Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer (original) (raw)
Introduction
Invasive epithelial ovarian cancer (EOC) has a strong heritable component1, with an approximate three-fold increased risk associated with a first-degree family history2. Much of the excess familial risk observed for EOC is unexplained3, and efforts to identify common susceptibility genes have proven to be difficult. Seven regions harbouring susceptibility single-nucleotide polymorphisms (SNPs) for ovarian cancer have been identified through genome-wide association studies4,5,6,7 thus far, but candidate gene studies have been largely unsuccessful8.
The Cancer Genome Atlas (TCGA) has fully characterized more than 500 serous EOC cases with respect to somatic mutation, DNA methylation, mRNA expression and germline genetic variants9. These data are publicly available and can be analysed to identify candidate genes for association studies of the disease.
We conducted such an analysis of TCGA data and found a unique expression and methylation pattern of HNF1B characterized by downregulation of expression in most cases, with epigenetic silencing in about half of the cases, suggesting it might have a role in the serous subtype of ovarian cancer. In contrast, HNF1B overexpression is common in clear cell ovarian cancer10. The HNF1B gene (formerly known as TCF2) encodes a POU-domain containing a tissue-specific transcription factor, and mutations in the gene cause maturity onset diabetes of the young type 5 (ref. 11). HNF1B is also a susceptibility gene for type II diabetes12,13, prostate cancer12,14,15,16 and uterine cancer17.
We report here on our comprehensive characterization of this gene in ovarian cancer and show evidence of a differential effect of HNF1B on the serous and clear cell subtypes of ovarian cancer. It appears that HNF1B has a loss-of-function role in serous and a gain-of-function role in clear cell ovarian cancers, and variants in this gene differentially affect genetic susceptibility to these subtypes.
Results
DNA methylation/expression analysis
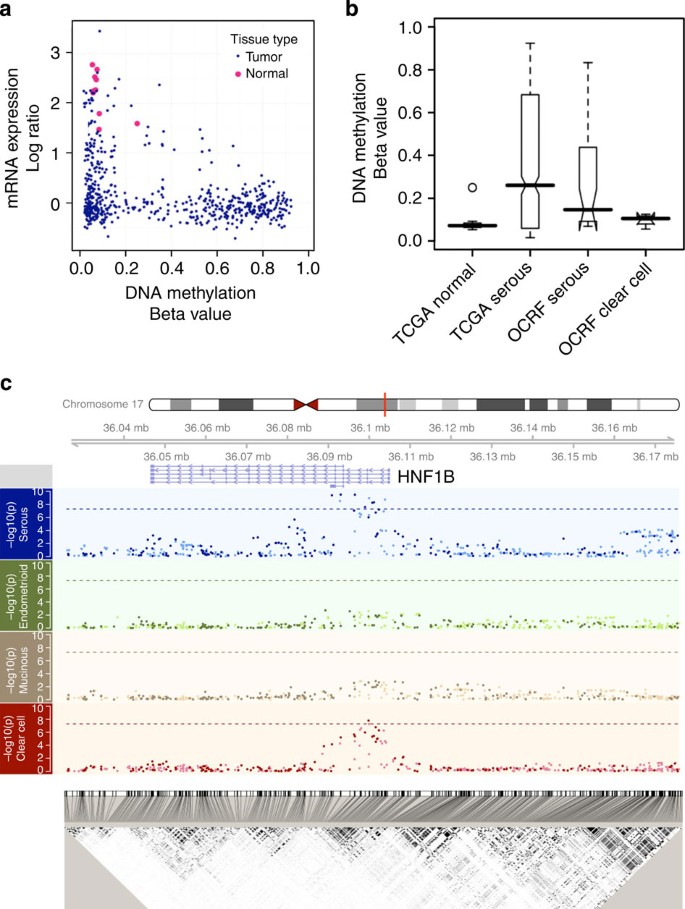
From TCGA data (see Methods), HNF1B was observed to be epigenetically silenced in approximately half of the 576 primary serous ovarian tumours and downregulated by another mechanism in most of the other tumours, whereas no evidence of methylation was seen in the normal fallopian tube samples (Fig. 1a, Supplementary Fig. S1) available from TCGA. We further assessed _HNF1B_-promoter methylation in an independent data set (OCRF panel; see Methods) and found the promoter region to be methylated in 42% of serous tumours and in none of the clear cell ovarian tumours (Fig. 1b). The pattern in serous tumours, in contrast to clear cell cancers, led to the evaluation of HNF1B as a candidate subtype-specific susceptibility gene for ovarian cancer.
Figure 1: Identification of HNF1B as a subtype-specific candidate gene for ovarian cancer and its establishment as a susceptibility gene.
(a) The scatterplot compares the mRNA expression (y axis) versus DNA methylation (x axis) in serous ovarian tumours from TCGA (see Methods). Each blue dot is a serous tumour sample, whereas each pink dot is one of the ten normal fallopian tube samples. The HNF1B promoter is silenced in the majority of these tumours, either by an epigenetic (bottom right, high DNA methylation and low mRNA expression) or an unknown alternative mechanism. The mRNA expression data were integrated from three platforms (online Methods) and interpreted as log ratios, and we observe the same pattern with each individual expression platform (Supplementary Fig. S1). (b) _HNF1B_-promoter DNA methylation differs by histological subtype. Although unmethylated in the normal fallopian tissue, this locus is hypermethylated (beta value >0.2) in approximately 50% of the TCGA (_n_=576; see Methods) serous cases as well as another independent set of 32 serous tumour samples (OCRF panel; see Methods), but remains unmethylated in clear cell tumours (OCRF panel; see Methods) (_n_=4). These data are consistent with reported HNF1B expression in the clear cell tumours. (c) Genetic variants in the HNF1B locus are associated with risk of ovarian cancer histological subtypes. Plotted in each panel is the −log10 (_P_-value) from the SNP association with risk for each subtype (Manhattan plots) located in the 150-kb region described in the text. Imputed SNPs are indicated with a relatively lighter colour, whereas the genotyped SNPs are indicated with a darker colour. Dashed lines indicate the genome-wide significance threshold (5 × 10−8). The linkage disequilibrium plot on the bottom shows the _r_2 between the SNPs. Genomic coordinates are based on hg19 (Build37).
SNP analysis
With all invasive cancer subtypes considered together, we found no genome-wide significant (P<5 × 10−8) HNF1B SNP associations among women of European ancestry (Table 1; Supplementary Data S1). However, when analyses were stratified by histological subtype, we observed genome-wide significant results for both serous and clear cell EOC subtypes, but with risk associations in opposite directions. The association was similar for high- and low-grade serous cancers. There was no evidence of association for mucinous or endometrioid subtypes (Fig. 1c). Associations in the non-European populations are shown in Supplementary Table S2.
Table 1 Association between invasive, serous and clear cell ovarian cancer for ten HNF1B SNPs that reached genome-wide significance in Whites.
Minor alleles at nine SNPs, six genotyped and three imputed, were associated with increased risk of invasive serous ovarian cancer at P<5 × 10−8 (Table 1). The risk signal spanned a 21.4-kb region from the 5′ untranslated region (UTR) through part of intron 4 of HNF1B (Fig. 1c). The most strongly associated SNP for invasive serous ovarian cancer (rs7405776, minor allele frequency (MAF) 36%) conferred a 13% increased risk per minor allele (_P_=3.1 × 10−10; Table 1, Supplementary Fig. S2A). The signals of this SNP and the eight other genome-wide significant SNPs were indistinguishable, given the linkage disequilibrium and resulting haplotype structure (Supplementary Figs S3, S4 and S5).
For the clear cell subtype, rs11651755 (MAF 45%) was associated with a 23% decreased risk of disease at a genome-wide significant level (_P_=2 × 10−8; Table 1, Supplementary Fig. S2B). This signal was distinct from the nine significant SNPs for invasive serous cancer (Table 1). The odds against the serous-associated SNP, rs7405776, as the true best hit for clear cell ovarian cancer were 244:1. Conversely, the odds against the clear cell SNP, rs11651755, as the true best hit for serous were 1808:1. Further, when rs11651755 and rs7405776 were jointly modelled, the signal for clear cell cancer was driven completely by rs11651755, whereas that for the serous disease was driven by rs7405776 (Table 1). The clear cell SNP (rs11651755) sits on five haplotypes, only three of which also contain the serous SNP (rs7405776; Supplementary Fig. S5). Thus, different SNPs in the HNF1B gene regions explain the associations observed for serous and clear cell ovarian cancer.
