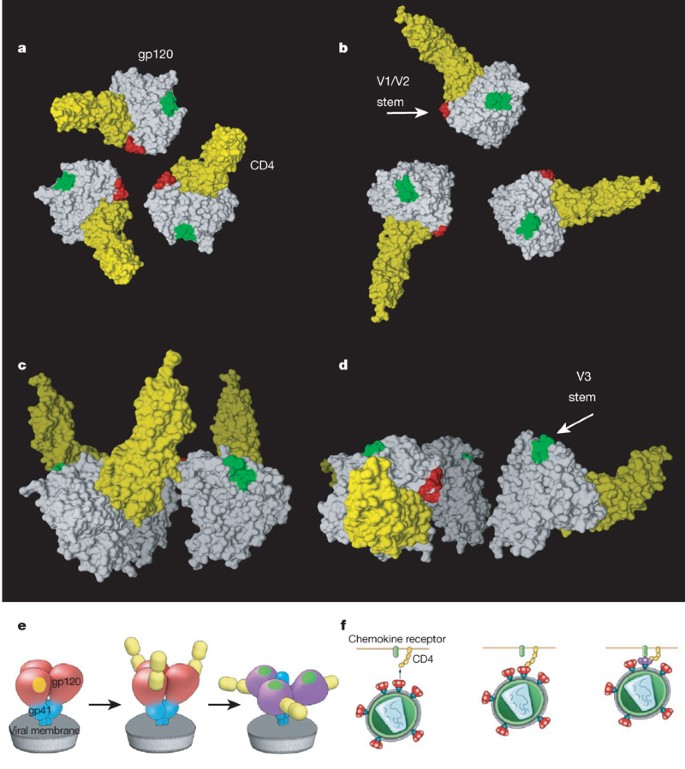
Comparison of the locations of the docked gp120 monomers in the free, b12-bound and CD4-bound states (Fig. 3c–e) provides insights into the overall quaternary structural changes that occur in the trimeric spike. The binding of b12 results in a partial opening of the spike, coupled with rotation of each monomer by ∼20°–25° around an axis perpendicular to the viral membrane (Fig. 3d). However, CD4 binding results in a rotation around this central axis in the same direction that is twice as large, in addition to an out-of-plane rotation (Fig. 3e), and slight vertical displacement of gp120. Thus, while the binding sites for CD4 and b12 are on roughly the same face of the gp120 monomer, they result in very different outcomes for the conformation of the Env trimer.
The observed fit of the gp120 core regions from CD4-liganded gp120 complexes (with X5 as well as 17b antibodies) into the corresponding regions in the density map of the native trimer has important implications for the nature and extent of structural differences in gp120 that occur upon CD4 binding. So far, there are no reports of the atomic structures of either monomeric or trimeric HIV-1 envelope glycoproteins determined using X-ray crystallography. Chen et al.7 reported a structure for a truncated, unliganded SIV gp120 monomer in which the conformation of gp120 is different from that of the HIV-1 gp120 monomer seen in either the CD4-liganded or b12-bound states7. Our findings show that the conformation of the HIV-1 gp120 monomer observed in the b12 and CD4-liganded states can be docked into density maps of the unliganded HIV-1 spike, whereas the conformation reported for the SIV gp120 monomer does not represent a good fit (Supplementary Figs 5 and 6). Further, the trimeric gp120 arrangement we have derived has the V1/V2 loop regions at the apex of the trimer, in contrast to the arrangement suggested by Chen et al.7, in which the V1/V2 loop regions are close to the base of the gp120 trimer. Probable explanations for these differences include the possibility that the three-dimensional (3D) crystals used to determine the structure of the truncated SIV gp120 core captured a conformation of gp120 that is different from the physiologically relevant conformation in the native trimer, or that there are fundamental differences in conformation between monomeric SIV and HIV-1 gp120. We note that the density map we have obtained for the HIV-1 spike has resemblance in the stalk region to the map reported by Zanetti et al.12 for the SIV spike, and the overall features of this map are comparable to a low-resolution version of the map we show in Fig. 2 (see Supplementary Fig. 3 for progressive improvement in map resolution with iterative refinement). However, our results are at variance with the conclusion of Zhu et al.13 that the membrane-proximal region of gp41 is splayed out into three distinct ‘legs’ separated by ∼80 Å from each other at the point of contact with the membrane (Supplementary Fig. 7).
