Beyond Cigarettes Per Day. A Genome-Wide Association Study of the Biomarker Carbon Monoxide (original) (raw)
Journal Article
Department of Psychiatry, and
Search for other works by this author on:
Department of Psychiatry, and
Search for other works by this author on:
Department of Psychology, University of Wisconsin, Madison, Wisconsin
Search for other works by this author on:
Department of Psychiatry, and
Search for other works by this author on:
Department of Psychology, University of Wisconsin, Madison, Wisconsin
Search for other works by this author on:
Department of Psychiatry, and
Search for other works by this author on:
Department of Psychiatry, and
Search for other works by this author on:
Department of Psychiatry, University of Minnesota, Minneapolis, Minnesota
Search for other works by this author on:
RTI International, Research Triangle Park, North Carolina; and
Search for other works by this author on:
Department of Biostatistics, University of Washington, Seattle, Washington
Search for other works by this author on:
Received:
08 January 2014
Published:
01 September 2014
Cite
A. Joseph Bloom, Sarah M. Hartz, Timothy B. Baker, Li-Shiun Chen, Megan E. Piper, Louis Fox, Maribel Martinez, Dorothy Hatsukami, Eric O. Johnson, Cathy C. Laurie, Nancy L. Saccone, Alison Goate, Laura J. Bierut, Beyond Cigarettes Per Day. A Genome-Wide Association Study of the Biomarker Carbon Monoxide, Annals of the American Thoracic Society, Volume 11, Issue 7, September 2014, Pages 1003–1010, https://doi.org/10.1513/AnnalsATS.201401-010OC
Close
Navbar Search Filter Mobile Enter search term Search
Abstract
Rationale
The CHRNA5-CHRNA3-CHRNB4 locus is associated with self-reported smoking behavior and also harbors the strongest genetic associations with chronic obstructive pulmonary disease (COPD) and lung cancer. Because the associations with lung disease remain after adjustment for self-reported smoking behaviors, it has been asserted that CHRNA5-CHRNA3-CHRNB4 variants increase COPD and lung cancer susceptibility independently of their effects on smoking.
Objectives
To compare the genetic associations of exhaled carbon monoxide (CO), a biomarker of current cigarette exposure, with self-reported smoking behaviors.
Methods
A total of 1,521 European American and 247 African American current smokers recruited into smoking cessation studies were assessed for CO at intake before smoking cessation. DNA samples were genotyped using the Illumina Omni2.5 microarray. Genetic associations with CO and smoking behaviors (cigarettes smoked per day, Fagerstrom test for nicotine dependence) were studied.
Measurements and Main Results
Variants in the CHRNA5-CHRNA3-CHRNB4 locus, including rs16969968, a nonsynonymous variant in CHRNA5, are genomewide association study–significantly associated with CO (β = 2.66; 95% confidence interval [CI], 1.74–3.58; P = 1.65 × 10−8), and this association remains strong after adjusting for smoking behavior (β = 2.18; 95% CI, 1.32–3.04; P = 7.47 × 10−7). The correlation between CO and cigarettes per day is statistically significantly lower (z = 3.43; P = 6.07 × 10−4) in African Americans (r = 0.14; 95% CI, 0.02–0.26; P = 0.003) than in European-Americans (r = 0.36; 95% CI, 0.31–0.40; P = 0.0001).
Conclusions
Exhaled CO, a biomarker that is simple to measure, captures aspects of cigarette smoke exposure in current smokers beyond the number of cigarettes smoked per day. Behavioral measures of smoking are therefore insufficient indices of cigarette smoke exposure, suggesting that genetic associations with COPD or lung cancer that persist after adjusting for self-reported smoking behavior may still reflect genetic effects on smoking exposure.
More than 85% of deaths from chronic obstructive pulmonary disease (COPD) and lung cancer and 25 to 30% of all cancer deaths are directly attributed to cigarette smoking (1, 2). Genetic studies of smoking behavior, COPD, and lung cancer have all identified the chromosome 15q25.1 locus, which includes the α5-α3-β4 nicotinic receptor subunit genes CHRNA5-CHRNA3-CHRNB4, as the genetic region most strongly associated with both smoking behavior and smoking-related illnesses. The reported genetic associations with COPD and lung cancer remain strong after adjusting for amount smoked estimated by self-reported cigarettes smoked per day (3, 4), and it has therefore been asserted that CHRNA5-CHRNA3-CHRNB4 variants directly influence COPD and lung cancer susceptibility beyond their effects on smoking behavior (5, 6). However, this interpretation has been challenged because the measure of cigarettes smoked per day does not fully capture the health risks of smoking (7, 8). The use of biomarkers of cigarette smoke exposure has been suggested as a route to elucidate the complex relation between smoking behavior and cancer (9).
Self-reported cigarettes per day is the most commonly used and easily collected measure of cigarette consumption, and clinicians use self-reported cigarettes per day over a person’s smoking lifetime history to estimate health risks for COPD, cancer, and other smoking-related diseases. However, number of cigarettes smoked does not capture smoking topography, such as depth of inhalation and the number of puffs per cigarette, and smokers vary these parameters to titrate nicotine dose (10–12). For example, saliva levels of nicotine’s primary metabolite, cotinine, can vary by more than an order of magnitude among smokers who report smoking the same number of cigarettes per day (13). In addition, self-reports of smoking amount contain errors of memory and other biases inherent in self-reports of behavior integrated over time (14, 15).
