Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960 (original) (raw)
- Letter
- Published: 02 October 2008
- Marlea Gemmel1,
- Dirk E. Teuwen2,3,
- Tamara Haselkorn1,
- Kevin Kunstman4,
- Michael Bunce5,
- Jean-Jacques Muyembe6,7,
- Jean-Marie M. Kabongo6,
- Raphaël M. Kalengayi6,
- Eric Van Marck8,
- M. Thomas P. Gilbert1 nAff9 &
- …
- Steven M. Wolinsky4
Nature volume 455, pages 661–664 (2008)Cite this article
- 16k Accesses
- 519 Citations
- 387 Altmetric
- Metrics details
Abstract
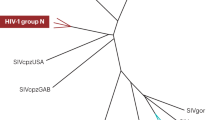
Human immunodeficiency virus type 1 (HIV-1) sequences that pre-date the recognition of AIDS are critical to defining the time of origin and the timescale of virus evolution1,2. A viral sequence from 1959 (ZR59) is the oldest known HIV-1 infection1. Other historically documented sequences, important calibration points to convert evolutionary distance into time, are lacking, however; ZR59 is the only one sampled before 1976. Here we report the amplification and characterization of viral sequences from a Bouin’s-fixed paraffin-embedded lymph node biopsy specimen obtained in 1960 from an adult female in Léopoldville, Belgian Congo (now Kinshasa, Democratic Republic of the Congo (DRC)), and we use them to conduct the first comparative evolutionary genetic study of early pre-AIDS epidemic HIV-1 group M viruses. Phylogenetic analyses position this viral sequence (DRC60) closest to the ancestral node of subtype A (excluding A2). Relaxed molecular clock analyses incorporating DRC60 and ZR59 date the most recent common ancestor of the M group to near the beginning of the twentieth century. The sizeable genetic distance between DRC60 and ZR59 directly demonstrates that diversification of HIV-1 in west-central Africa occurred long before the recognized AIDS pandemic. The recovery of viral gene sequences from decades-old paraffin-embedded tissues opens the door to a detailed palaeovirological investigation of the evolutionary history of HIV-1 that is not accessible by other methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
Similar content being viewed by others
HIV infection
Article 17 August 2023
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
The sequences reported in this study have been deposited in GenBank under accession numbers EU580739 – EU580854 and EU589211 – EU589236.
References
- Zhu, T. F. et al. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391, 594–597 (1998)
Article ADS CAS Google Scholar - Korber, B. et al. Timing the ancestor of the HIV-1 pandemic strains. Science 288, 1789–1796 (2000)
Article ADS CAS Google Scholar - Gilbert, M. T. P. et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues — which methods are useful when? PLoS ONE 2, e537 (2007)
Article ADS Google Scholar - Worobey, M. Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus. J. Virol. 82, 3769–3774 (2008)
Article CAS Google Scholar - Huelsenbeck, J. P. & Ronquist, F. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755 (2001)
Article CAS Google Scholar - Worobey, M. A novel approach to detecting and measuring recombination: insights into evolution in viruses, bacteria, and mitochondria. Mol. Biol. Evol. 18, 1425–1434 (2001)
Article CAS Google Scholar - Lemey, P. et al. The molecular population genetics of HIV-1 group O. Genetics 167, 1059–1068 (2004)
Article CAS Google Scholar - Gilbert, M. T. P. et al. The emergence of HIV-1 in the Americas and beyond. Proc. Natl Acad. Sci. USA 104, 18566–18570 (2007)
Article ADS CAS Google Scholar - Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007)
Article Google Scholar - Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006)
Article Google Scholar - Salemi, M., de Oliveira, T., Ciccozzi, M., Rezza, G. & Goodenow, M. M. High-resolution molecular epidemiology and evolutionary history of HIV-1 subtypes in Albania. PLoS ONE 3, e1390 (2008)
Article ADS Google Scholar - Suchard, M. A., Weiss, R. E. & Sinsheimer, J. S. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18, 1001–1013 (2001)
Article CAS Google Scholar - Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005)
Article CAS Google Scholar - Sharp, P. M. et al. The origins of acquired immune deficiency syndrome viruses: where and when? Phil. Trans. R. Soc. Lond. B 356, 867–876 (2001)
Article CAS Google Scholar - Salemi, M. et al. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15, 276–278 (2001)
Article CAS Google Scholar - Yusim, K. et al. Using human immunodeficiency virus type 1 sequences to infer historical features of the acquired immune deficiency syndrome epidemic and human immunodeficiency virus evolution. Phil. Trans. R. Soc. Lond. B 356, 855–866 (2001)
Article CAS Google Scholar - Vidal, N. et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74, 10498–10507 (2000)
Article CAS Google Scholar - Rambaut, A., Robertson, D. L., Pybus, O. G., Peeters, M. & Holmes, E. C. Human immunodeficiency virus phylogeny and the origin of HIV-1. Nature 410, 1047–1048 (2001)
Article ADS CAS Google Scholar - Keele, B. F. et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526 (2006)
Article ADS CAS Google Scholar - Worobey, M. in Global HIV/AIDS Medicine (eds Volberding, P. A., Sande, M. A., Lange, J. & Greene, W. C.) 13–21 (Saunders Elsevier, 2008)
Book Google Scholar - Hahn, B. H., Shaw, G. M., De Cock, K. M. & Sharp, P. M. AIDS as a zoonosis: scientific and public health implications. Science 287, 607–614 (2000)
Article ADS CAS Google Scholar - Hance, W. A. Population, Migration, and Urbanization in Africa 209–297 (Columbia Univ. Press, 1970)
Book Google Scholar - Chitnis, A., Rawls, D. & Moore, J. Origin of HIV type 1 in colonial French Equatorial Africa? AIDS Res. Hum. Retrov. 16, 5–8 (2000)
Article CAS Google Scholar - Taubenberger, J. K. et al. Characterization of the 1918 influenza virus polymerase genes. Nature 437, 889–893 (2005)
Article ADS CAS Google Scholar - Tumpey, T. M. et al. Characterization of the reconstructed 1918 Spanish Influenza pandemic virus. Science 310, 77–80 (2005)
Article ADS CAS Google Scholar - Leitner, T. et al. HIV Sequence Compendium 〈http://www.hiv.lanl.gov〉 (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, 2005)
Google Scholar - Drummond, A., Pybus, O. G. & Rambaut, A. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 54, 331–358 (2003)
Article Google Scholar
Acknowledgements
We thank J. Wertheim and M. Sanderson for computational assistance, and L. Jewel for providing the Canadian control specimen. The NIH/NIAID and the David and Lucile Packard Foundation funded the research.