The dramatic ‘opening’ of the trimer induced by CD4 has profound consequences for the disposition of the various key regions of the spike relative to the viral membrane and the target cell (Fig. 4). Previous measurements of the energetics of the CD4 binding have suggested the existence of a large entropic contribution (conformational ‘fixation’ of gp120) that results from CD4 binding10,14. At the present resolution of our maps (∼19, 22 and 23 Å respectively for the maps shown in Figs 1–3, as determined by the 0.5 FSC criterion, see Supplementary Fig. 1), we cannot yet directly determine the nature of conformational changes in the monomer, but our analysis implies that in addition to any such changes in the monomer, there are large, additional contributions from quaternary changes in the structure of the trimer. The lever-like opening of the trimer upon CD4 binding makes way for exposure of the central gp41 stalk. The V3 loop region is released from the lateral edge of the apex of the spike to directly point towards the target cell, while the V1/V2 regions as well as the CD4 binding sites move away from the centre of the spike (Fig. 4b).
Figure 4: Description of the conformational change in the gp120 trimer induced by CD4 binding.
a–d, Model for the conformational change from the unliganded (a, c) to the CD4-bound state (b, d) shown as top (a, b) and front (c, d) views. The gp120 core, CD4, V1/V2 and V3 stems are shown in white, yellow, red and green colours, respectively. e, Schematic description of the gp41 (blue) and gp120 (red/purple) regions of the trimeric spike and the conformational changes that occur upon CD4 binding. The yellow patch near the apex marks the location of the CD4 binding site in the unliganded spike and the green patch at the apex marks the location of the V3 loop region in the spike after CD4 binding. f, Schematic view of the consequence of the CD4-induced conformational changes for viral attachment to the target cell and interaction with chemokine receptors (green at top). Colours in f have same meaning as in e.
The determination of molecular models for trimeric gp120 in the free and CD4-complexed states could represent a useful starting point for the development of rationally designed vaccines to counter the AIDS epidemic that take into account the trimeric structure of the spike. In the native state, the trimer is held together by strong contacts at the gp41 base and at the apex, which appear to have significant contributions from the V1/V2 loop regions, and are adjacent to the V3 loop region and chemokine receptor binding sites15. The locations of these regions in the unliganded spike at the critical interface between virus and the target cell are consistent with observations documenting the altered susceptibility of variants with deletions in the V1 and V2 loop regions to neutralization16,17,18 as well as the identification of antibodies to unique quaternary epitopes on Env19. There appears to be relatively little contact between most other regions of the neighbouring gp120 monomers, resulting in a spike architecture that is held together somewhat tenuously at the top and bottom, poised to be sprung open upon CD4 binding. The CD4 binding site is recessed by about 20 Å from the top of the spike, with the V1/V2 regions and associated carbohydrate moieties forming a sheath at the top. The recessed site implies that cell surface CD4 must delve into the spike to achieve binding. The outward movement of gp120 results in a steep change in the orientation of the two outermost domains (D1D2) of CD4 (Fig. 4e), implying that this motion must draw the virus closer to the target cell membrane by virtue of the flexibility between the D1D2 and D3D4 domains of membrane-anchored CD4 (Fig. 4f). Indeed, recent cryo-electron tomographic studies of the complex of native viral gp120 with D1D2-IgP suggest that the highly potent neutralizing activity of D1D2-IgP probably arises from its flexible, polyvalent nature, which allows its binding to multiple spikes on the same virus and across neighbouring viruses20. A prediction of our model is that the hinge region between the second and third extracellular domains of CD4 is critical for viral entry, potentially explaining observations that antibodies directed against epitopes close to this hinge region block fusion and HIV infection21, and that binding to gp120 induces a dramatic bend in CD4 at this hinge region22.
