Human papilloma virus is associated with breast cancer (original) (raw)
Main
Although the association of human papilloma virus (HPV) with cervical cancer, and head and neck cancers is well established, the involvement of the virus in breast cancer is more controversial. Previous studies have demonstrated the presence of HPV high-risk types 16, 18 and 33 in breast cancer specimens from diverse populations around the world: Italy, Norway, China, Japan, USA, Austria, Brazil, Australia, Taiwan, Turkey, Greece, Korea, Mexico, Hungary and Syria (Lonardo et al, 1992; Hennig et al, 1999; Yu et al, 1999; Liu et al, 2001; Damin et al, 2004; Widschwendter et al, 2004; de Villiers et al, 2005; Kan et al, 2005; Tsai et al, 2005; Gumus et al, 2006; Kroupis et al, 2006; Lawson et al, 2006; Choi et al, 2007; Akil et al, 2008; Khan et al, 2008; Kulka et al, 2008; Mendizabal-Ruiz et al, 2009). The prevalence of HPV positive breast cancer in these studies was reported to vary from 4% in Mexican to 86% in American women. In all studies, high-risk HPV was found in tumour tissue only and not in surrounding normal tissue, with the exception of the study from Turkey, in which the virus was also detected in normal tissue but at a lower level than in the cancer (Gumus et al, 2006). Although the route of transmission for the virus has not been determined, women positive for both breast and cervical cancers were found to be infected with the same HPV type in both tumours (Hennig et al, 1999; Widschwendter et al, 2004).
The controversy surrounding the role of HPV in breast cancer may be because of the difficulty that has been encountered in detecting the virus in breast specimens, in contrast to the relative ease of detection in cervical cancers (Lindel et al, 2007; Khan et al, 2008). Indeed, in a previous study from our group, we demonstrated that it was necessary to use SYBR Green I (Molecular Probes, Carlsbad, CA, USA) for polymerase chain reaction (PCR) detection of virus in breast cancer in DNA extracted from breast tissue (Kan et al, 2005). This is considered to be because of the fact that a considerable proportion of breast cancer specimens is non-cancerous and that the levels of virus are low in breast cancer. One solution to the detection of such low levels of HPV is the use of in situ PCR.
The oncogenic mechanisms by which HPV induces cervical cancer have been intensively studied (zur Hausen, 2002). In this study, we have used HPV-associated cervical cancer as a model. High-risk HPV encodes a series of proteins, designated as early (E1–E7) or late (L1 and L2). Although all of the viral proteins have a role in viral replication, only a small number of the viral early proteins have a role in cellular transformation. Key to transformation are the E6 and E7 oncoproteins, which work in concert to disrupt cell-cycle regulation, inhibit apoptosis and stimulate cell-cycle progression by binding/inhibiting the p53 and p110RB tumour suppressor genes, respectively. In addition, and relevant to this study, HPV E5 and E6 act early in transformation (before integration) and are known to disrupt cytokeratin causing perinuclear cytoplasmic clearing and nuclear enlargement, which leads to the appearance of a koilocyte (Krawczyk et al, 2008; Thomison et al, 2008).
If HPV is oncogenic in human breast cancer, we hypothesise that high-risk HPV should be present in (some) human breast cancers and in some normal and pre-cancerous tissue (although at a lower proportion), and that koilocytosis should be apparent in HPV-associated breast cancers, and that HPV will be detected in some proportion of breast cancer cell lines.
In this paper we report on the presence of HPV in breast cancer cell lines, in the nuclei of cells within the cancerous regions of breast cancer specimens and correlate the presence of HPV with the histopathological features of HPV-induced transformation.
Materials and methods
Archival specimens
Unselected formalin-fixed breast cancer specimens and non-cancer specimens from women who had breast reduction surgery, were analysed using in situ PCR. All the specimens were from women living in Australia. A total of 28 breast cancer specimens and 28 unselected non-cancerous breast specimens were suitable for use in this in situ PCR study. Fifteen of the fixed specimens were ductal carcinoma in situ (dcis) and 13 were predominantly invasive ductal carcinomas (idc).
