Color transitions in coral's fluorescent proteins by site-directed mutagenesis (original) (raw)
- Research article
- Published: 10 July 2001
- Alexander P Savitsky2,
- Yurii G Yanushevich1,
- Sergey A Lukyanov1 &
- …
- Konstantin A Lukyanov1
BMC Biochemistry volume 2, Article number: 6 (2001)Cite this article
- 20k Accesses
- 36 Citations
- 3 Altmetric
- Metrics details
Abstract
Background
Green Fluorescent Protein (GFP) cloned from jellyfish Aequorea victoria and its homologs from corals Anthozoa have a great practical significance as in vivo markers of gene expression. Also, they are an interesting puzzle of protein science due to an unusual mechanism of chromophore formation and diversity of fluorescent colors. Fluorescent proteins can be subdivided into cyan (~ 485 nm), green (~ 505 nm), yellow (~ 540 nm), and red (>580 nm) emitters.
Results
Here we applied site-directed mutagenesis in order to investigate the structural background of color variety and possibility of shifting between different types of fluorescence. First, a blue-shifted mutant of cyan amFP486 was generated. Second, it was established that cyan and green emitters can be modified so as to produce an intermediate spectrum of fluorescence. Third, the relationship between green and yellow fluorescence was inspected on closely homologous green zFP506 and yellow zFP538 proteins. The following transitions of colors were performed: yellow to green; yellow to dual color (green and yellow); and green to yellow. Fourth, we generated a mutant of cyan emitter dsFP483 that demonstrated dual color (cyan and red) fluorescence.
Conclusions
Several amino acid substitutions were found to strongly affect fluorescence maxima. Some positions primarily found by sequence comparison were proved to be crucial for fluorescence of particular color. These results are the first step towards predicting the color of natural GFP-like proteins corresponding to newly identified cDNAs from corals.
Background
Since the introduction of Green Fluorescent Protein (GFP) into biotechnology many attempts have been made to change its fluorescence color by means of mutagenesis. Finally, only two positions on protein sequence were demonstrated to considerably influence emission maximum [1,2,3]. First, a replacement of fluorophore's Tyr-66 with any aromatic residue (Trp, Phe, or His) results in a strong blue shift of emission up to λmax = 442 nm [4, 5]. Second, a substitution S203Y,H leads to a red shift of fluorescence up to λmax = 529 nm [6]. These blue and yellow mutants (called BFP and YFP respectively) proved to be extremely handy for multicolor labeling and FRET-based applications.
Recently we have isolated several GFP-like proteins that determine fluorescent or non-fluorescent body color in corals Anthozoa [7,8,9]. Absorption-emission maxima of these proteins are distributed loosely along the wavelength axis. These proteins fall into four groups: cyan (~ 485 nm), green (~ 505 nm), yellow (~ 540 nm), and red (>580 nm) emitters. Obviously, understanding the relation between protein structure and its fluorescent properties is of great scientific and practical importance. However, in spite of the abundance of sequences available, it is difficult to determine amino acid positions that are responsible for a particular type of fluorescence.
In present work, site-directed mutagenesis was used to change the emission color of some Anthozoa fluorescent proteins (FP). To search presumptive amino acid positions important for particular color we compared sequences of FP belonging to the different groups. Special attention was focused on positions that are probably lie in the close proximity of fluorophore. We checked several suspicious positions and found some substitutions that affected fluorescence maxima. To simplify discussion, we used amino acid numbering in accordance with GFP (see alignment in Fig. 1).