DNA methylation and protein expression
The identification of HNF1B as a susceptibility gene for serous and clear cell ovarian cancer led us to further evaluate the relationship between _HNF1B_-promoter DNA methylation, protein expression and histological subtype. Immunohistochemistry (IHC) analysis for HNF1B protein expression in 1,149 ovarian cancers from the Ovarian Tumor Tissue Analysis Consortium, and DNA-methylation analysis on 269 of these tumours, revealed that the majority of clear cell tumours expressed the HNF1B protein and were unmethylated at the HNF1B promoter, whereas the majority of serous tumours lacked HNF1B protein expression and displayed frequent _HNF1B_-promoter methylation (Fig. 2, Supplementary Fig. S6).
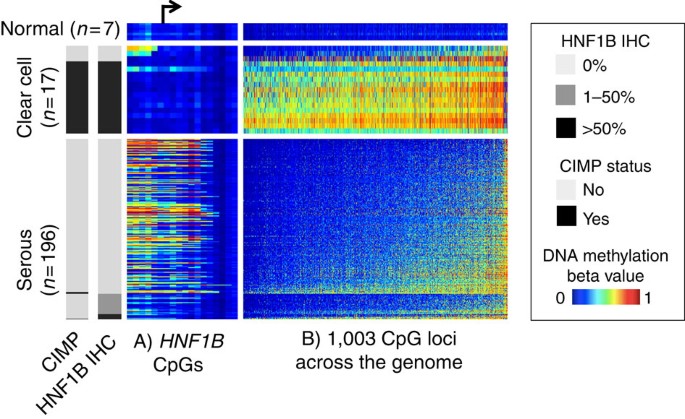
Figure 2: _HNF1B_-promoter DNA methylation, protein expression and global DNA-methylation pattern by subtype.
Each row is a tissue sample collected at the Mayo Clinic that belongs to one of the three categories: normal ovarian tissue (_n_=7), clear cell ovarian tumours (_n_=17) or serous ovarian tumours (_n_=196). Endometrioid (_n_=49) and mucinous (_n_=7) tumours are not included in this figure. Each column represents a CpG locus, either from the region flanking the HNF1B transcription start site (panel A, ordered by genomic locations with an arrow indicating the transcription start site) or from a global panel of 1,003 CpG loci mapped to autosomal CpG island regions that distinguish clear cell and serous subtypes (panel B, ordered by average DNA methylation across the samples). For each horizontal panel group, the samples (rows) are ordered by HNF1B IHC status. The heatmap shows the DNA-methylation beta value, with blue indicating low DNA methylation and red indicating high methylation. Clear cell tumours showed less DNA methylation at the _HNF1B_-promoter region and correspondingly higher HNF1B protein expression. The clear cell tumours generally show a CIMP where there is extensive gain of aberrant promoter methylation in a correlated manner. CIMP status (left side bar, defined as methylated at >80% of the 1,003 loci) is highly correlated HNF1B expression. Also noteworthy is that the _HNF1B_-promoter DNA methylation (panel a) is the opposite from the global pattern (panel b, Supplementary Fig. S8). This suggests HNF1B DNA methylation is not a passenger event of global DNA-methylation changes.
Although most clear cell tumours were devoid of _HNF1B_-promoter methylation, they revealed a surprisingly high frequency of CpG island hypermethylation at other sites across the genome, indicative of a CpG island methylator phenotype (CIMP). The few clear cell tumours lacking HNF1B expression exhibited _HNF1B_-promoter methylation, and a correspondingly low frequency of CpG island methylation throughout the genome, similar to the serous subtype (Fig. 2). HNF1B expression and CIMP methylation are strongly associated (_P_=3 × 10−16; Fig. 2). Further, minimal hypermethylation is observed in serous tumours overall, but HNF1B is one notable exception (Supplementary Fig. S7).
DNA methylation and genotype
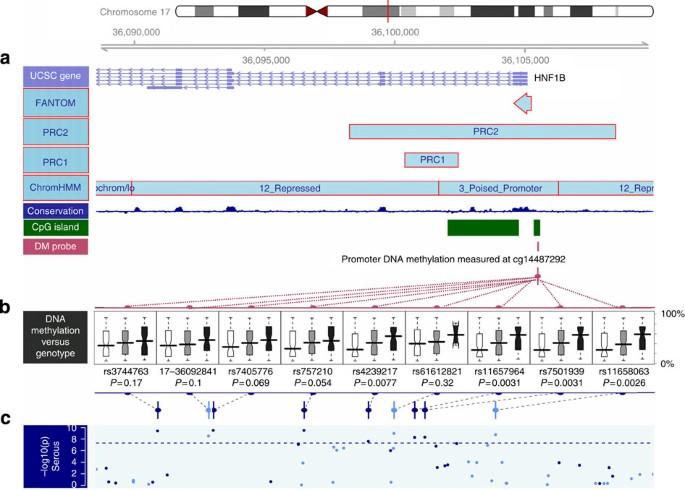
We further investigated the relationship between risk allele genotypes and HNF1B DNA methylation in 231 serous ovarian cancers. The top serous risk SNP, rs7405776, showed only a borderline association with increased promoter methylation (_P_=0.07; Fig. 3). Intriguingly, the association between SNPs in HNF1B and _HNF1B_-promoter DNA methylation strengthened as their location approached the promoter region, and the strongest signal came from a few SNPs, exemplified by rs11658063, overlapping with a polycomb repressive complex 2 (PRC2) mark in embryonic stem cells (_P_=0.003; Fig. 3, Supplementary Fig. S8). We validated this SNP–methylation association in the TCGA data (Supplementary Fig. S9; see Methods). None of the probes used contained common SNPs in the sequence, excluding technical artifact as a confounder of this association.
Figure 3: Correlation of serous risk-associated SNPs with _HNF1B_-promoter DNA-methylation level.
Plotted is the linkage disequilibrium region defined as _r_2>0.2 with the top serous SNP rs7405776. (a) Annotation of the region in terms of (from top to bottom:) UCSC genes, FANTOM mark, PRC marks (PRC2 and PRC1)32, the chromatin status determined in stem cells33, the conservation score across this region and the CpG island information, on top of the location of the HM450 probe used in b boxplots of promoter DNA-methylation level of HNF1B (cg14487292) by SNP genotype with position indicated in c. This DNA-methylation probe was selected based on inverse association with mRNA expression for HNF1B, and does not contain any SNP with MAF >1% in its probe sequence. Each boxplot shows the distribution of DNA-methylation level by genotype (homozygous major—white; heterozygous—grey; and homozygous minor—black, where the minor alleles are the risk alleles). Two-sided _P_-values testing for trend are presented, and are computed for 231 Mayo Clinic high-grade, high-stage serous tumours to avoid confounding by histological subtypes, and also to be consistent with the TCGA data (primarily high-grade, high-stage serous). Results were similar with all subtypes combined. The risk alleles are associated with significantly increased DNA methylation. The association of rs11658063 genotype with promoter methylation is consistent across the entire region flanking HNF1B transcription start site, and stronger for the upstream promoter region (Supplementary Fig. S8).
Overexpression of HNF1B
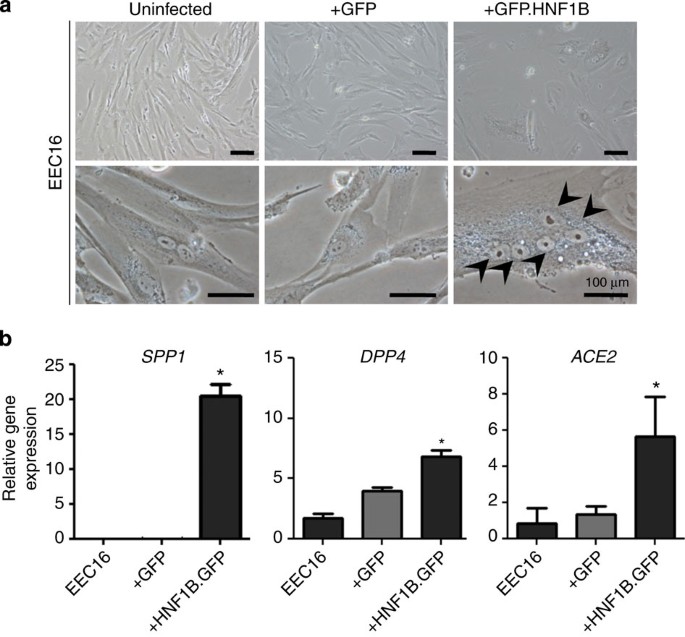
Given the proposed role of HNF1B in clear cell tumorigenesis, we stably overexpressed the gene in immortalized endometriosis epithelial cells (EECs), which are hypothesized to be a cell of origin for clear cell ovarian cancers (Supplementary Fig. S10)18. EECs overexpressing HNF1B acquired an enlarged, flattened morphology and multi-nucleated cells accumulated in the cultures (Fig. 4a). Also, significant upregulation of HNF1B-associated genes SPP1, DPP4, and ACE2 was observed upon HNF1B overexpression in EECs (Fig. 4b).