Breast cancer cell lines
Standard PCR techniques were used to determine if HPV DNA genetic material was present in the human breast cancer cell lines: MCF-7, T47D, BT-549, HBL-100, Hs578.T, MDA-MB-453, MDA-MB-468, MDA-MB-175-VII and SK-BR-3.
In situ PCR
Four-micron thick sections of breast tumour tissue were cut and placed on silanised slides. Positive controls (known virus-positive cervical cancer tissue), and negative controls (known virus-negative tissue), and controls omitting DIG-11-dUTP (to confirm incorporation of DIG label), omitting primers (to confirm the signal was the result of specific amplification rather than self priming of degraded tissues), or omitting Taq polymerase, were also undertaken for each specimen. The primers used for HPV in situ PCR for the L1 region of HPV-16 and HPV-18 were forward (5′-GCMCAGGGWCATAAYAATGG-3′) and reverse (5′-CGTCCMARRGGAWACTGATC-3′). The outcomes were assessed by the staining of the DIG label in the tissues by light microscopy.
A number of specimens gave a false-positive signal when the primers were omitted. The use of exo-zap, initial filling in with Klenow, and doing two rounds of PCR – the first with no DIG-11-dUTP and the second with DIG-11-dUTP – gave the same false-positive results with these particular specimens. Consequently, we carried out a (no-primer) negative control for each sample at the same time as the in situ screening PCR and eliminated the specimens that gave false-positive colour signals. Any specimens that produced a signal in the in situ PCR negative controls (omission of the DNA primers and separately, omission of the Taq polymerase) were eliminated from the study. False positives indicated that the DNA was self-priming and were unsuitable for in situ work (this is probably because of fragmented DNA, caused by formalin fixation, acting as primers). In addition, tissues were screened by in situ PCR for _β_-globin to confirm their suitability for this study.
To confirm the validity of the outcomes of in situ PCR analyses, DNA was extracted from the same formalin-fixed specimens and standard PCR analyses were conducted. The methods used are outlined below. The products were sequenced and the identity of the sequences was determined using the BLAST alignment system.
Standard PCR
Genomic DNA preparation
Previously described protocols were used to extract genomic DNA from the breast cancer specimens (Dawkins et al, 1993). The DNA quality was tested by the amplification of a 268-bp fragment of the _β_-globin gene using HotStarTaq DNA polymerase (Qiagen, Dusseldorf, Germany) and primers G073 (5′-GAAGAGCCAAGGACAGGTAC-3′) and G074 (5′-CAACTTCATCCACGTTCACC-3′). The cycling conditions were 95°C for 9 min; followed by 35 cycles of 95°C, 30 s; 55°C, 30 s; 72°C, 1 min; and a final extension at 72°C, 10 min. The amplified products were visualised on 2% agarose gel.
Screening for HPV sequences
Standard PCR was carried out in a total volume of 50 _μ_l using HotStarTaq DNA polymerase (Qiagen) with the following conditions: 95°C for 9 min; followed by 35 cycles of 95°C, 30 s; 55°C, 40 s; 72°C, 40 s; and a final extension at 72°C, 10 min. Cell lines were screened using the MY/GP HPV consensus primers as described by Kroupis et al (2006), omission of the DNA template was used as a negative control. Both MY and GP primers sets amplify the L1 region of the HPV genome and are used as a preliminary step for HPV screening. The amplified products were visualised on a 1.5% agarose gel and sequenced to confirm the type of HPV. The PCR was independently repeated for each sample.
Histology
The presence of koilocytosis in the fixed breast cancer series was assessed by light microscopy with koilocyte positive cervical cancer specimens used for comparison. Koilocytes were best characterised by (Koss and Durfee, 1956) the presence of large cells with relatively small, but irregular and hyperchromatic nuclei surrounded by clear and transparent cytoplasm. Koilocytosis is restricted to the replicating basal cells and multinucleation is common in these cells. The histological features of HPV positive breast tumours are similar to cervical koilocytosis and have previously been reported by de Villiers et al (2005).