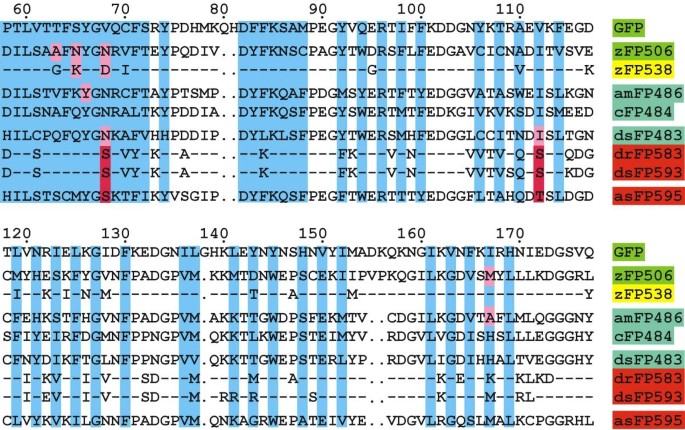
Figure 1
Multiple alignment of GFP and Anthozoa FPs. The numbering is according to GFP, the N- and C-termini of the proteins are not shown. Coral FPs are named using a nomenclature suggested in [7]. Each protein is designated by lowercase letters identifying the species (e.g., "z" for Zoanthus sp., "am" for Anemonia majano, "c" for Clavularia sp., "ds" for Discosoma sp., "dr" for Discosoma sp. red, "as" for Anemonia sulcatd), FP for fluorescent protein, and a number corresponding to the major emission maximum. Two proteins from Zoanthus and three from Discosoma are grouped and compared inside the groups: in the second/third protein of the group the amino acids identical to the corresponding ones in the first protein are represented by dashes. The protein names are shaded in colors corresponding to its fluorescence. The residues whose side chains form the interior of the β-can are shaded in blue (according to [6]). Introduced gaps are represented by dots. The residues that were mutated are shaded in pink. Red FP-specific Ser-68 and Ser/Thr-112 are shaded in red.
Results and discussion
Blue shift for cyan FP amFP486
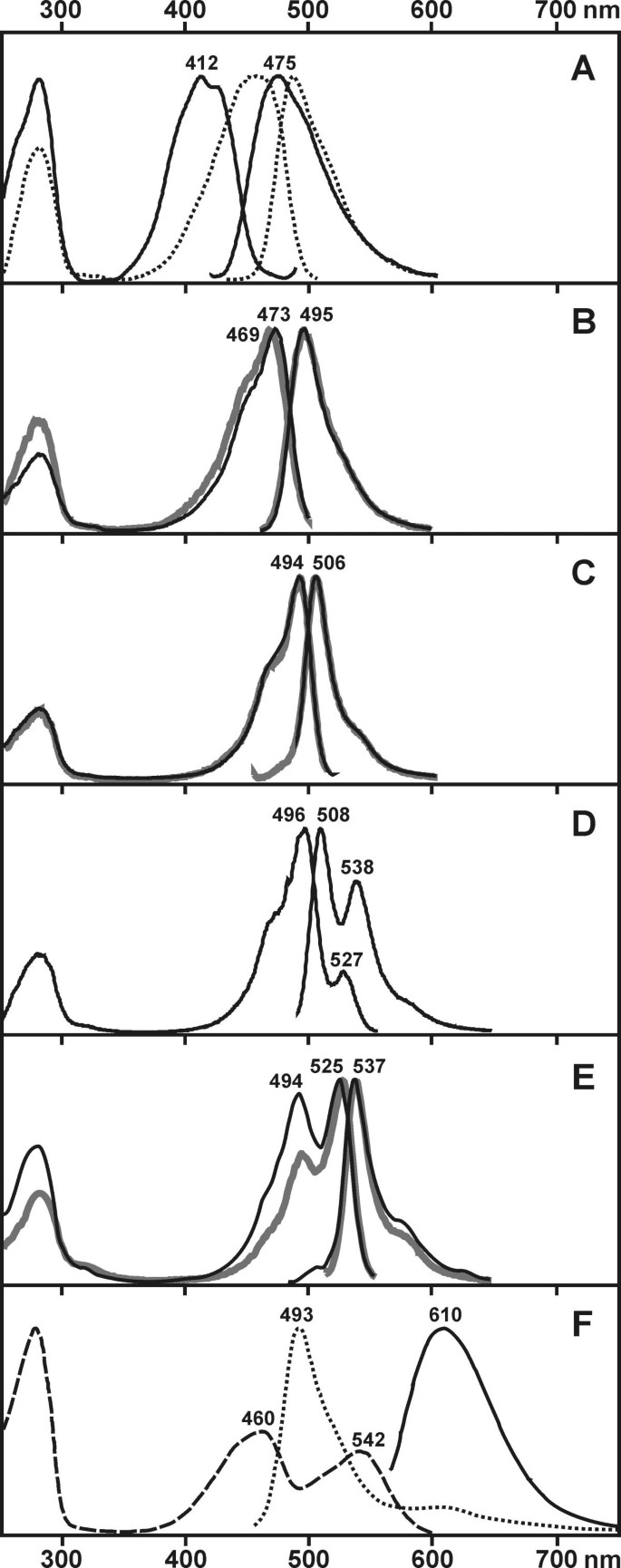
First of all, we reproduced mutations known to be responsible for the blue shift in GFP [4]. As amFP486 is already blue shifted in comparison with GFP, we expected to obtain an even more blue protein than BFP. However, the results of mutagenesis were rather modest. Of two substitutions tested, Y66H resulted in a complete loss of