Figure 4: Phenotypic effects and downstream targets of HNF1B overexpression in immortalized EECs.
(a) Morphological changes in EECs expressing a HNF1B GFP fusion protein (EECGFP.HNF1B). GFP-positive cells were sorted using flow cytometry. The arrows indicate five nuclei contained within a single EECGFP.HNF1B cell, showing the aberrant polynucleation that we observed in these cells. Using flow cytometry, we quantified the increase in polynucleation in EECGFP.HNF1B to be around eightfold compared with controls (data not shown). (b) Gene-expression analysis of _HNF1B_-target genes and clear cell ovarian cancer associated genes. *_P_>0.01.
Discussion
HNF1B appears to have a prominent role in ovarian cancer aetiology. It is the first clear cell ovarian cancer-susceptibility gene identified, and variation in the gene is also associated with risk of serous ovarian cancer at a genome-wide significance level. The gene is overexpressed in clear cell tumours and silenced in serous tumours. The strong association between HNF1B expression and CIMP methylation (_P_=3 × 10−16), and the reciprocal nature of DNA methylation at the _HNF1B_-promoter CpG islands, versus other CpG islands across the genome, suggests that _HNF1B_-promoter methylation is not merely a CIMP passenger event; in fact, HNF1B expression may even contribute to the hypermethylation phenotype. Taken together, these data indicate differing roles for HNF1B in these invasive EOC subtypes: a potential gain-of-function in clear cell ovarian cancer and loss-of-function in serous ovarian cancer, underscoring the heterogeneity of this disease.
Different SNPs in the HNF1B gene regions explain the associations observed for serous and clear cell ovarian cancers. These different effects provide further support for the growing view that the histological subtypes of ovarian cancer represent distinct diseases18,19,20,21,22,23,24, with endometriosis as a proposed cell of origin for clear cell disease18 and fallopian tube fimbriae as one for serous disease22. Interestingly, no association was observed between HNF1B genotypes and endometrioid ovarian cancer despite the view that, like clear cell, endometriosis is also a cell of origin for this subtype. The lack of association may be due to a different transformation mechanism from endometriosis for the endometrioid subtype, given that although the HNF1B promoter remains unmethylated in the endometrioid subtype, the endometrioid subtype does not overexpress HNF1B. Alternatively, misclassification of high-grade serous EOC as high-grade endometrioid could result in a bias towards the null for the endometrioid subtype.
Variation in the 5′ UTR through the intron 4 region of HNF1B is also associated with susceptibility to prostate12,14,15,16 and uterine cancer17 (where minor alleles of certain SNPs are associated with decreased risk) and type II diabetes12,13 (increased risk for the same or correlated SNP alleles; Supplementary Fig. S4). The opposing directions of these associations mirror the differential effects seen here in ovarian cancer. The most strongly associated SNP for both prostate14 and uterine cancer17 is rs4430796, correlated at _r_2=0.94 with the top clear cell ovarian cancer SNP, rs11651755, suggesting a common risk variant. Although increased risk of type II diabetes has been reported with rs4430796 (ref. 12), Winckler et al.13 have suggested that the best marker of diabetes risk is rs757210, which correlated at _r_2=0.97 with our top serous SNP. Thus, the evidence suggests that a specific variant in HNF1B predisposes to clear cell ovarian, uterine and prostate cancers and that a different variants is associated with diabetes and serous ovarian cancer.
We were able to completely fine-map the HNF1B region, localize the signal and identify a handful of potentially causal SNPs. This is quite different from other regions of the genome where it is not uncommon to identify hundreds of candidate causal SNPs. Further, an important link, often missing when susceptibility loci are identified, is the functional role that the variant has in disease. In the case of serous ovarian cancer, the SNP–_HNF1B_-promoter DNA methylation association strengthens as it approaches the promoter region, particularly where it overlaps with a PRC2 mark. PRC2–DNA methyltransferase cross-talk has been proposed to be a mechanism of predisposition to cancer-specific hypermethylation25. Our DNA-methylation data indicate that the causal risk alleles for the serous subtype may predispose the promoter to acquiring aberrant methylation, thereby promoting the development of serous but not clear cell tumours. This predisposition could be a direct functional effect of the SNP on the DNA-methylation machinery, or could act indirectly through differential binding affinity for PRC2 or one or more transcription factors. Given that we were able to fine-map the HNF1B region, it is unlikely that an unidentified common variant explains these associations. For serous ovarian cancer, the methylation signal suggests that the causal variant is most likely to be among those located within the region with the PRC2 mark for which we identified five SNPs with genome-wide significance.
This is the first study investigating the effects of overexpression of HNF1B in endometriosis, and the results support the hypothesis that HNF1B may have an oncogenic role in the initiation of clear cell ovarian cancers, as speculated by Gounaris et al.23 as a key step of endometriosis transformation. The observation in our data that HNF1B induces a polynucleated phenotype in EEC cells is intriguing, as clear cell ovarian cancers are often tetradiploid, more so than other ovarian cancer subtypes26. The polynucleated phenotype may suggest that HNF1B overexpression in EECs perturbs cytokinesis, causing aneuploidy in some cells.
Histology re-review of the three clear cell tumours that do not express HNF1B revealed two scenarios: two samples with inconsistent evaluations between pathologists, and one consistently called clear cell. They might be cases that are especially difficult to classify, and therefore a molecular signature, for example, CIMP or HNF1B status, would be of great help in correctly classifying those tumours. The one sample that is called consistently clear cell tumour but does not express HNF1B might represent a rare subtype of clear cell carcinoma. With a larger cohort of clear cell ovarian cancers, these possibilities can be investigated.
To our knowledge, this is the first report of tumour DNA-methylation patterns leading to the identification of a germline susceptibility locus, underscoring the value of TCGA. Recent studies suggest a strong genetic component to inter-individual variation in tumour DNA methylation, and demonstrate both cis- and _trans_- associations between genotypes and DNA methylation27. In addition, methylation quantitative trait loci were found to be enriched for expression quantitative trait loci. It has also been shown that epimutation is associated with genetic variation, for example, associations have been demonstrated between 5′ UTR MLH1 variants and MLH1 epigenetic silencing28. Moreover, we have for the first time demonstrated the existence of a CIMP phenotype in ovarian cancer, highlighting the complicated nature of the disease.
In summary, variation in HNF1B is associated with serous and clear cell subtypes of ovarian cancer in opposite manner at genetic, epigenetic and protein expression levels. These observations are compatible with a tumour suppressor role in serous cancer and an oncogenic role in clear cell disease. Future efforts should focus on understanding these mechanisms as they could have major clinical implications for ovarian cancer, based on better subtype stratification, potential novel treatment approaches and a better understanding of disease aetiology. Currently, effective chemotherapeutics for clear cell ovarian cancer is lacking, but our study reveals that HNF1B-expressing clear cell tumours have extensive epigenetic alterations that potentially make them good candidates for epigenetic therapies.
Methods
Molecular aspects
TCGA data access
We downloaded the TCGA serous ovarian cancer data packages from the TCGA public-access ftp (https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumour/ov/). Data generated with the following platforms were used: Affymetrix HT Human Genome U133 Array Plate Set; Agilent 244K Custom Gene Expression G4502A-07-3; Affymetrix Human Exon 1.0 ST Array; and Illumina Infinium HumanMethylation27 Beadchip (a full list of the packages is provided in Supplementary Methods).The Illumina Human1M-Duo DNA Analysis BeadChip Genotype data were downloaded from the controlled access data tier.
DNA methylation data production for the OCRF tumour panel
The Illumina Infinium HumanMethylation27 assay was performed as described9 on 32 serous and 4 clear cell ovarian tumours from USC Norris Comprehensive Cancer Center and Duke University (‘OCRF tumour panel’). The beta values for each sample and locus were calculated with mean non-background corrected methylated (M) and unmethylated (U) signal intensities with the formula M/(M+U), representing the percentage of methylated alleles. Detection _P_-values were calculated by comparing the set of analytical probe replicates for each locus to the set of 16 negative control probes. Data points with detection _P_-values >0.05 were masked.
DNA methylation data production for the Mayo tumour panel
We also performed the Infinium HumanMethylation450 BeadChip assay on an independent set of tumour DNA in the Mayo Clinic Genotyping Shared Facility using recommended Illumina protocol29. 1 μg of tumour DNA was bisulfite-converted using the Zymo EZ96 DNA Methylation Kit. Three samples failing quality control were removed, leaving DNA-methylation data on 333 ovarian cancer cases, including 254 serous and 17 clear cell tumours. Plate normalization was done with a linear model on the logit-transformed beta values, following back-transformation to the (0,1) range.