Results
Breast cancer cell lines
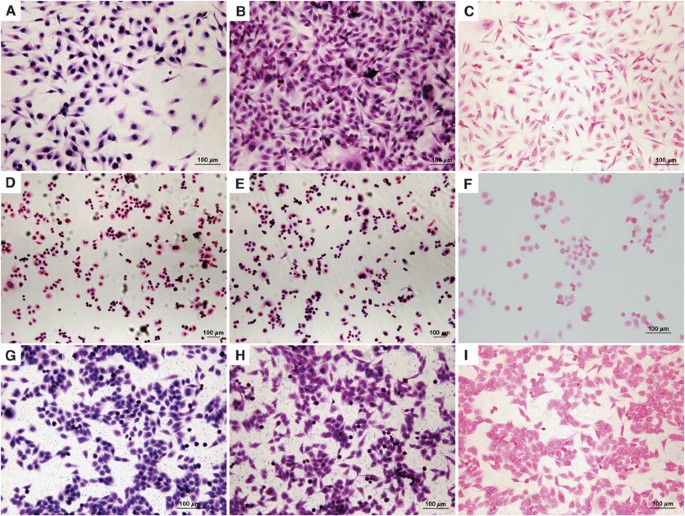
In this study, we demonstrate the application of in situ PCR to identify HPV sequences within breast cancer cells. We initially screened nine breast cancer cell lines (as listed in Materials and Methods) for HPV in situ PCR. High-risk HPV gene sequences were identified in two of the cell lines (MDA-MB-175-VII and SK-BR-3) of the nine lines tested. In situ PCR demonstrated that HPV DNA was confined within the nucleus of the cells (Figure 1). The presence and type (HPV type 18) was confirmed by automated sequencing (Figure 2) of the PCR products shown in Figure 3.
Figure 1
Human papilloma virus (HPV) in situ polymerase chain reaction (PCR) of breast cancer cell lines. (A–C) Cell line MBA-MB-175V11, (D–F) cell lines BR-SK3 and (G–I) cell lines HeLa (HPV-18 containing cervical cancer cell line: positive control). (A, D and G) In situ PCR with HPV E6 primers. (B, E and H) In situ PCR with HPV L1 primers. (C, F and I) No primer (negative) in situ PCR control.
Figure 2
Polymerase chain reaction (PCR) product nucleotide sequences of human papilloma virus (HPV)-positive patient samples and breast cancer cell lines. Four HPV-positive breast cancer samples were identified in Figure 4 below (specimens 2, 4, 5 and 6) and two breast cancer cell lines (SK-BR3 and MDA-MB-175) were identified as HPV type 18, whereas one breast cancer specimen (specimen 7) was identified as HPV type 16. Minor sequence variations were observed in three samples (specimens 2, 4 and 6) and in both cell lines (SK-BR and MDA-MB-175) when matched against reference sequence HPV-18 positive HeLa. There is no sequence variation in specimen 7 when matched against HPV-type 16 genome (accession FJ006723).
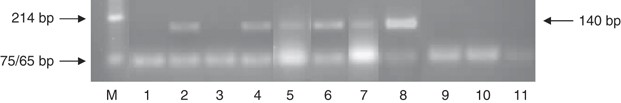
Figure 3
Human papilloma virus (HPV) screen of patient samples using MY and GP primers. Lane M is the Puc/Hinf ladder marker. Lanes 1–7 are patient samples (breast cancer specimens 1–7). Lane 8 is HeLa DNA as the positive control. Lanes 9–11 are negative controls (water in place of DNA in reaction). Lanes 2 (specimen 2), 4 (specimen 4), 5 (specimen 5), 6 (specimen 6) and 7 (specimen 7) show positive bands of 140 bp. Both samples in lanes 1 (specimen 1) and 3 (specimen 3) are negative for HPV.