The nature and extent of the CD4-induced structural change that we have identified here provides a structural foundation to interpret and refine the plethora of biochemical studies on Env and to better understand mechanisms that are important for virus neutralization and entry4,23,24. We propose from our results that CD4 binding draws the spike closer to the target cell membrane as a result of the hinge-like motion of the D1D2 domains resulting from the change in trimer structure. It is clear from this sequence of events that the exposure of the V3 loop and other antigenic determinants important for viral attachment occur in the protected milieu of the interface formed between the viral and target cell membrane, providing a mechanism for seclusion of these epitopes from antibodies whose binding could potentially neutralize HIV-1.
Given the limited accessibility of key antigenic sites on gp120, the observed rearrangement of the gp120 trimer upon binding of the broadly neutralizing antibody b12 is a surprising and potentially important finding. The outward displacement of gp120 in the b12-bound state appears to be along the same general trajectory observed upon CD4 binding (Fig. 3d, e), but appears to lock gp120 and trimeric Env in a state that prevents further conformational changes that could lead to exposure of the V3 loop or rearrangement of gp41. It is possible that the observed outward movement of gp120 is driven by conformational changes at the gp120–b12 interface3 that occur after initial contact with b12 in order to accommodate the steric consequences of the b12 binding in the context of an intact trimer. Knowledge of the trimeric structure of the HIV-1 spike in strains with differing levels of neutralization sensitivity, and at various stages of activation that ultimately culminate in formation of the viral entry claw25, will be important to further understand the range of variation in spike structure. Approaches for intervention that target regions on Env that are critical for the conformational change could provide a new addition to the arsenal of strategies to combat HIV/AIDS.
Methods Summary
Purified viruses in the presence or absence of added reagents (antibodies, CD4) were deposited on home-made holey carbon grids and plunge-frozen in liquid ethane maintained at about -180° to prepare vitrified specimens for cryo-electron tomography. Specimens were imaged in a Polara transmission electron microscope equipped with an energy filter, with the specimen maintained at liquid nitrogen temperatures. Typically, a series of 141 low dose images of each frozen hydrated virus was recorded at 1° tilt intervals in the range of ±70°. The tilt series were aligned and back-projected to reconstruct 3D volumes (tomograms) of individual viruses. Viral spikes protruding from the membrane surface were readily identified in the tomograms (Supplementary Fig. 1), and were extracted for further processing. In total, we extracted 4,741 spikes from 382 virions of HIV-1 strain BaL, 4,323 spikes from 306 virions of HIV-1 BaL complexed with b12-Fab, and 4,900 spikes from 503 virions of HIV-1 BaL complexed with CD4 and 17b-Fab. Alignment, classification and 3D averaging of the extracted subvolumes for determination of the 3D structure were carried out based on the framework described in ref. 26 (see Supplementary Fig. 1a, b for representative examples of a tomographic slice and segmented virus, respectively, and Supplementary Figs 2 and 3 for an illustration of classes and class averages at early and late stages of refinement). No external references were used for alignment and classification, and the presence of the missing wedge of information in each volume was taken into account for 3D alignment. Iterative 3D classification and alignment runs were executed starting with the raw images until no further changes were observed in the final density maps. Fitting of coordinates into the density maps (Supplementary Figs 4–6) was carried out using automated procedures implemented in the software package Chimera27.
Online Methods
Reagents
Samples of HIV-1 strain BaL (estimated concentration ∼1011 virions ml-1), purified by sucrose gradient centrifugation and inactivated by treatment with Aldrithiol-2 (AT-2), were contributed by J. Bess and J. Lifson. AT-2 treated viruses are capable of supporting viral entry at levels comparable to untreated viruses, and have a similar antigenic profile to untreated viruses28. Purified soluble CD4 (sCD4; 1–183 containing fragment) was obtained from the NIH AIDS reagent program, while purified Fab fragments from b12 and 17b antibodies were provided by P. Kwong.