fluorescence, and Y66W led to a comparatively small shift of emission maximum up to 475 nm (Fig. 2A). Interestingly, in spite of dramatic differences in spectra of wild type GFP and amFP486, their W66 mutants provide similar excitation-emission spectra, especially if one compares amFP486-Y66W with an improved blue-shifted mutant of GFP called W7 (Y66W/N146I/M153T/V163A/N212K) [5]. The latter two excitation spectra share common dual-peak shape and differ only in the position of maxima (at 412 nm for amFP486-Y66W and at 433 nm for W7). Emission spectra of these mutants display the same maxima at 475 nm.
Figure 2
Excitation-emission spectra for mutants with altered fluorescent color. In some cases spectra for the wild type proteins are shown for comparison. Within each pair of lines the emission spectrum is the one at longer wavelengths. (A) Spectra for amFP486 wild type (dotted lines) and its blue shifted mutant Y66W (solid lines). (B) Spectra for mutants amFP486-A167M (solid lines) and zFP506-M167A (gray thick lines). (C) Spectra for green mutant zFP538-K65M (solid lines) and zFP506 wild type (gray thick lines). (D) Spectra for dual-color mutant zFP538-D68N. (E) Spectra for yellow mutant zFP506-A63G/N65K/N68D (solid lines) and zFP538 wild type (gray thick lines). (F) Spectral characteristics of the dual-color mutant protein dsFP483-N68S/I112S. Dotted line - double-peak fluorescence (excitation at 280 nm). Solid line - red fluorescence (excitation at 540 nm). Dashed line - excitation spectrum for red emission (at 620 nm).