IHC assay
Previously built tissue microarrays, triplicate core, measuring 0.6 mm were cut at 4-μm thickness and mounted on superfrost slides. Slides were stained on a Ventana Benchmark XT using the manufacturer’s pretreatment protocol CC1 standard (Supplementary Methods). A pathologist (MK) evaluated the IHC staining, and assigned the sample a score 0 in the absence of any nuclear staining, score 1 for any nuclear staining >1–50% or score 2 for >50% tumour cell nuclei-positive for HNF1B.
Genotype and DNA methylation association
We assessed the correlation of germline genotype at the nine genome-wide significant SNPs in serous cancer, with HNF1B DNA promoter methylation status using the Mayo Tumour Panel. Probe cg14487292 was used as it was most inversely correlated with mRNA expression. The nominal _P_-values are from two-sided tests for linear trend in the DNA-methylation beta values across the three genotypes for each locus. Bonferroni adjustment was not done for multiple comparisons as the SNPs are highly correlated. Validation was done with the TCGA data (Supplementary Appendix).
In vitro model of HNF1B overexpression
An immortalized EEC line was generated by lentiviral transduction of hTERT (Addgene plasmid 12245) into primary EECs (Supplementary Fig. S10). _TERT-_immortalized EECs were transduced with lentiviral HNF1B-green fluorescent protein (GFP) or GFP (Genecopoeia) supernatants and positive cells selected with 400 ng ml−1 puromycin (Sigma). GFP expression was confirmed by fluorescent microscopy; HNF1B expression was confirmed by real-time PCR (Supplementary Fig. S10).
For gene-expression studies, RNA was collected from cells using the Qiagen RNeasy kit with on-column DNase I digestion. An amount of 1 μg RNA was reverse transcribed using an MMLV reverse transcriptase enzyme (Promega), and relative mRNA level was assayed using the ABI 7900HT Fast Real-Time PCR system utilizing the delta-delta Ct method. Statistical analyses were performed using Prism. Two-tailed paired _t_-tests with significance level of 0.05 were used.
Genetic association study
Study design
The genetic susceptibility aspect of this study was organized by the Collaborative Oncological Gene-Environment Study, an ovarian, breast and prostate cancer consortium. The ovarian cancer part of this effort on which the current report is based is led by the Ovarian Cancer Association Consortium and included 43 studies (Supplementary Table S1). Following sample quality control, 44,308 subjects, including 16,111 patients with invasive EOC, 2,063 with low malignant potential (borderline) disease and 26,134 controls, were available for analysis; results presented here are restricted to invasive cancers. All studies obtained approval from their respective human research ethics committees, and all participants provided written informed consent.
Selection of SNPs
Data for 174 SNPs in this region were available from the Collaborative Oncological Gene-Environment Study genotyping effort and provided full fine-mapping information in the 150-kb region surrounding HNF1B (hg18 coordinates 33,100,000–33,250,000). In addition, phase I haplotype data from the 1000 Genomes Project (January 2012) were used to impute genotypes for SNPs across this region, resulting in available data on an additional 307 SNPs with MAF >0.02 in European Whites and imputation _r_2>0.30 (IMPUTE 2.2).
SNP genotyping
The Ovarian Cancer Association Consortium genotyping was conducted by McGill University and Génome Québec Innovation Centre (_n_=19,806) and the Mayo Clinic Medical Genome Facility (_n_=27,824) using an Illumina Infinium iSelect BeadChip. Genotypes were called using GenCall. Sample and SNP quality-control measures are described in the Supplementary Methods.
Statistical analysis
We used the program LAMP30 for principal components analysis to assign intercontinental ancestry based on the HapMap (release no. 22) genotype frequency data for European, African and Asian populations (Supplementary Methods). For LAMP-derived European ancestry groups for all patients of invasive cancer and for those with serous invasive cancer, we carried out unconditional logistic regression analyses within each study site, adjusted for the first five eigenvalues from the principal components analysis for European ancestry and then used a fixed-effects meta-analytic approach to obtain the summary OR estimate, 95% confidence interval and _P_-value. Details on analysis for the non-European groups are provided in the Supplementary Methods. Log-additive mode of inheritance was modelled (that is, co-dominant), treating each SNP as an ordinal variable.
For haplotype analysis, we used the tagSNPs program31 to obtain the haplotype dosage for each subject for the LAMP-derived European ancestry group for haplotypes with a frequency of ≥1%. The associations between haplotype and risks of serous and clear cell ovarian cancer were modelled by meta-analysis relative to the most common haplotype.
Additional information
How to cite this article: Shen, H., Fridley, B. L., Song, H. et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat. Commun. 4:1628 doi: 10.1038/ncomms2629 (2013).
References
- Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000).
Article CAS Google Scholar - Auranen, A. et al. Cancer incidence in the first-degree relatives of ovarian cancer patients. Br. J. Cancer 74, 280–284 (1996).
Article CAS Google Scholar - Antoniou, A. C. & Easton, D. F. . Risk prediction models for familial breast cancer. Future Oncol. 2, 257–274 (2006).
Article Google Scholar - Pharaoh, P. D. P. et al. GWAS meta-analysis and replication identifies three novel common susceptibility loci for ovarian cancer. Nat. Genet (e-pub ahead of print 27 March 2013; doi:10.1038/ng2564) (2013).
- Goode, E. L. et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 42, 874–879 (2010).
Article CAS Google Scholar - Bolton, K. L. et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 42, 880–884 (2010).
Article CAS Google Scholar - Song, H. et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 41, 996–1000 (2009).
Article CAS Google Scholar - Bolton, K. L., Ganda, C., Berchuck, A., Pharaoh, P. D. & Gayther, S. A. . Role of common genetic variants in ovarian cancer susceptibility and outcome: progress to date from the Ovarian Cancer Association Consortium (OCAC). J. Intern. Med. 271, 366–378 (2012).
Article CAS Google Scholar - Cancer Genome Atlas Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
- Tsuchiya, A. et al. Expression profiling in ovarian clear cell carcinoma: identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. Am. J. Pathol. 163, 2503–2512 (2003).
Article CAS Google Scholar - Horikawa, Y. et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat. Genet. 17, 384–385 (1997).
Article CAS Google Scholar - Gudmundsson, J. et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 39, 977–983 (2007).
Article CAS Google Scholar - Winckler, W. et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes 56, 685–693 (2007).
Article CAS Google Scholar - Berndt, S. I. et al. Large-scale fine mapping of the HNF1B locus and prostate cancer risk. Hum. Mol. Genet. 20, 3322–3329 (2011).
Article CAS Google Scholar - Sun, J. et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet. 40, 1153–1155 (2008).
Article CAS Google Scholar - Thomas, G. et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 40, 310–315 (2008).
Article CAS Google Scholar - Spurdle, A. B. et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat. Genet. 43, 451–454 (2011).
Article CAS Google Scholar - Pearce, C. L. et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 13, 385–394 (2012).
Article Google Scholar - Kurman, R. J. & Shih, IeM. . The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol. 34, 433–443 (2010).
Article Google Scholar - Gilks, C. B. . Molecular abnormalities in ovarian cancer subtypes other than high-grade serous carcinoma. J. Oncol. 2010, 740968 (2010).
Article Google Scholar - Risch, H. A., Marrett, L. D., Jain, M. & Howe, G. R. . Differences in risk factors for epithelial ovarian cancer by histologic type. Results of a case-control study. Am. J. Epidemiol. 144, 363–372 (1996).
Article CAS Google Scholar - Crum, C. P. et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr. Opin. Obstet. Gynecol. 19, 3–9 (2007).
Article Google Scholar - Gounaris, I., Charnock-Jones, D. S. & Brenton, J. D. . Ovarian clear cell carcinoma—bad endometriosis or bad endometrium? J. Pathol. 225, 157–160 (2011).
Article CAS Google Scholar - Kobel, M. et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 5, e232 (2008).
Article Google Scholar - Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007).
Article CAS Google Scholar - Skirnisdottir, I., Seidal, T., Karlsson, M. G. & Sorbe, B. . Clinical and biological characteristics of clear cell carcinomas of the ovary in FIGO stages I-II. Int. J. Oncol. 26, 177–183 (2005).
PubMed Google Scholar - Bell, J. T. et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 12, R10 (2011).
Article CAS Google Scholar - Hitchins, M. P. et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5' UTR. Cancer Cell 20, 200–213 (2011).
Article CAS Google Scholar - Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Article CAS Google Scholar - Sankararaman, S., Sridhar, S., Kimmel, G. & Halperin, E. . Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 82, 290–303 (2008).