Specimen preparation
Purified viral suspensions were pre-incubated at 4 °C for 15–30 min in buffer alone, or in the presence of (1) the Fab fragment of the b12 antibody, and (2) sCD4 and the Fab fragment of 17b. All ligands were added at a concentration corresponding to an estimated fivefold molar ratio with Env trimers. Samples were then mixed with 5-nm colloidal gold (used as fiducial makers in initial image alignment) and deposited on home-made, carbon-coated holey carbon grids. Excess liquid was blotted with filter paper from both sides of the grid to form a thin layer of buffer which was then rapidly frozen by plunging the grid in a liquid/solid ethane slush (about -180 °C). This procedure, which results in the embedding of the viruses in a ∼150 nm layer of amorphous ice spanning holes in the carbon layer, was carried out using a Vitrobot rapid freezing device (FEI).
Cryo-electron tomography
Frozen virus specimens were imaged at liquid nitrogen temperatures using a Polara field emission gun electron microscope (FEI) equipped with a 2k × 2k CCD placed at the end of GIF 2002 energy filter (Gatan), operated in the zero-energy-loss mode with a slit width of 20 eV. The microscope was operated at 200 kV and a magnification of ×34,000, resulting in an effective pixel size of 4.1 Å. Tilt series were collected in automatic batch mode. Low dose single-axis tilt series were collected from each virus specimen at nominal underfocus settings of -2 µm. Since the defocus was determined in regions on the carbon film, and ∼3–4 µm away from the imaged viruses located in the vitreous ice, we estimate that the actual defocus of the collected data ranges from about -1.5 to -2.5 µm. Under these conditions, the first zero of the contrast transfer function (CTF) is at ∼22 Å. The angular range of the tilt series was from -70° to +70°, typically at tilt increments of 1°, and at a cumulative dose of ∼80 electrons Å-2. Tilt series were initially aligned with gold markers using Inspect3D (FEI), and reconstructed after further refinement using weighted back-projection as implemented in the software packages IMOD29 and Protomo30. Visualization of tomograms was carried out using software tools implemented in the program Amira (TGS Inc) and UCSF Chimera.
Classification and 3D averaging
Volumes corresponding to reconstructions of individual viruses were extracted from the tomograms. Locations of surface spikes on each virion were identified by manual inspection, and the corresponding subvolumes (128 × 128 × 128 voxels) were computationally extracted. Densities on the surface much smaller than the expected size of the trimer (∼120 Å high and ∼150 Å wide) were not selected. The approximate local orientations of the long axis of the spike were determined by fitting an ellipsoidal surface to the picked spike positions, with the surface normals at each measured point providing initial estimates for two of the three Euler angles. The remaining in-plane rotation was initially randomized to eliminate possible bias in initial alignment. After application of the Euler angles, sub-volumes were translationally aligned to their cylindrically averaged global average so that all sub-volumes in a given data set shared the same centre of mass. Alignment and classification of the spike volumes was carried out using the framework described in ref. 26. Early stages of refinement clearly showed the inherent three-fold symmetry in the spike structure (see Supplementary Figs 2 and 3), and once this was visually ascertained, three-fold symmetry was imposed for subsequent rounds of refinement. At each round, the classes that showed the most clearly delineated features in all regions of the spike (typically ∼50%–60%) were selected to be used as references for the next round (for example, the top five classes in Supplementary Fig. 2 that show well-resolved densities for both the spike and the Fab fragment). All images were retained until the final iteration to allow for movement between classes. The final maps shown in Figs 1–3 were obtained after ∼10–12 refinement rounds and include contributions from ∼50% of the sub-volumes in each case. Fourier shell correlation coefficients were estimated by comparing the correlation between two randomly divided halves of the aligned images used to generate the final map. No temperature factor sharpening was applied to the final maps, and no correction for CTF was applied. Note that the resolution obtained is roughly the same as the resolution corresponding to the first zero of the CTF under the conditions used for data collection.