Cyan-green transitions
When it comes to human eye, cyan and green sorts of fluorescence look rather similar. However, their spectral properties are clearly different. The three existing cyan emitters - amFP486, dsFP483, and cFP484 - have similar spectra that are characterized by an emission maximum at about 485 nm and a broad peak of fluorescence with spectral width (width of the peak at half of maximal intensity) of about 50 nm. Green FPs - zFP506 and EGFP (F64L/S65T mutant of GFP) - emit at about 505 nm and have a narrow peak of fluorescence with spectral width of about 25 nm. These two sorts of fluorescence having characteristic maxima and spectral widths (with no intermediate spectra) were revealed by spectral measurements of coral's fluorescence in their natural habitats [10]. This implies that cyan and green FPs differ by some distinct feature(s) of the protein structure. Sequence comparison of proteins mentioned above revealed only one significant mismatch between cyan FPs on the one hand and green FPs on the other. Position 167 is occupied by Ala or His in the cyan emitters and by Met or Ile in the green emitters (Fig. 1). According to X-ray crystal structure of GFP, Ile-167 lies in the closest proximity of the fluorophore [6, 11]. It is well known that polarizability of a fluorophore's environment affects strongly the spectral width of fluorescence: the more polar the environment the broader the emission peak is. Also, residues at position 167 can be discussed in terms of rigidity of the fluorophore: the tighter its environment the narrower the fluorescence spectrum is. We hypothesized that bulky hydrophobic residues (Met, Ile) at position 167 stand for a narrow peak of fluorescence, while smaller and more polar residues (His, Ala) leads to the broadening of the peak. Also, small Ala residue in amFP486 may vacate space for an additional water molecule that polarizes the fluorophore's environment. To test this presumption we generated mutants zFP506-M167A,H and amFP486-A167M,I and characterized corresponding proteins. Surprisingly, all four mutants produced very similar curves that were intermediate between cyan and green FPs in terms of position of the maxima and spectral widths of the emission and excitation peaks (Fig. 2B and Table 1). These data clearly demonstrated the importance of position 167 for fluorescence color of cyan and green FPs. However, as the mutants mentioned above display an intermediate variant of fluorescence, one may conclude that some other residues are also important for cyan or green emission typical for the wild type proteins. Our results are in line with mutagenetic studies on GFP, where substitutions I167V,T led to a slight blue shift together with a considerable widening of the fluorescence curve [4].
Table 1 Comparison of the spectral properties of zFP506, amFP486, and their mutants at position 167.
Green-yellow transitions
In one particular case, two closely related FPs of different colors coexist in a single organism, Zoanthus sp. [7]. One of them, zFP506, displayed green fluorescence and the other, zFP538, - yellow fluorescence. Having 87% identity to each other, these proteins are an excellent model for hunting amino acids required for green-yellow transitions. Most of 31 amino acids that distinguish zFP506 from zFP538 are external and only 8 of these residues are buried, assuming that structures of Anthozoa FPs are similar to the β-can fold of GFP [6, 11]. Four out of eight buried amino acid disparities are grouped in proximity of chromophore-forming residues at positions 66 and 67 (Fig. 1). It is reasonable to suppose that some of these residues are the cause of the color difference.
Accidentally, our other mutagenesis project on randomization of position 65 in several FPs (this work will be published elsewhere) revealed Lys-65 in zFP538 as a key residue responsible for the yellow fluorescence. Among seven randomly tested amino acid substitutions at position 65 (Leu, Ile, Met, His, Gln, Asn, or Lys) only the wild type variant Lys-65 emitted yellow light. Other mutants displayed green fluorescence or lacked fluorescent properties. We characterized in detail the brightest green-emitting mutant containing a single amino acid substitution K65M. In terms of brightness, shape and maxima of excitation-emission spectra this mutant protein was estimated to be practically undistinguishable from zFP506 (Fig. 2C). The finding that Lys-65 determines the yellow color of fluorescence was unexpected as Lys in this position is not unique for zFP538: one cyan emitter, amFP486, hosts Lys-65 (Fig. 1). Thus, one can conclude that only a simultaneous action of Lys-65 together with some other residue(s) results in a red shift of fluorescence.
Then, we paid attention to Asp-68 that is unique for zFP538. A substitution D68N resulted in dual-peak (green and yellow) fluorescence (Fig. 2D). So, Asp-68 in zFP538 is important but not vital for yellow fluorescence.
Finally, we attempted to convert zFP506 into a yellow emitter. Unexpectedly, two of the replacements discussed above were not sufficient for this transition. A mutant protein N65K/N68D possessed green fluorescence similar to the wild type protein's (not shown). Only a triple mutant of zFP506 carrying substitutions A63G/N65K/N68D displayed emission maximum in yellow region at 537 nm (Fig. 2E). In comparison with the natural yellow emitter zFP538 this mutant protein had slightly modified excitation spectrum: the minor peak at 494 nm became more pronounced.