Article CAS Google Scholar - Stram, D. O. et al. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum. Hered. 55, 179–190 (2003).
Article Google Scholar - Ku, M. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 (2008).
Article Google Scholar - Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Article ADS CAS Google Scholar
Acknowledgements
We thank all the individuals who took part in this study and all the researchers, clinicians and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, P.M. Webb, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissière and F Robidoux (Génome Québec); U. Eilber and T. Koehler (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T.-W. Park-Simon, K. Beer-Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WY (NHS); L. Paddock, M. King, U. Chandran, A. Samoila and Y. Bensman (NJO); M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini, P. Harrington, the SEARCH team and ECRIC (SEA); the Scottish Gynaecological Clinical Trials group and SCOTROC1 investigators (SRO); W.-H. Chow and Y.-T. Gao (SWH); I. Jacobs, M. Widschwendter, N. Balogun, A. Ryan and J. Ford (UKO); and Carole Pye (UKR). The Collaborative Oncological Gene-Environment Study (COGS) project is funded through a European Commission’s Seventh Framework Programme grant (agreement number 223175–HEALTH-F2-2009-223175). The Ovarian Cancer Association Consortium (OCAC) is supported by a grant from the Ovarian Cancer Research Fund, thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding of this project were (in part) supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network: a NCI Cancer Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. Funding for the project was provided by the Wellcome Trust under award 076113. The results published here are in part based upon data generated by The Cancer Genome Atlas Pilot Project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA, and the investigators and institutions who constitute the TCGA research network, can be found at http://cancergenome.nih.gov/. G.C.T. is a Senior Principal Research Fellow of the National Health and Medical Research Council, Australia. D.F.E. is a Principal Research Fellow of Cancer Research UK. P.A.F. is supported by the Deutsche Krebshilfe. B.K. holds an American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN). L.E.K. is supported by a Canadian Institutes of Health Research Investigator award (MSH-87734). H.S.1 is supported by a National Institutes of Health training grant (T32GM067587), ‘Training in Cellular, Biochemical and Molecular Sciences’. Funding of the constituent studies was provided by the American Cancer Society (CRTG-00-196-01-CCE); the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes for Health Research (MOP-86727); Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124, C536/A13086 and C536/A6689); the Celma Mastry Ovarian Cancer Foundation; the Danish Cancer Society (94-222-52); the Norwegian Cancer Society, Helse Vest, the Norwegian Research Council, ELAN Funds of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Imperial Experimental Cancer Research Centre (C1312/A15589); the Ovarian Cancer Research Fund (PPD/USC.06); Nationaal Kankerplan of Belgium; Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA143047, K07-CA80668, K22-CA138563, N01-CN55424, N01-PC067001, N01-PC035137, P01-CA017054, P01-CA087696, P50-CA105009, P50-CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063682, R01-CA064277, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA076016, R01-CA080742, R01-CA080978, R01-CA087538, R01-CA092044, R01-095023, R01-CA106414, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA149429, R01-CA141154, R03-CA113148, R03-CA115195, R37-CA070867, R37-CA70867, U01-CA069417, U01-CA071966, P30-CA15083, R01CA83918, U24 CA143882 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17-98-1-8659, DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669 and W81XWH-10-1-0280); the National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01 GB 9401); the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Phi Beta Psi Sorority; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital ‘Womens Health Theme’ and the Royal Marsden Hospital; and WorkSafeBC. We acknowledge the contributions of Kyriaki Michailidou, Ali Amin Al Olama and Karoline Kuchenbaecker to the iCOGS statistical analyses and Shahana Ahmed, Melanie J. Maranian and Catherine S Healey for their contributions to the iCOGS genotyping quality-control process. US National Health Institute/National Center for Research Resources/General Clinical Research Center M01- RR000056.
Author information
Author notes
- Hui Shen, Brooke L. Fridley and Honglin Song: These authors contributed equally to this work.
Authors and Affiliations
- USC Epigenome Center, Keck School of Medicine, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, 90033, California, USA
Hui Shen & Peter W. Laird - Department of Biostatistics, University of Kansas Medical Center, Kansas City, 66160, Kansas, USA
Brooke L. Fridley - Department of Oncology, University of Cambridge, Cambridge, CB1 8RN, UK
Honglin Song, Jonathan Tyrer, Douglas F. Easton & Paul D.P. Pharoah - Department of Preventive Medicine, Keck School of Medicine, University of Southern California Norris Comprehensive Cancer Center, Los Angeles, 90033, California, USA
Kate Lawrenson, Susan J. Ramus, Douglas Stram, Anna H. Wu, David Van Den Berg, Daniel O. Stram, Malcolm C. Pike, Alice W. Lee, Rod Karevan, Simon A. Gayther & Celeste Leigh Pearce - Division of Experimental Pathology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, 55905, Minnesota, USA
Julie M. Cunningham & Viji Shridhar - Division of Epidemiology, Department of Health Science Research, Mayo Clinic, Rochester, 55905, Minnesota, USA
Mine S. Cicek & Ellen L. Goode - Division of Biomedical Statistics and Informatics, Department of Health Science Research, Mayo Clinic, Rochester, 55905, Minnesota, USA
Melissa C. Larson, Robert A. Vierkant & Sebastian M. Armasu - Department of Pathology and Laboratory Medicine, Calgary Laboratory Services, University of Calgary, Calgary, T2N 2T9, Alberta, Canada
Martin Köbel - Department of Epidemiology, Center for Cancer Genetics Research and Prevention, School of Medicine, University of California Irvine, Irvine, 92697, California, USA
Argyrios Ziogas, Xiao-Ou Shu & Hoda Anton-Culver - Vanderbilt Epidemiology Center, Vanderbilt-Ingram Cancer Center, Vanderbilt University School of Medicine, Vanderbilt University, Nashville, 37232, Tennessee, USA
Wei Zheng - Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, 20892, Maryland, USA
Hannah P. Yang, Nicolas Wentzensen & Louise A. Brinton - Gynaecological Cancer Research Centre, UCL EGA Institute for Women's Health, London, NW1 2BU, UK
Eva L. Wozniak, Usha Menon & Aleksandra Gentry-Maharaj - Department of Obstetrics and Gynaecology, Faculty of Medicine, Affiliated to UM Cancer Research Institute, University of Malaya, Kuala Lumpur, 59100, Malaysia
Yin Ling Woo, Siti Zawiah Omar & Noor Azmi Mat Adenan - Department of Obstetrics and Gynecology, Mayo Clinic, Rochester, 55905, Minnesota, USA
Boris Winterhoff - Department of Gynecology and Obstetrics, Haukeland University Hospital, Bergen, HB 5006, Norway
Elisabeth Wik, Helga B. Salvesen, Camilla Krakstad & Mari K. Halle - Department of Clinical Science, University of Bergen, Bergen, HB 5006, Norway
Elisabeth Wik, Helga B. Salvesen, Camilla Krakstad & Mari K. Halle - Department of Health Research and Policy, Stanford University School of Medicine, Stanford, 94305, California, USA
Alice S. Whittemore, Weiva Sieh & Valerie McGuire - Department of Community and Family Medicine, Duke University Medical Center, Durham, 27708, North Carolina, USA
Rachel Palmieri Weber & Joellen M. Schildkraut - Obstetrics and Gynecology Epidemiology Center, Brigham and Women's Hospital, Harvard Medical School, Boston, 02115, Massachusetts, USA
Allison F. Vitonis, Kathryn L. Terry & Daniel W. Cramer - Génome Québec, Montréal, H3A 0G1, Québec, Canada
Daniel Vincent, Daniel C. Tessier & François Bacot - Vesalius Research Center, VIB, Leuven, 3000, Belgium
Ignace Vergote, Sandrina Lambrechts, Diether Lambrechts & Evelyn Despierre - Division of Gynecologic Oncology, Department of Obstetrics and Gynaecology and Leuven Cancer Institute, University Hospitals Leuven, Leuven, 3000, Belgium
Ignace Vergote, Sandrina Lambrechts & Evelyn Despierre - Department of Gynaecology, Radboud University Medical Centre, Nijmegen, HB 6500, The Netherlands
Anne M. Van Altena & Leon F.A.G Massuger - Channing Division of Network Medicine, Harvard Medical School, Brigham and Women's Hospital, Boston, 02115, Massachusetts, USA