Fitting of coordinates into map
Surface renderings of all the maps, and automated fitting of atomic coordinates, was carried out in the environment of the software package Chimera27. The gp120 complexes with b12 and CD4/17b were docked as rigid bodies, that is, the coordinates used are identical to the structures of the complex derived by X-ray crystallography. The coordinates were filtered to 20 Å resolution before carrying out the fits to match the resolution of the experimental density maps; however, the use of coordinates without filtering, or with filtering to intermediate resolutions, had little effect on the overall fit. The fits shown in Fig. 2 were done directly using 2NY7 (Fig. 2d, e) and 2B4C (Fig. 2f) coordinates, after verifying that the results were similar to those obtained using 1GC1 coordinates (used to derive the molecular model presented in Fig. 4). To obtain the superposition shown in Fig. 3c–e, the three density maps were first aligned to each other to establish a single frame of reference for the three sets of fitted coordinates. The coordinates of the gp120 component of 1GC1 (CD4/17b complex) in Fig. 3e were then directly fitted into the map of the unliganded spike to obtain Fig. 3c, and aligned to gp120 coordinates 2NY7 (b12 complex) to obtain Fig. 3d. The hypervariable loop regions V1, V2 and V3 were not considered for arriving at the final fits, and inclusion or exclusion of the residues present in the V3 loop region (2B4C coordinates) or the stump of the V1/V2 loop region (1GC1 coordinates) did not alter the results. The best fit of HIV-1 gp120 coordinates into the map is unambiguous (Supplementary Fig. 4). This is further supported by the geometric fitting exercises shown in Supplementary Fig. 5 that illustrate comparative analysis of fits to different sets of HIV-1 and SIV gp120 coordinates, and in Supplementary Fig. 6 showing comparative analysis of fit quality visualized over a wide range of thresholds. To arrive at the molecular model shown in Fig. 4, the coordinates of gp120 in the CD4/17b complex were fitted to the map of the unliganded spike (as in Fig. 3c) and the coordinates for CD4 were then placed in the same relative orientation to gp120 as observed in the X-ray structure of the gp120/CD4/17b complex to derive Fig. 4a and c.
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Data are available as accession numbers EMD-5018 to EMD-5023 in the EM Data Bank, with associated PDB entries 3DNL, 3DNN and 3DNO in the Protein Data Bank.
References
- Dalgleish, A. G. et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767 (1984)
Article ADS CAS Google Scholar - Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998)
Article ADS CAS Google Scholar - Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007)
Article ADS CAS Google Scholar - Burton, D. R. et al. HIV vaccine design and the neutralizing antibody problem. Nature Immunol. 5, 233–236 (2004)
Article CAS Google Scholar - Huang, C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005)
Article ADS CAS Google Scholar - Kwong, P. D. et al. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74, 1961–1972 (2000)
Article CAS Google Scholar - Chen, B. et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841 (2005)
Article ADS CAS Google Scholar - Subramaniam, S. Bridging the imaging gap: Visualizing subcellular architecture with electron tomography. Curr. Opin. Microbiol. 8, 316–322 (2005)
Article Google Scholar - Sattentau, Q. J. & Moore, J. P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174, 407–415 (1991)
Article CAS Google Scholar - Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002)
Article ADS CAS Google Scholar - Chan, D. C. & Kim, P. S. HIV entry and its inhibition. Cell 93, 681–684 (1998)
Article CAS Google Scholar - Zanetti, G. et al. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2, e83 (2006)
Article Google Scholar - Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006)
Article ADS CAS Google Scholar - Myszka, D. G. et al. Energetics of the HIV gp120–CD4 binding reaction. Proc. Natl Acad. Sci. USA 97, 9026–9031 (2000)
Article ADS CAS Google Scholar - Wyatt, R. et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711 (1998)
Article ADS CAS Google Scholar - Cao, J. et al. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71, 9808–9812 (1997)