Identification of HPV DNA sequences in formalin-fixed breast cancer specimens by in situ PCR and sequencing
Having established the method and demonstrated the presence of HPV in breast cancer cell lines, we screened a series of fixed breast cancer and normal breast tissue specimens using in situ PCR. As discussed in Materials and Methods section, we eliminated those samples, which potentially could give false-positive HPV results, identified by positive in situ PCR without primers. An unknown number of these eliminated specimens would have been true positives. Accordingly, the data cannot be used to make estimates of prevalence of the presence of these viruses.
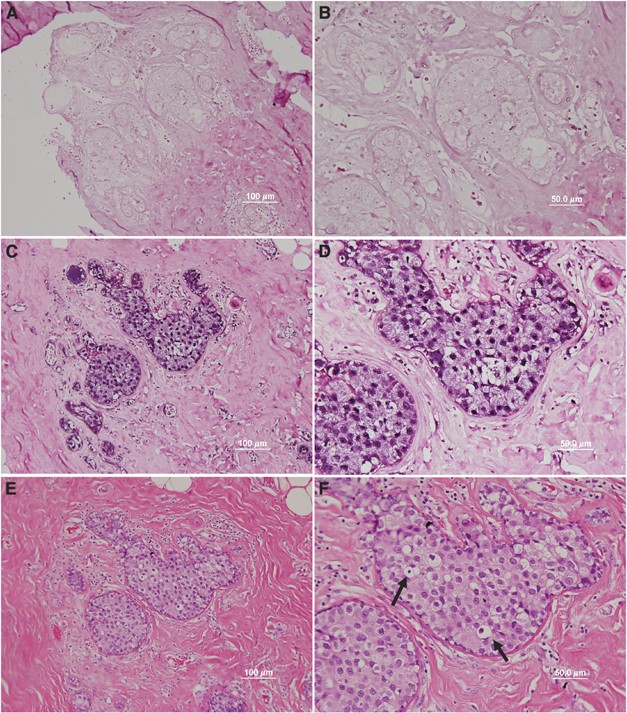
High-risk HPV DNA sequences were identified in the nuclei of breast cancer epithelial cells in 5 (39%) of 13 dcis and 3 (23%) of 13 idc breast cancer specimens (Figure 4, summarised in Table 1). Unexpectedly, we saw HPV containing cells in the surrounding normal tissue of some samples (Figure 4D). In all 3 (18%) of 17 normal breast specimens (from breast reduction surgery) were HPV positive by in situ PCR (Table 1). The presence of HPV in normal breast tissues is consistent with the requirement for HPV infection in the breast tissue before HPV-induced tumourigenic transformation of a single clone.
Figure 4
Human papilloma virus (HPV) in cancer cells of ductal carcinoma in situ breast cancer demonstrated by in situ polymerase chain reaction (PCR) (same specimen in all panels). (A and B) No primer in situ PCR control ( × 20 and × 40 objective, respectively), (C and D) HPV L1 primer in situ PCR ( × 20 and × 40 objective, respectively), Dark purple stain in panels A–D indicated amplification of HPV L1 sequences by in situ PCR. (E and F) Haematoxylin and eosin stain ( × 20, × 40 objective, respectively). The appearance of koilocytes in the HPV-18 containing cells shown in panel F (selected koilocytes shown by arrows) is indicated by the clearing of the cytoplasm and condensed, hyperchromatic nuclei.
Table 1 In situ PCR screening for HPV in fixed breast tissue specimens and breast cancer cell lines
Seven breast cancer specimens and three normal specimens showed false-positive outcomes when the PCR primer was omitted from the in situ PCR analysis. These specimens were not considered further.
Confirmation of in situ PCR analyses
Deoxyribonucleic acid was extracted from the HPV-positive specimens and analysed by standard PCR (Figure 3) to confirm the findings based on in situ PCR and to type the HPV by sequencing (Figure 2). Not all gels and sequences are shown in Figures 2 and 3. The outcome was confirmed in all the three HPV-positive normal breast specimens. The outcome was confirmed in five of the eight HPV-positive breast cancer specimens. Minor sequence variations indicate that contamination was unlikely.