Shelley S. Tworoger & Elizabeth M. Poole - Department of Epidemiology, Harvard School of Public Health, Boston, 02115, Massachusetts, USA
Shelley S. Tworoger, Kathryn L. Terry, Elizabeth M. Poole & Daniel W. Cramer - Cancer Epidemiology Program, University of Hawaii Cancer Center, Hawaii, 96813, USA
Pamela J. Thompson, Yurii B. Shvetsov & Galina Lurie - Cancer Research Initiatives Foundation, Sime Darby Medical Centre, Subang Jaya, 47500, Malaysia
Soo-Hwang Teo - University Malaya Cancer Research Institute, University Malaya Medical Centre, University of Malaya, Kuala Lumpur, 59100, Malaysia
Soo-Hwang Teo - Department of Obstetrics and Gynecology, Keck School of Medicine, University of Southern California, Los Angeles, 90033, California, USA
Claire Templeman & Doerthe Brueggmann - Department of Pathology, Genetic Epidemiology Laboratory, University of Melbourne, Melbourne, VIC 3053, Victoria, Australia
Melissa C. Southey - Department of Gynaecological Oncology, Glasgow Royal Infirmary, Glasgow, G4 0SF, UK
Nadeem Siddiqui - Department of Obstetrics and Gynecology, University of Ulm, Ulm, 89091, Germany
Shan Wang-Gohrke - Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, VIC 3053, Victoria, Australia
Gianluca Severi, Graham G. Giles & Laura Baglietto - Centre for Molecular, Environmental, Genetic and Analytical Epidemiology, University of Melbourne, Melbourne, VIC 3010, Victoria, Australia
Gianluca Severi, Graham G. Giles & Laura Baglietto - Institut für Humangenetik Wiesbaden, Wiesbaden, 65187, Germany
Ira Schwaab - Department of Molecular Pathology, Maria Sklodowska-Curie Memorial Cancer Center, Institute of Oncology, Warsaw, 02-781, Poland
Iwona K. Rzepecka, Joanna Moes-Sosnowska, Jolanta Kupryjanczyk & Agnieszka Dansonka-Mieszkowska - Department of Gynecology and Obstetrics, Jena University Hospital, Jena, 07743, Germany
Ingo B. Runnebaum & Matthias Dürst - Division of Public Health Sciences, Program in Epidemiology, Fred Hutchinson Cancer Research Center, Seattle, 98109, Washington, USA
Mary Anne Rossing - Department of Epidemiology, University of Washington, Seattle, 98109, Washington, USA
Mary Anne Rossing - Cancer Institute of New Jersey, Robert Wood Johnson Medical School, New Brunswick, 08901, New Jersey, USA
Lorna Rodriguez-Rodriguez & Elisa V. Bandera - Department of Chronic Disease Epidemiology, Yale School of Public Health, New Haven, 06520, Connecticut, USA
Harvey A. Risch - Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center, Erlangen, 91054, Germany
Stefan P. Renner, Peter A. Fasching & Matthias W. Beckmann - Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, 10065, New York, USA
Malcolm C. Pike, Irene Orlow & Sara H. Olson - Division of Population Sciences, Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, 33612, Florida, USA
Catherine M. Phelan, Alvaro N.A. Monteiro & Thomas A. Sellers - Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Central Hospital, Helsinki, 00530, Finland
Liisa M. Pelttari, Heli Nevanlinna, Arto Leminen & Ralf Butzow - Department of Obstetrics and Gynecology, Oregon Health and Science University, Portland, 97239, Oregon, USA
Tanja Pejovic - Knight Cancer Institute, Oregon Health and Science University, Portland, 97239, Oregon, USA
Tanja Pejovic - Beatson West of Scotland Cancer Centre, Glasgow, G12 0YN, UK
James Paul - Department of Gynecologic Oncology, Roswell Park Cancer Institute, Buffalo, 14263, New York, USA
Kunle Odunsi - Division of Cancer Epidemiology, German Cancer Research Center, Heidelberg, 69120, Germany
Stefan Nickels, Rebecca Hein & Jenny Chang-Claude - School of Public Health, University of Texas, Houston, 77030, Texas, USA
Roberta B. Ness - Women’s College Research Institute, University of Toronto, Toronto, M5G IN8, Ontario, Canada
Steven A. Narod - Department of Gynecologic Oncology, Aichi Cancer Center Central Hospital, Nagoya, 464-8681, Japan
Toru Nakanishi - Department of Cancer Prevention and Control, Roswell Park Cancer Institute, Buffalo, 14263, New York, USA
Kirsten B. Moysich - Department of Obstetrics, Gynecology and Reproductive Sciences, University of Pittsburgh, Pittsburgh, 15213, Pennsylvania, USA
Francesmary Modugno - Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, 15213, Pennsylvania, USA
Francesmary Modugno & Clareann H. Bunker - Women’s Cancer Research Program, Magee-Womens Research Institute, University of Pittsburgh Cancer Institute, Pittsburgh, 15213, Pennsylvania, USA
Francesmary Modugno - Dalla Lana School of Public Health, Faculty of Medicine, University of Toronto, Toronto, M5T 3M7, Ontario, Canada
John R. McLaughlin - Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, M5G IX5, Ontario, Canada
John R. McLaughlin - Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, 464-8681, Japan
Keitaro Matsuo & Satoyo Hosono - Gynecologic Clinic, The Juliane Marie Center, Copenhagen University Hospital, Copenhagen, DK-2100, Denmark
Lene Lundvall, Susanne Krüger Kjaer & Claus K. Høgdall - Department of Genetics and Pathology, International Hereditary Cancer Center, Pomeranian Medical University, Szczecin, 70-115, Poland
Jan Lubiński, Anna Jakubowska, Jacek Gronwald & Cezary Cybulski - Department of Cancer Epidemiology and Prevention, Maria Sklodowska-Curie Memorial Cancer Center, Warsaw, 02-781, Poland
Jolanta Lissowska - Department of Surgery, Gynecology Service, Memorial Sloan-Kettering Cancer Center, New York, 10021, New York, USA
Douglas A. Levine - Cancer Control Research, BC Cancer Agency, Vancouver, G12 0YN, British Columbia, Canada
Nhu D. Le - Department of Oncology, Laboratory for Translational Genetics, University of Leuven, Leuven, 3000, Belgium
Sandrina Lambrechts, Diether Lambrechts & Evelyn Despierre - Division of Hematology and Oncology, Department of Medicine, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, 90095, California, USA
Gottfried E. Konecny & Peter A. Fasching - Virus, Lifestyle and Genes, Danish Cancer Society Research Center, Copenhagen, DK-2100, Denmark
Susanne Krüger Kjaer, Allan Jensen & Estrid Høgdall - Department of Epidemiology, Biostatistics and HTA, Radboud University Medical Centre, Nijmegen, HB 6500, Netherlands
Lambertus A. Kiemeney & Katja K. Aben - Department of Urology, Radboud University Medical Centre, Nijmegen, HB 6500, The Netherlands
Lambertus A. Kiemeney - Comprehensive Cancer Center, Utrecht, 1066CX, The Netherlands
Lambertus A. Kiemeney & Katja K. Aben - Department of Population Health Research, Alberta Health Services-Cancer Care, Calgary, T2N 2T9, Alberta, Canada
Linda E. Kelemen - Department of Medical Genetics and Oncology, University of Calgary, Calgary, T2N 2T9, Alberta, Canada
Linda E. Kelemen - Division of Anatomic Pathology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, 55905, Minnesota, USA
Gary L. Keeney - Women’s Cancer Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, 90048, California, USA
Beth Y. Karlan - Department of Medical Oncology, Mayo Clinic, Rochester, 55905, Minnesota, USA
Kimberly R. Kalli & Matthew S. Block - Department of Obstetrics and Gynecology, Nagoya University Graduate School of Medicine, Nagoya, 464-8601, Japan
Hiroaki Kajiyama - Division of Cancer Etiology and Genetics, National Cancer Institute, Bethesda, 20892, Maryland, USA
Bu-Tian Ji - Department of Statistical Science, Duke University, Durham, 27708, North Carolina, USA
Edwin Iversen - Department of Pathology, Molecular Unit, Herlev Hospital, University of Copenhagen, Copenhagen, 2730, Denmark
Estrid Høgdall - Department of Biochemistry and Molecular Biology, Oregon Health and Science University, Portland, 97239, Oregon, USA
Maureen Hoatlin - Clinics of Obstetrics and Gynaecology, Hannover Medical School, Hannover, 30625, Germany
Peter Hillemanns - Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte, Essen, 45136, Germany
Florian Heitz, Philipp Harter & Andreas du Bois - Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Klinik, Wiesbaden, 65199, Germany
Florian Heitz, Philipp Harter & Andreas du Bois - Department of Child and Adolescent Psychiatry and Psychotherapy, PMV Research Group, University of Cologne, Cologne, 50923, Germany
Rebecca Hein - Department of Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, 171-77, Sweden
Per Hall - Gynecological Oncology Unit, Royal Marsden Hospital, London, SW3 6JJ, UK
Martin Gore - Cedars Sinai Medical Center, Samuel Oschin Comprehensive Cancer Center Institute, Los Angeles, 90048, California, USA
Marc T. Goodman - Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, VIC 3806, Victoria, Australia
Graham G. Giles - Division of Genetics and Epidemiology, Breakthrough Breast Cancer Research Centre, Institute of Cancer Research, London, SW7 3RP, UK