CAS PubMed PubMed Central Google Scholar - Johnson, W. E. et al. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76, 2075–2086 (2002)
Article CAS Google Scholar - Saunders, C. J. et al. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79, 9069–9080 (2005)
Article CAS Google Scholar - Gorny, M. K. et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79, 5232–5237 (2005)
Article CAS Google Scholar - Bennett, A. E. et al. Cryo electron tomographic analysis of an HIV neutralizing protein and its complex with native viral gp120. J. Biol. Chem. 282, 27754–27759 (2007)
Article CAS Google Scholar - Healey, D. et al. Novel anti-CD4 monoclonal antibodies separate human immunodeficiency virus infection and fusion of CD4+ cells from virus binding. J. Exp. Med. 172, 1233–1242 (1990)
Article CAS Google Scholar - Ashish et al. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J. Biol. Chem. 283, 2761–2772 (2008)
Article CAS Google Scholar - Montefiori, D. et al. Antibody-based HIV-1 vaccines: Recent developments and future directions. PLoS Med. 4, e348 (2007)
Article Google Scholar - Zwick, M. B. & Burton, D. R. HIV-1 neutralization: Mechanisms and relevance to vaccine design. Curr. HIV Res. 5, 608–624 (2007)
Article CAS Google Scholar - Sougrat, R. et al. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 3, e63 (2007)
Article Google Scholar - Bartesaghi, A. et al. Classification and 3D averaging with missing wedge correction in biological electron tomography. J. Struct. Biol. 162, 436–450 (2008)
Article CAS Google Scholar - Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Article CAS Google Scholar - Rossio, J. L. et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72, 7992–8001 (1998)
CAS PubMed PubMed Central Google Scholar - Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996)
Article CAS Google Scholar - Winkler, H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol. 157, 126–137 (2007)
Article CAS Google Scholar
Acknowledgements
We thank J. Bess and J. Lifson for providing purified, AT-2 treated HIV-1 BaL virus; P. Kwong for providing purified preparations of Fab fragments of the neutralizing antibodies b12 and 17b; S. Fellini and colleagues for assistance with use of the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland (http://biowulf.nih.gov); E. Tyler for assistance with figures; T. M. Chou for assistance with microscope maintenance; and A. Bennett, J. Milne and T. White for comments on the manuscript. This work was supported by funds from the intramural programme of the National Cancer Institute (to S.S.), and from the National Science Foundation and the Department of Defense (to G.S.).
Author information
Author notes
- Jun Liu, Alberto Bartesaghi and Mario J. Borgnia: These authors contributed equally to this work.
Authors and Affiliations
- Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, Maryland 20892, USA ,
Jun Liu, Alberto Bartesaghi, Mario J. Borgnia & Sriram Subramaniam - Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, USA,
Guillermo Sapiro
Authors
- Jun Liu
You can also search for this author inPubMed Google Scholar - Alberto Bartesaghi
You can also search for this author inPubMed Google Scholar - Mario J. Borgnia
You can also search for this author inPubMed Google Scholar - Guillermo Sapiro
You can also search for this author inPubMed Google Scholar - Sriram Subramaniam
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toSriram Subramaniam.
Supplementary information
Supplementary Information 1
The file contains Supplementary Figures S1-S7 and Legends. (PDF 5432 kb)
Supplementary Information 2
The file contains Supplementary Movie 1. The movie shows density map of unliganded HIV-1 spike determined by cryo-electron tomography combined with 3D averaging. The density includes contributions from three gp120 and three gp41 molecules. The coordinates of monomeric gp120 as determined in the complexe with CD4 and the X5 antibody (2B4C coordinates) are shown docked into the map, highlighting the location of the V3 loop regions at the top of the spike. (MOV 4563 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Liu, J., Bartesaghi, A., Borgnia, M. et al. Molecular architecture of native HIV-1 gp120 trimers.Nature 455, 109–113 (2008). https://doi.org/10.1038/nature07159
- Received: 09 March 2008
- Accepted: 10 June 2008
- Published: 30 July 2008
- Issue Date: 04 September 2008
- DOI: https://doi.org/10.1038/nature07159