All HPVs were type 18 except for one specimen in which HPV was type 16. The identification of two HPV types is a further indication that contamination is unlikely. Known HPV-positive (cervical cancer specimens and HeLa cervical cancer cultured cells) and negative (no PCR primers) were used as controls. All controls gave the expected outcomes.
HPV-associated morphological characteristics
The features of koilocytosis were observed in 18 (66%) of 28 breast cancer specimens (Figure 4). Koilocyte-like cells were also observed in some HPV-positive normal breast tissue specimens. The koilocyte-like cells were all HPV positive as shown by in situ PCR. These features are very similar to koilocytosis present in HPV-positive cervical cancer.
Discussion
In this report, we have confirmed the presence of HPV in the nuclei of cells in breast cancer tissue. The use of in situ PCR localises the HPV to the nuclei of cells within the cancerous tissue, and substantially decreases the possibility that detection of HPV in breast cancer is a contamination. In addition, the detection of HPV was consistent with the cell morphology and standard PCR/sequencing results.
The detection of HPV in breast cancer is consistent with 15 (of 17) previous publications reporting the presence of HPV in breast cancer world-wide with a prevalence of ranging from 4–86% (Lawson et al, 2006). Given such a high proportion of studies reporting HPV in breast cancer and breast tissue, the question is why some (2 of 17) studies report that HPV is not present in breast cancer. Potential explanations include difficulties in detection due to low viral load and low frequency of HPV in breast cancers in some populations. The in situ PCR results demonstrate just how much non-cancerous tissue is present in breast cancer specimens, which may explain the difficulty in detection of HPV by standard PCR (upon DNA extraction from the whole tissue sample).
The observation that HPV type 18 was by far the most common type in these Australian breast cancers is meaningful as most breast cancers can be regarded as originating from breast-milk epithelial cells (despite the historically misleading terminology of ‘ductal’ breast carcinomas). Therefore, most breast cancers are ‘glandular’. HPV type 18 has an affinity or tropism to glandular as compared with squamous epithelial cells (Clifford and Franceschi, 2008).
It is important to note that we report the presence on high-risk HPV in normal breast tissue. This supports the previous report of HPV in normal tissue in Turkish breast samples (Gumus et al, 2006). The presence of HPV in some normal breast samples (normal breast and normal surrounding breast tissue as shown in Figure 4) is consistent with a tumourigenic role for HPV in some breast cancers. If HPV has a causal role in breast cancer, it is reasonable to expect that this would be an early event, similarly with cervical cancerogenesis, and that HPV would be found in at least some normal tissue. Furthermore, it is expected that the presence of high-risk HPV would not be sufficient for full tumourigenic transformation and further changes would accumulate over time in a step-wise manner.
Significance of these findings
Although the proportion of HPV-positive breast cancer specimens in this study is higher than HPV-positive normal breast tissues, the overall number of specimens is small and definitive conclusions cannot be made. However, given the propensity for HPV to oncogenically transform human epithelial cells, including breast epithelial cells (Band et al, 1990), plus the unambiguous evidence that high-risk HPVs are present in the nuclei of breast cancer cells and in breast cancer cultured cell lines, and that HPV-positive koilocytes are present on many normal and breast cancer specimens, suggests that HPV may have a causal role in many breast cancers.
Establishing an oncogenic role for HPV in some breast cancers leads to the possibility of primary prevention of some breast cancers by vaccination against HPV, as current vaccines are known to be effective against HPV types 16 and 18 (Rambout et al, 2007).
