Estimating rates of alternative splicing in mammals and invertebrates (original) (raw)
- Correspondence
- Published: 01 September 2004
Nature Genetics volume 36, pages 915–916 (2004)Cite this article
- 1867 Accesses
- 75 Citations
- 3 Altmetric
- Metrics details
To the editor:
The sequencing of the human genome showed that humans have ∼30,000 genes. This finding raised the possibility that alternative splicing, rather than an increased number of expressed genomic loci, was responsible for the functional complexity of vertebrates relative to invertebrates1. It has been estimated that 40–60% of all human genes1,2,3,4 and 74% of multiexon human genes5 are alternatively spliced. These estimates do not take into account how many different alternative splice forms exist for a given gene. Brett et al. examined alternative splicing in seven species, including human, using large-scale expressed-sequence tag (EST) analysis6. They concluded that vertebrates and invertebrates had similar rates of alternative splicing, not only with respect to the proportion of the genes affected but also with respect to the number of alternative splicing forms per gene. The method they used depends on the extent of EST coverage in the underlying data sets.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
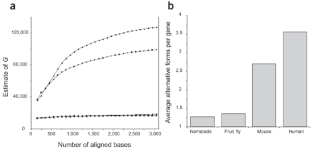
Figure 1: Extent of alternative splicing in four organisms.
References
- Modrek, B. & Lee, C. Nat. Genet. 30, 13–19 (2002).
Article CAS PubMed Google Scholar - Mironov, A.A. et al. Genome Res. 12, 1288–1293 (1999).
Article Google Scholar - Modrek, B. et al. Nucleic Acids Res. 29, 2850–2859 (2001).
Article CAS PubMed PubMed Central Google Scholar - Kan, Z. et al. Genome Res. 11, 889–900 (2001).
Article CAS PubMed PubMed Central Google Scholar - Johnson, J.M. et al. Science 302, 2141–2144 (2003).
Article CAS PubMed Google Scholar - Brett, D. et al. Nat. Genet. 30, 29–30 (2002).
Article CAS PubMed Google Scholar - Ewing, B. & Green, P. Nat. Genet. 25, 232–234 (2000).
Article CAS PubMed Google Scholar - Wheeler, D.L. et al. Nucleic Acids Res. 31, 28–33 (2003).
Article CAS PubMed PubMed Central Google Scholar - Pruitt, K.D. & Maglott, D.R. Nucleic Acids Res. 29, 137–140 (2001).
Article CAS PubMed PubMed Central Google Scholar - Quackenbush, J. et al. Nucleic Acids Res. 28, 141–145 (2000).
Article CAS PubMed PubMed Central Google Scholar
Acknowledgements
This work was supported by a grant from BioGreen 21 Program of the Korean Rural Development Administration (to H.K.), by the Brain Korea 21 Project of the Ministry of Education (to H.K.), by a grant from the US National Human Genome Research Institute (to J.O.) and by a grant from the US National Institute of Mental Health (to J.O.).
Author information
Authors and Affiliations
- School of Agricultural Biotechnology, Seoul National University San 56-1, Sillim-dong, Gwanak-gu, Seoul, 151-742, Korea
Heebal Kim - Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, 10021, New York, USA
Robert Klein, Jacek Majewski & Jurg Ott
Authors
- Heebal Kim
- Robert Klein
- Jacek Majewski
- Jurg Ott
Corresponding author
Correspondence toHeebal Kim.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, H., Klein, R., Majewski, J. et al. Estimating rates of alternative splicing in mammals and invertebrates.Nat Genet 36, 915–916 (2004). https://doi.org/10.1038/ng0904-915
- Issue Date: 01 September 2004
- DOI: https://doi.org/10.1038/ng0904-915