Author Contributions M.W., D.E.T., S.M.W. and M.T.P.G. designed the study. M.G., T.H., K.K. and M.T.P.G. performed digestion and extraction, PCR, quantitative PCR, cloning and sequencing experiments. M.T.P.G., M.G. and M.B. optimized DNA/RNA isolation methods and designed PCR assays. D.E.T., J.-J.M., E.V.M., J.-M.M.K. and R.M.K. organized and provided samples. M.W. analysed the data, performed the phylogenetic analyses, and wrote the paper. S.M.W. contributed to the analyses and writing. All authors discussed the results and commented on the manuscript.
Author information
Author notes
- M. Thomas P. Gilbert
Present address: Present address: Centre for Ancient Genetics, Biological Institute, University of Copenhagen, Copenhagen DK-2100, Denmark.,
Authors and Affiliations
- Ecology and Evolutionary Biology, University of Arizona, Tucson, Arizona 85721, USA ,
Michael Worobey, Marlea Gemmel, Tamara Haselkorn & M. Thomas P. Gilbert - Sanofi Pasteur, F-69367 Lyon Cedex 07, France
Dirk E. Teuwen - UCB SA Pharma, Braine l’Alleud, BE-1420, Belgium ,
Dirk E. Teuwen - The Feinberg School of Medicine, Northwestern University, Chicago, Illinois 60611, USA ,
Kevin Kunstman & Steven M. Wolinsky - Ancient DNA Laboratory, School of Biological Sciences and Biotechnology, Murdoch University, Perth, Western Australia 6150, Australia ,
Michael Bunce - Department of Anatomy and Pathology, University of Kinshasa, Kinshasa B.P. 864, Democratic Republic of the Congo
Jean-Jacques Muyembe, Jean-Marie M. Kabongo & Raphaël M. Kalengayi - National Institute for Biomedical Research, National Laboratory of Public Health, Kinshasa B.P. 1197, Democratic Republic of the Congo
Jean-Jacques Muyembe - Department of Pathology, University Hospital, University of Antwerp, Antwerp B-2610, Belgium
Eric Van Marck
Authors
- Michael Worobey
- Marlea Gemmel
- Dirk E. Teuwen
- Tamara Haselkorn
- Kevin Kunstman
- Michael Bunce
- Jean-Jacques Muyembe
- Jean-Marie M. Kabongo
- Raphaël M. Kalengayi
- Eric Van Marck
- M. Thomas P. Gilbert
- Steven M. Wolinsky
Corresponding author
Correspondence toMichael Worobey.
Supplementary information
Supplementary Information
This file contains Authenticity of Data, Supplementary Table S1 and Supplementary Figures S1-S3 with Legends (PDF 873 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Worobey, M., Gemmel, M., Teuwen, D. et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960.Nature 455, 661–664 (2008). https://doi.org/10.1038/nature07390
- Received: 21 May 2008
- Accepted: 08 September 2008
- Issue date: 02 October 2008
- DOI: https://doi.org/10.1038/nature07390
This article is cited by
Editorial Summary
HIV/AIDS then and now
A histological specimen from the University of Kinshasa archives has been used to obtain HIV gene sequences dating back to the pre-AIDS era. From a lymph node biopsy taken in 1960 from an adult female in Léopoldville in the Belgian Congo (now Kinshasa, Democratic Republic of the Congo), sample 'DRC60' makes possible the first evolutionary analysis of pre-AIDS 'fossil' HIV-1 sequences, via comparison with the one other viral sequence from the period, from a plasma sample taken in 1959, also in Kinshasa. The analysis supports the idea that diversification of HIV-1 in west-central Africa occurred long before the recognized AIDS pandemic. Almost fifty years on, a major concern in HIV epidemiology is China. Here, HIV-1 infection was largely confined to high-risk groups but it is now breaking out into the general population. Lin Lu et al. report on efforts to contain the epidemic in Yunnan Province, where there has been a dramatic increase in sexual transmission of HIV.