Montserrat Garcia-Closas - Department of Surgery and Cancer, Imperial College London, London, SW7 2AZ, UK
James M. Flanagan & Robert Brown - Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Erlangen, 91054, Germany
Arif B. Ekici - Maggee Women’s Hospital, Pittsburg, 15213, Pennsylvania, USA
Robert Edwards - Faculty of Medicine, University of Southampton, University Hospital Southampton, Southampton, S017 1BJ, UK
Diana Eccles - Gynaecology Research Unit, Hannover Medical School, Hannover, 30625, Germany
Thilo Dörk & Natalia Bogdanova - Section of Biostatistics and Epidemiology, Geisel School of Medicine at Dartmouth, Lebanon, 03755, New Hampshire, USA
Jennifer A. Doherty - Division of Epidemiology and Biostatistics, Department of Internal Medicine, University of New Mexico, Albuquerque, 87131, New Mexico, USA
Linda S. Cook & Bridget Charbonneau - Department of Genetics, Queensland Institute of Medical Research, Herston, QLD 4006, Queensland, Australia
Xiaoqing Chen, Jonathan Beesley & Georgia Chenevix-Trench - Research Division, Peter MacCallum Cancer Centre, Cancer Genetics Laboratory, Melbourne, VIC 3002, Victoria, Australia
Ian Campbell - Department of Pathology, University of Melbourne, Parkville, VIC 3053, Victoria, Australia
Ian Campbell - Sir Peter MacCallum Department of Oncology, University of Melbourne, Parkville, VIC 3002, Victoria, Australia
Ian Campbell - Department of Pathology, Helsinki University Central Hospital, Helsinki, 00530, Finland
Ralf Butzow - Genome Sciences Centre, BC Cancer Agency, Vancouver, V52 1L3, British Columbia, Canada
Angela Brooks-Wilson - Department of Pathology, Cancer Institute, University College London, London, WC1E 6JJ, UK
Elizabeth Benjamin - Belarusian Institute for Oncology and Medical Radiology Aleksandrov N.N., Minsk, 223040, Belarus
Natalia Antonenkova - College of Pharmacy and Health Sciences, Texas Southern University, Houston, 77044, Texas, USA
Dong Liang - Department of Epidemiology, University of Texas MD Anderson Cancer Center, Houston, 77030, Texas, USA
Xifeng Wu & Michelle A.T. Hildebrandt - Department of Gynecologic Oncology, University of Texas MD Anderson Cancer Center, Houston, 77030, Texas, USA
Karen Lu - Cancer Prevention, Detection and Control Research Program, Duke Cancer Institute, Durham, 27708, North Carolina, USA
Joellen M. Schildkraut - Department of Pathology, Vancouver General Hospital, BC Cancer Agency, Vancouver, V5Z 4E6, British Columbia, Canada
David Huntsman - Gynecologic Cancer Program, Duke Cancer Institute, Durham, 27708, North Carolina, USA
Andrew Berchuck - Department of Public Health and Primary Care, University of Cambridge, Cambridge, CB1 8RN, UK
Paul D.P. Pharoah
Authors
- Hui Shen
You can also search for this author inPubMed Google Scholar - Brooke L. Fridley
You can also search for this author inPubMed Google Scholar - Honglin Song
You can also search for this author inPubMed Google Scholar - Kate Lawrenson
You can also search for this author inPubMed Google Scholar - Julie M. Cunningham
You can also search for this author inPubMed Google Scholar - Susan J. Ramus
You can also search for this author inPubMed Google Scholar - Mine S. Cicek
You can also search for this author inPubMed Google Scholar - Jonathan Tyrer
You can also search for this author inPubMed Google Scholar - Douglas Stram
You can also search for this author inPubMed Google Scholar - Melissa C. Larson
You can also search for this author inPubMed Google Scholar - Martin Köbel
You can also search for this author inPubMed Google Scholar - Argyrios Ziogas
You can also search for this author inPubMed Google Scholar - Wei Zheng
You can also search for this author inPubMed Google Scholar - Hannah P. Yang
You can also search for this author inPubMed Google Scholar - Anna H. Wu
You can also search for this author inPubMed Google Scholar - Eva L. Wozniak
You can also search for this author inPubMed Google Scholar - Yin Ling Woo
You can also search for this author inPubMed Google Scholar - Boris Winterhoff
You can also search for this author inPubMed Google Scholar - Elisabeth Wik
You can also search for this author inPubMed Google Scholar - Alice S. Whittemore
You can also search for this author inPubMed Google Scholar - Nicolas Wentzensen
You can also search for this author inPubMed Google Scholar - Rachel Palmieri Weber
You can also search for this author inPubMed Google Scholar - Allison F. Vitonis
You can also search for this author inPubMed Google Scholar - Daniel Vincent
You can also search for this author inPubMed Google Scholar - Robert A. Vierkant
You can also search for this author inPubMed Google Scholar - Ignace Vergote
You can also search for this author inPubMed Google Scholar - David Van Den Berg
You can also search for this author inPubMed Google Scholar - Anne M. Van Altena
You can also search for this author inPubMed Google Scholar - Shelley S. Tworoger
You can also search for this author inPubMed Google Scholar - Pamela J. Thompson
You can also search for this author inPubMed Google Scholar - Daniel C. Tessier
You can also search for this author inPubMed Google Scholar - Kathryn L. Terry
You can also search for this author inPubMed Google Scholar - Soo-Hwang Teo
You can also search for this author inPubMed Google Scholar - Claire Templeman
You can also search for this author inPubMed Google Scholar - Daniel O. Stram
You can also search for this author inPubMed Google Scholar - Melissa C. Southey
You can also search for this author inPubMed Google Scholar - Weiva Sieh
You can also search for this author inPubMed Google Scholar - Nadeem Siddiqui
You can also search for this author inPubMed Google Scholar - Yurii B. Shvetsov
You can also search for this author inPubMed Google Scholar - Xiao-Ou Shu
You can also search for this author inPubMed Google Scholar - Viji Shridhar
You can also search for this author inPubMed Google Scholar - Shan Wang-Gohrke
You can also search for this author inPubMed Google Scholar - Gianluca Severi
You can also search for this author inPubMed Google Scholar - Ira Schwaab
You can also search for this author inPubMed Google Scholar - Helga B. Salvesen
You can also search for this author inPubMed Google Scholar - Iwona K. Rzepecka
You can also search for this author inPubMed Google Scholar - Ingo B. Runnebaum
You can also search for this author inPubMed Google Scholar - Mary Anne Rossing
You can also search for this author inPubMed Google Scholar - Lorna Rodriguez-Rodriguez
You can also search for this author inPubMed Google Scholar - Harvey A. Risch
You can also search for this author inPubMed Google Scholar - Stefan P. Renner
You can also search for this author inPubMed Google Scholar - Elizabeth M. Poole
You can also search for this author inPubMed Google Scholar - Malcolm C. Pike
You can also search for this author inPubMed Google Scholar - Catherine M. Phelan
You can also search for this author inPubMed Google Scholar - Liisa M. Pelttari
You can also search for this author inPubMed Google Scholar - Tanja Pejovic
You can also search for this author inPubMed Google Scholar - James Paul
You can also search for this author inPubMed Google Scholar - Irene Orlow
You can also search for this author inPubMed Google Scholar - Siti Zawiah Omar
You can also search for this author inPubMed Google Scholar - Sara H. Olson
You can also search for this author inPubMed Google Scholar - Kunle Odunsi
You can also search for this author inPubMed Google Scholar - Stefan Nickels
You can also search for this author inPubMed Google Scholar - Heli Nevanlinna
You can also search for this author inPubMed Google Scholar - Roberta B. Ness
You can also search for this author inPubMed Google Scholar - Steven A. Narod
You can also search for this author inPubMed Google Scholar - Toru Nakanishi
You can also search for this author inPubMed Google Scholar - Kirsten B. Moysich
You can also search for this author inPubMed Google Scholar - Alvaro N.A. Monteiro
You can also search for this author inPubMed Google Scholar - Joanna Moes-Sosnowska
You can also search for this author inPubMed Google Scholar - Francesmary Modugno
You can also search for this author inPubMed Google Scholar - Usha Menon
You can also search for this author inPubMed Google Scholar - John R. McLaughlin
You can also search for this author inPubMed Google Scholar - Valerie McGuire
You can also search for this author inPubMed Google Scholar - Keitaro Matsuo
You can also search for this author inPubMed Google Scholar - Noor Azmi Mat Adenan
You can also search for this author inPubMed Google Scholar - Leon F.A.G Massuger
You can also search for this author inPubMed Google Scholar - Galina Lurie
You can also search for this author inPubMed Google Scholar - Lene Lundvall