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
- Akil N, Yasmeen A, Kassab A, Ghabreau L, Darnel AD, Al Moustafa AE (2008) High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study. Br J Cancer 99: 404–407
Article CAS Google Scholar - Band V, Zajchowski D, Kulesa V, Sager R (1990) Human papilloma virus DNAs immortalize human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci USA 87: 463–467
Article CAS Google Scholar - Choi YL, Cho EY, Kim JH, Nam SJ, Oh YL, Song SY, Yang JH, Kim DS (2007) Detection of human papillomavirus DNA by DNA chip in breast carcinomas of Korean women. Tumour Biol 28: 327–332
Article CAS Google Scholar - Clifford G, Franceschi S (2008) Members of the human papillomavirus type 18 family (alpha-7 species) share a common association with adenocarcinoma of the cervix. Int J Cancer 122: 1684–1685
Article CAS Google Scholar - Damin A, Karam R, Zettler CG, Caleffi M, Alexandre CO (2004) Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Re Treat 84: 131–137
Article CAS Google Scholar - Dawkins HJ, Robbins PD, Sarna M, Carrello S, Harvey JM, Sterrett GF (1993) c-erbB-2 amplification and overexpression in breast cancer: evaluation and comparison of Southern blot, slot blot, ELISA and immunohistochemistry. Pathology 25: 124–132
Article CAS Google Scholar - de Villiers E-M, Sandstrom RE, zur Hausen H, Buck CE (2005) Presence of papillomatous sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast. Breast Cancer Res 7: R1–R11
Article CAS Google Scholar - Gumus M, Yumuk PF, Salepci T, Aliustaoglu M, Dane F, Ekenel M, Basaran G, Kaya H, Barisik N, Turhal NS (2006) HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. J Exp Clin Cancer Res 25: 515–521
CAS Google Scholar - Hennig EM, Suo Z, Thoresen S, Holm R, Kvinnsland S, Nesland JM (1999) Human papillomavirus 16 in breast cancer of women treated for high grade cervical intraepithelial neoplasia (CIN III). Breast Cancer Res Treat 53: 121–135
Article CAS Google Scholar - Kan CY, Iacopetta BJ, Lawson JS, Whitaker J (2005) Identification of human papillomavirus DNA gene sequences in human breast cancer. Br J Cancer 93: 946–948
Article CAS Google Scholar - Khan NA, Castillo A, Koriyama C, Kijima Y, Umekita Y, Ohi Y, Higashi M, Sagara Y, Yoshinaka H, Tsuji T, Natsugoe S, Douchi T, Eizuru Y, Akiba S (2008) Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer 99: 408–414
Article CAS Google Scholar - Koss LG, Durfee GR (1956) Unusual paterns of squamous epithelium of the uterine cervix: cytologic and pathologic study of koilocytic atypia. Ann NY Acad Sci 63: 1245–1261
Article CAS Google Scholar - Krawczyk E, Suprynowicz FA, Liu X, Dai Y, Hartmann DP, Hanover J, Schlegel R (2008) Koilocytosis: a cooperative interaction between the human papillomavirus E5 and E6 oncoproteins. Am J Pathol 173: 682–688
Article CAS Google Scholar - Kroupis C, Markou A, Vourlidis N, Dionyssiou-Asteriou A, Lianidou ES (2006) Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clin Biochem 39: 727–731
Article CAS Google Scholar - Kulka J, Kovalszky I, Svastics E, Berta M, Füle T (2008) Lymphoepithelioma-like carcinoma of the breast: not Epstein-Barr virus, but human papilloma virus-positive. Hum Pathol 39: 298–301
Article CAS Google Scholar - Lawson JS, Guenzburg WH, Whitaker NJ (2006) Viruses and breast cancer. Review. Future Microbiol 1: 33–51
Article CAS Google Scholar - Lindel K, Forster A, Altermatt HJ, Greiner R, Gruber G (2007) Breast cancer and human papillomavirus (HPV) infection: no evidence of a viral etiology in a group of Swiss women. Breast 16: 172–177
Article Google Scholar - Liu Y, Klimberg VS, Andrews NR, Hicks CR, Peng H, Chiriva-Internati M, Henry-Tillman R, Hermonat PL (2001) Human papillomavirus DNA is present in a subset of unselected breast cancers. J Hum Virol 4: 329–334
CAS Google Scholar - Lonardo DA, Venuti A, Marcante ML (1992) Human papillomavirus in breast cancer. Breast Cancer Res Treat 21: 95–100