Summing up, we performed the following transitions of fluorescent colors for zFP506 and zFP538 proteins: (i) yellow to green, (ii) yellow to dual color (green and yellow), and (iii) green to yellow. Switch-like color transitions as well as dual-color fluorescence may indicate that the green and yellow fluorescence represent chemically distinct fluorophores. Our hypothesis is that a positive charge of Lys-65 and a negative charge of Asp-68 are involved in catalysis at yellow fluorophore formation, while Gly-63 provides the required steric freedom.
Cyan-red transition
One of the most challenging goals when modifying fluorescent proteins is the transformation of a green emitter into a red. An alignment of all known fluorescent proteins revealed only two suspicious amino acid disparities between red emitters on the one hand and cyan, green and yellow FPs on the other (Fig. 1). In contrast to all other FPs, positions 68 and 112 in red proteins are occupied by Ser (excepting the Thr-112 in asFP595 that should be viewed as a conservative substitution). We experimentally evaluated these positions introducing Ser-68 and Ser-112 into a cyan emitter dsFP483 that is a close homolog of two red FPs from Discosoma species (about 60% identity). Three variants of dsFP483 were generated: N68S, I112S, and a double mutant N68S/I112S. The single mutants N68S and II 12S displayed ordinary cyan fluorescence but were much dimmer than the wild type protein (data not shown). However, the double mutant carrying both Ser residues produced dual-color fluorescence. When viewed by fluorescence microscopy, E. coli colonies expressing the mutant protein clearly displayed both green (under FITC filter set) and red (under TRITC filter set) fluorescence (Fig. 3). Spectroscopy of purified dsFP483-N68S/I112S showed dual-peak emission spectrum where the peak of red fluorescence at 610 nm is much lower than the peak at 493 nm (Fig. 2F). However, red fluorescence is definitely detected when exciting the protein at 530-560 nm. Although this mutant has no practical significance, it should be emphasized that this mutagenesis resulted in the strongest red shift (127 nm) ever-achieved with GFP-like proteins.
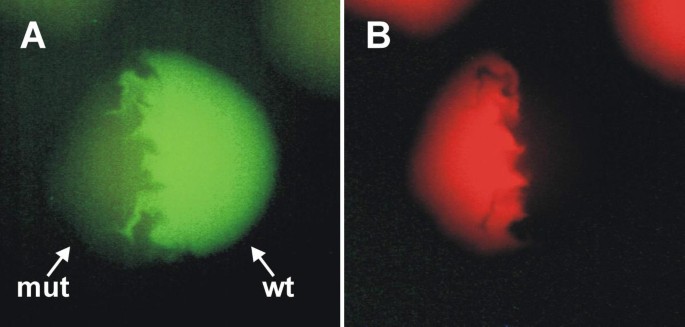
Figure 3
Colonies of E. coli expressing wild type dsFP483 (wt) or its mutant N68S/I112S (mut) as viewed by fluorescence microscopy. (A) - using FITC filter set. (B) - using TRITC filter set. Colony expressing the wild type protein possesses only green fluorescence. In contrast, colony expressing the mutant protein is clearly visible in both filter set due to dual color (green and red) fluorescence of dsFP483-N68 S/I112S.