You can also search for this author inPubMed Google Scholar - Jan Lubiński
You can also search for this author inPubMed Google Scholar - Jolanta Lissowska
You can also search for this author inPubMed Google Scholar - Douglas A. Levine
You can also search for this author inPubMed Google Scholar - Arto Leminen
You can also search for this author inPubMed Google Scholar - Alice W. Lee
You can also search for this author inPubMed Google Scholar - Nhu D. Le
You can also search for this author inPubMed Google Scholar - Sandrina Lambrechts
You can also search for this author inPubMed Google Scholar - Diether Lambrechts
You can also search for this author inPubMed Google Scholar - Jolanta Kupryjanczyk
You can also search for this author inPubMed Google Scholar - Camilla Krakstad
You can also search for this author inPubMed Google Scholar - Gottfried E. Konecny
You can also search for this author inPubMed Google Scholar - Susanne Krüger Kjaer
You can also search for this author inPubMed Google Scholar - Lambertus A. Kiemeney
You can also search for this author inPubMed Google Scholar - Linda E. Kelemen
You can also search for this author inPubMed Google Scholar - Gary L. Keeney
You can also search for this author inPubMed Google Scholar - Beth Y. Karlan
You can also search for this author inPubMed Google Scholar - Rod Karevan
You can also search for this author inPubMed Google Scholar - Kimberly R. Kalli
You can also search for this author inPubMed Google Scholar - Hiroaki Kajiyama
You can also search for this author inPubMed Google Scholar - Bu-Tian Ji
You can also search for this author inPubMed Google Scholar - Allan Jensen
You can also search for this author inPubMed Google Scholar - Anna Jakubowska
You can also search for this author inPubMed Google Scholar - Edwin Iversen
You can also search for this author inPubMed Google Scholar - Satoyo Hosono
You can also search for this author inPubMed Google Scholar - Claus K. Høgdall
You can also search for this author inPubMed Google Scholar - Estrid Høgdall
You can also search for this author inPubMed Google Scholar - Maureen Hoatlin
You can also search for this author inPubMed Google Scholar - Peter Hillemanns
You can also search for this author inPubMed Google Scholar - Florian Heitz
You can also search for this author inPubMed Google Scholar - Rebecca Hein
You can also search for this author inPubMed Google Scholar - Philipp Harter
You can also search for this author inPubMed Google Scholar - Mari K. Halle
You can also search for this author inPubMed Google Scholar - Per Hall
You can also search for this author inPubMed Google Scholar - Jacek Gronwald
You can also search for this author inPubMed Google Scholar - Martin Gore
You can also search for this author inPubMed Google Scholar - Marc T. Goodman
You can also search for this author inPubMed Google Scholar - Graham G. Giles
You can also search for this author inPubMed Google Scholar - Aleksandra Gentry-Maharaj
You can also search for this author inPubMed Google Scholar - Montserrat Garcia-Closas
You can also search for this author inPubMed Google Scholar - James M. Flanagan
You can also search for this author inPubMed Google Scholar - Peter A. Fasching
You can also search for this author inPubMed Google Scholar - Arif B. Ekici
You can also search for this author inPubMed Google Scholar - Robert Edwards
You can also search for this author inPubMed Google Scholar - Diana Eccles
You can also search for this author inPubMed Google Scholar - Douglas F. Easton
You can also search for this author inPubMed Google Scholar - Matthias Dürst
You can also search for this author inPubMed Google Scholar - Andreas du Bois
You can also search for this author inPubMed Google Scholar - Thilo Dörk
You can also search for this author inPubMed Google Scholar - Jennifer A. Doherty
You can also search for this author inPubMed Google Scholar - Evelyn Despierre
You can also search for this author inPubMed Google Scholar - Agnieszka Dansonka-Mieszkowska
You can also search for this author inPubMed Google Scholar - Cezary Cybulski
You can also search for this author inPubMed Google Scholar - Daniel W. Cramer
You can also search for this author inPubMed Google Scholar - Linda S. Cook
You can also search for this author inPubMed Google Scholar - Xiaoqing Chen
You can also search for this author inPubMed Google Scholar - Bridget Charbonneau
You can also search for this author inPubMed Google Scholar - Jenny Chang-Claude
You can also search for this author inPubMed Google Scholar - Ian Campbell
You can also search for this author inPubMed Google Scholar - Ralf Butzow
You can also search for this author inPubMed Google Scholar - Clareann H. Bunker
You can also search for this author inPubMed Google Scholar - Doerthe Brueggmann
You can also search for this author inPubMed Google Scholar - Robert Brown
You can also search for this author inPubMed Google Scholar - Angela Brooks-Wilson
You can also search for this author inPubMed Google Scholar - Louise A. Brinton
You can also search for this author inPubMed Google Scholar - Natalia Bogdanova
You can also search for this author inPubMed Google Scholar - Matthew S. Block
You can also search for this author inPubMed Google Scholar - Elizabeth Benjamin
You can also search for this author inPubMed Google Scholar - Jonathan Beesley
You can also search for this author inPubMed Google Scholar - Matthias W. Beckmann
You can also search for this author inPubMed Google Scholar - Elisa V. Bandera
You can also search for this author inPubMed Google Scholar - Laura Baglietto
You can also search for this author inPubMed Google Scholar - François Bacot
You can also search for this author inPubMed Google Scholar - Sebastian M. Armasu
You can also search for this author inPubMed Google Scholar - Natalia Antonenkova
You can also search for this author inPubMed Google Scholar - Hoda Anton-Culver
You can also search for this author inPubMed Google Scholar - Katja K. Aben
You can also search for this author inPubMed Google Scholar - Dong Liang
You can also search for this author inPubMed Google Scholar - Xifeng Wu
You can also search for this author inPubMed Google Scholar - Karen Lu
You can also search for this author inPubMed Google Scholar - Michelle A.T. Hildebrandt
You can also search for this author inPubMed Google Scholar - Joellen M. Schildkraut
You can also search for this author inPubMed Google Scholar - Thomas A. Sellers
You can also search for this author inPubMed Google Scholar - David Huntsman
You can also search for this author inPubMed Google Scholar - Andrew Berchuck
You can also search for this author inPubMed Google Scholar - Georgia Chenevix-Trench
You can also search for this author inPubMed Google Scholar - Simon A. Gayther
You can also search for this author inPubMed Google Scholar - Paul D.P. Pharoah
You can also search for this author inPubMed Google Scholar - Peter W. Laird
You can also search for this author inPubMed Google Scholar - Ellen L. Goode
You can also search for this author inPubMed Google Scholar - Celeste Leigh Pearce
You can also search for this author inPubMed Google Scholar
Consortia
PRACTICAL Consortium
Australian Ovarian Cancer Study Group
Australian Cancer Study
Contributions
H. Shen, B.L.F., H. Song, K.L., M.K., G.C.T., S.A.G., P.D.P.P., P.W.L., E.L.G. and C.L.P. contributed to the preparation of the manuscript. All authors read and approved the final version. H. Shen, B.L.F., M.S.C., K.L., J.T., D.S., M.C.L., M.K., P.D.P.P., P.W.L., E.L.G. and C.L.P. carried out data analysis. S.J.R. and C.M.P. collated and prepared samples for genotyping. S.A.G. and K.L. performed functional analyses.
Corresponding author
Correspondence toEllen L. Goode.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of consortium members appears in Supplementary Note 1
A list of consortium members appears in Supplementary Note 2
A list of consortium members appears in Supplementary Note 3
Supplementary information
Supplementary Information
Supplementary Figures S1-S11, Supplementary Tables S1-S2, Supplementary Notes 1-3, Supplementary Methods and Supplementary References (PDF 2792 kb)
Supplementary Data
Supplementary Data 1: Association between all non-genome-wide significant SNPs in HNF1B and risk of ovarian cancer by histological subtype in European-Whites. (XLSX 113 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit Creative Commons Attribution-Noncommercial-No Derivative 3.0 license
About this article
Cite this article
Shen, H., Fridley, B., Song, H. et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer.Nat Commun 4, 1628 (2013). https://doi.org/10.1038/ncomms2629
- Received: 28 November 2012
- Accepted: 21 February 2013
- Published: 27 March 2013
- DOI: https://doi.org/10.1038/ncomms2629