Article Google Scholar - Mendizabal-Ruiz AP, Morales JA, Ramírez-Jirano LJ, Padilla-Rosas M, Morán-Moguel MC, Montoya-Fuentes H (2009) Low frequency of human papillomavirus DNA in breast cancer tissue. Breast Cancer Res Treat 114: 189–194
Article CAS Google Scholar - Rambout L, Hopkins L, Hutton B, Fergusson D (2007) Prophylactic vaccination against human papillomavirus infection and disease in women: a systematic review of randomized controlled trials. CMAJ 177: 469–479
Article Google Scholar - Thomison J, Thomas LK, Shroyer KR (2008) Human papillomavirus: molecular and cytologic/histologic aspects related to cervical intraepithelial neoplasia and carcinoma. Hum Pathol 39: 154–166
Article CAS Google Scholar - Tsai J, Tsai CH, Cheng MH, Lin SJ, Xu FL, Yang CC (2005) Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma and thyroid tumor tissues. J Med Virol 75: 276–281
Article Google Scholar - Widschwendter A, Brunhuber T, Wiedemair A, Mueller-Holzner E, Marth C (2004) Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J Clin Virol 31: 292–297
Article CAS Google Scholar - Yu Y, Morimoto T, Sasa M, Okazaki K, Harada Y, Fujiwara T, Irie Y, Takahashi E, Tanigami A, Izumi K (1999) HPV 33 DNA in premalignant and malignant breast lesions in Chinese and Japanese populations. Anticancer Res 19: 5057–5061
CAS Google Scholar - Zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nature Rev Cancer 2: 342–350
Article CAS Google Scholar
Acknowledgements
The Cooper Medical Research Foundation of Sydney, Australia and the Komen for the Cure Foundation of Dallas, TX, USA, gave crucial financial support. David Gillett and Laurence Gluch and colleagues of the Strathfield Breast Centre, Sydney, Australia, collected and donated specimens and offered valuable advice. Wendy K Glenn provided help with the co-ordination and conduct of laboratory work, development of concepts and writing of manuscript. Yulan Ye, Benjamen Heng and Bao Tran were were involved with the laboratory work. Warick Delprado conducted the histopathological assessments. Louise Lutze-Mann was involved with the co-ordination of laboratory work, development of concepts and writing of manuscript. Noel J Whitaker supported with the co-ordination of laboratory work, development of concepts and writing of manuscript. James S Lawson was involved with the co-ordination of project, development of concepts, conduct of immunohistochemistry analyses, statistical analyses and writing of manuscript. All authors have agreed to all the content in the manuscript, including the data as presented.
Author information
Author notes
- N J Whitaker and J S Lawson: NJW and JSL are co-senior investigators
Authors and Affiliations
- School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, Australia
B Heng, W K Glenn, Y Ye, B Tran, L Lutze-Mann, N J Whitaker & J S Lawson - Douglass Hanley Moir Pathology, Sydney, Australia
W Delprado
Authors
- B Heng
You can also search for this author inPubMed Google Scholar - W K Glenn
You can also search for this author inPubMed Google Scholar - Y Ye
You can also search for this author inPubMed Google Scholar - B Tran
You can also search for this author inPubMed Google Scholar - W Delprado
You can also search for this author inPubMed Google Scholar - L Lutze-Mann
You can also search for this author inPubMed Google Scholar - N J Whitaker
You can also search for this author inPubMed Google Scholar - J S Lawson
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toJ S Lawson.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/.
About this article
Cite this article
Heng, B., Glenn, W., Ye, Y. et al. Human papilloma virus is associated with breast cancer.Br J Cancer 101, 1345–1350 (2009). https://doi.org/10.1038/sj.bjc.6605282
- Received: 30 June 2009
- Revised: 21 July 2009
- Accepted: 01 August 2009
- Published: 01 September 2009
- Issue Date: 20 October 2009
- DOI: https://doi.org/10.1038/sj.bjc.6605282