Recently, the structure of drFP583 "red" chromophore was determined [12]. Crystallographic studies of drFP583 supported this model of chromophore structure [13, 14]. In comparison with GFP-type green chromophore, red one was found to contain a more extended π-system due to the presence of a double bond between the α-carbon and the nitrogen of Gln-65. Thus, the red chromophore matures through a green GFP-like intermediate by means of additional autocatalytic step of dehydrogenation. Mutagenetic studies of red FPs, drFP583 and asFP595, revealed many point mutations that transform them into dual-color (green and red) or even pure green emitters [9, 15, [16](/articles/10.1186/1471-2091-2-6#ref-CR16 "Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P: "Fluorescent Timer": protein that changes color with time. Science. 2000, 290: 1585-1588. 10.1126/science.290.5496.1585.")]. So, red fluorescent color can be very easily converted into green by a disruption of the chromophore's environment required for the last autocatalytic step. Here we attempted to create such "red" environment in a cyan FP. Serine residues at positions 68 and 112 were found to be essential for red chromophore formation, but are not sufficient to completely convert cyan FP into red: only a small portion of the mutant protein becomes red emitting. Obviously, an elaborate set of amino acids should surround the chromophore to ensure the maturation of the "red" structure. According to the crystal structure of drFP583, Ser-68 is located in the closest proximity to the double bond between α-carbon and nitrogen of Gln-65 specific for the red fluorophore [13, 14]. One is tempted to suppose that Ser-68 plays some catalytic role in formation of this double bond. It is more difficult to explain significance of Ser-112 because this residue is situated rather far from chromophore. However, Ser-112 is bound through water molecules with oxygen atoms of both Gly-67 and the imidazolidinone ring. These hydrogen bonds could be important for proper chromophore positioning or could be a pathway for proton transfer.
Based on the results reported here we believe it is possible to predict the color of natural GFP-like proteins corresponding to newly identified cDNAs from corals. The most characteristic point is position 68. At this position cyan and green FPs should contain Asn or a hydrophobic residue; yellow FPs should contain Asp or possibly Glu (negatively charged residues); red FPs and asFP595-like chromoproteins should contain Ser. Then, position 167 probably allows to discriminate cyan and green emitters: bulky aliphatic residues (e.g. Met, He, Leu) are characteristic for green FPs and polar or small residues (e.g. His, Ala) are specific for cyan FPs. However, a more extended list of FPs of different colors is required to prove or contradict our conclusions.
Materials and Methods
Site-directed mutagenesis was performed by PCR using the overlap extension method, with primers containing appropriate target substitutions [17]. All mutants were cloned into pQE30 vector (Qiagen) using BamHI and SalI restriction sites. Recombinant proteins contained 6xHis tag on N-terminus. E. coli clones were grown at 37°C in 50 ml to an optical density of (OD) 0.6. At that point, the expression of recombinant FP was induced with 0.2 mM IPTG. The cultures were then incubated overnight. The following day, cells were harvested by centrifugation, resuspended in buffer (20 mM Tris-HCl, pH 8.0; 100 mM NaCl), and disrupted by sonication. Fluorescent proteins were purified from the soluble fraction using TALON Metal Affinity Resin (CLONTECH). Proteins were at least 95% pure according to SDS-PAGE. Perkin-Elmer LS50B spectrometer was used for spectral measurements of the purified fluorescent proteins diluted in buffer (20 mM Tris-HCl, pH 8.0; 100 mM NaCl) up to about 50 μg/ml. The excitation and emission slits were set at 5 nm. The spectra were corrected for photomultiplier response and monochromator transmittance.
References
- Tsien RY, Prasher D: Molecular biology and mutation of green fluorescent protein. In: Green fluorescent protein. Chalfie, M., Kain, S., Eds. Willey-Liss, New York,. 1998, : 97-118.
Google Scholar - Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, Wlodawer A: The structural basis for spectral variations in green fluorescent protein. Nature Struct. Biol. 1997, 4: 361-365.
Article CAS PubMed Google Scholar - Remington SJ: Structural basis for understanding spectral variations in green fluorescent protein. Methods Enzymol. 2000, 305: 196-211.
Article CAS PubMed Google Scholar - Heim R, Prasher DC, Tsien RY: Wavelength mutations and post-translational autooxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1994, 91: 12501-12504.
Article PubMed Central CAS PubMed Google Scholar - Heim R, Tsien RY: Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Current Biol. 1996, 6: 178-182.
Article CAS Google Scholar - Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ: Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996, 273: 1392-1395.
Article PubMed Google Scholar - Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA: Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotech. 1999, 17: 969-973. 10.1038/13657.
Article CAS Google Scholar - Fradkov AF, Chen Y, Ding L, Barsova EV, Matz MV, Lukyanov SA: Novel fluorescent protein from Discosoma coral and its mutants possesses a unique far-red fluorescence. FEBS Lett. 2000, 479: 127-130. 10.1016/S0014-5793(00)01895-0.
Article CAS PubMed Google Scholar - Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, Savitsky AP, Markelov ML, Zaraisky AG, Zhao X, Fang Y, Tan W, Lukyanov SA: Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J. Biol. Chem. 2000, 275: 25879-25882. 10.1074/jbc.C000338200.
Article CAS PubMed Google Scholar - Mazel CH: Spectral measurements of fluorescence emission in Caribbean cnidarians. Mar. Ecol. Prog. Ser. 1995, 120: 185-191.
Article Google Scholar - Yang F, Moss LG, Phillips GN: The molecular structure of green fluorescent protein. Nature Biotechnol. 1996, 14: 1246-1251.
Article CAS Google Scholar - Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY: The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA. 2000, 97: 11990-11995. 10.1073/pnas.97.22.11990.
Article PubMed Central CAS PubMed Google Scholar - Wall MA, Socolich M, Ranganathan R: The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nature Struct. Biol. 2000, 7: 1133-1138. 10.1038/81992.
Article CAS PubMed Google Scholar - Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ: Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0 A resolution. Proc. Natl. Acad. Sci. USA. 2001, 98: 462-467. 10.1073/pnas.98.2.462.
Article PubMed Central CAS PubMed Google Scholar - Baird GS, Zacharias DA, Tsien RY: Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA. 2000, 97: 11984-11989. 10.1073/pnas.97.22.11984.
Article PubMed Central CAS PubMed Google Scholar - Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P: "Fluorescent Timer": protein that changes color with time. Science. 2000, 290: 1585-1588. 10.1126/science.290.5496.1585.
Article CAS PubMed Google Scholar - Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR: Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989, 77: 51-59. 10.1016/0378-1119(89)90358-2.
Article CAS PubMed Google Scholar
Acknowledgments
We are grateful to M. V. Matz for fruitful discussion, M. E. Bulina for the help in the manuscript preparation, and S. J. Remington and R. Y. Tsien for communication of data prior to publication. This work was supported by CLONTECH Laboratories Inc. and the Russian Foundation for Fundamental Research (grant 01-04-49037).
Author information
Authors and Affiliations
- Shemiakin and Ovchimiikov Institute of Bioorganic Chemistry RAS, Miklukho-Maklaya 16/10, 117871, Moscow, Russia
Nadya G Gurskaya, Yurii G Yanushevich, Sergey A Lukyanov & Konstantin A Lukyanov - Institute of Biochemistry RAS, Leninsky pr. 33, 117071, Moscow, Russia
Alexander P Savitsky
Authors
- Nadya G Gurskaya
You can also search for this author inPubMed Google Scholar - Alexander P Savitsky
You can also search for this author inPubMed Google Scholar - Yurii G Yanushevich
You can also search for this author inPubMed Google Scholar - Sergey A Lukyanov
You can also search for this author inPubMed Google Scholar - Konstantin A Lukyanov
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toKonstantin A Lukyanov.
Authors’ original submitted files for images
Rights and permissions
About this article
Cite this article
Gurskaya, N.G., Savitsky, A.P., Yanushevich, Y.G. et al. Color transitions in coral's fluorescent proteins by site-directed mutagenesis.BMC Biochem 2, 6 (2001). https://doi.org/10.1186/1471-2091-2-6
- Received: 04 May 2001
- Accepted: 10 July 2001
- Published: 10 July 2001
- DOI: https://doi.org/10.1186/1471-2091-2-6