Exhaled carbon monoxide (CO), a byproduct of cigarette combustion, varies with smoking topography (16–18). Nonsmokers have levels of CO less than or equal to 5 ppm (19), whereas the level in current smokers varies based on intensity of smoking and number of cigarettes smoked. Importantly, compared with number of cigarettes smoked, CO is more correlated with exposure to a key carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (20), indicating that CO more fully captures aspects of smoking behavior relevant to adverse health effects.
In this study we evaluate and compare genetic associations of the biomarker CO, cigarettes smoked per day, and nicotine dependence as measured by the Fagerstrom Test for Nicotine Dependence (FTND) (21) to better understand the relations between genetic variation, biomarkers, and smoking behavior and how they may relate to smoking-related health risks. This study is the first to report a genomewide significant association with exhaled CO as a measure of cigarette smoke exposure.
Methods
Subjects were participants in the University of Wisconsin Transdisciplinary Tobacco Use Research Center randomized, placebo-controlled smoking cessation trials (22–24). The University of Wisconsin-Madison Institutional Review Board approved these trials, and all subjects provided written informed consent. Participants were at least 18 years of age, smoked 10 or more cigarettes per day on average for at least the prior 6 months, and were motivated to quit smoking. Subject characteristics are provided in Table 1.
Table 1.
Sample characteristics by self-reported ethnicity
| Characteristic | European American (N = 1,521) | African American (N = 247) |
|---|---|---|
| Sex, % (N) | ||
| Male | 42.9 (653) | 32.8 (81) |
| Female | 57.1 (868) | 67.2 (166) |
| Age, mean (SD), yr | 43.0 (11.5) | 45.2 (9.3) |
| CO, mean (SD), ppm | 26.5 (12.3) | 21.4 (9.5) |
| CPD, mean (SD) | 22.4 (9.0) | 18.6 (7.7) |
| Categorized baseline CPD, % (N) | ||
| ≤10 | 4.5 (69) | 10.1 (25) |
| 11–20 | 51.2 (777) | 63.2 (156) |
| 21– 30 | 30.4 (461) | 19.8 (49) |
| 31+ | 14.0 (212) | 6.9 (17) |
| FTND, mean (SD) | 5.4 (2.2) | 5.5 (2.0) |
| CHRNA5-CHRNA3-CHRNB4 | ||
| rs16969968,* % (N) | ||
| AA | 12.8 (193) | 0.4 (1) |
| AG | 48.7 (737) | 10.5 (26) |
| GG | 38.5 (582) | 89.1 (220) |
| # of A alleles, mean (SD) | 0.74 (0.67) | 0.11 (0.33) |
| CHRNA6-CHRNB3 | ||
| rs4950,* % (N) | ||
| AA | 63.0 (955) | 8.1 (20) |
| AG | 32.9 (498) | 41.3 (102) |
| GG | 4.2 (63) | 50.6 (125) |
| # of G alleles, mean (SD) | 0.41 (0.57) | 1.43 (0.64) |
| CYP2A6 | ||
| rs28399442,* % (N) | ||
| AA | 0 (0) | 0 (0) |
| AG | 5.6 (85) | 0.8 (2) |
| GG | 94.2 (1,433) | 99.2 (245) |
| # of A alleles, mean (SD) | 0.56 (0.23) | 0.01 (0.09) |
| CYP2A6 | ||
| Metabolism metric,† mean (SD) | 0.86 (0.07) | 0.87 (0.06) |
| Characteristic | European American (N = 1,521) | African American (N = 247) |
|---|---|---|
| Sex, % (N) | ||
| Male | 42.9 (653) | 32.8 (81) |
| Female | 57.1 (868) | 67.2 (166) |
| Age, mean (SD), yr | 43.0 (11.5) | 45.2 (9.3) |
| CO, mean (SD), ppm | 26.5 (12.3) | 21.4 (9.5) |
| CPD, mean (SD) | 22.4 (9.0) | 18.6 (7.7) |
| Categorized baseline CPD, % (N) | ||
| ≤10 | 4.5 (69) | 10.1 (25) |
| 11–20 | 51.2 (777) | 63.2 (156) |
| 21– 30 | 30.4 (461) | 19.8 (49) |
| 31+ | 14.0 (212) | 6.9 (17) |
| FTND, mean (SD) | 5.4 (2.2) | 5.5 (2.0) |
| CHRNA5-CHRNA3-CHRNB4 | ||
| rs16969968,* % (N) | ||
| AA | 12.8 (193) | 0.4 (1) |
| AG | 48.7 (737) | 10.5 (26) |
| GG | 38.5 (582) | 89.1 (220) |
| # of A alleles, mean (SD) | 0.74 (0.67) | 0.11 (0.33) |
| CHRNA6-CHRNB3 | ||
| rs4950,* % (N) | ||
| AA | 63.0 (955) | 8.1 (20) |
| AG | 32.9 (498) | 41.3 (102) |
| GG | 4.2 (63) | 50.6 (125) |
| # of G alleles, mean (SD) | 0.41 (0.57) | 1.43 (0.64) |
| CYP2A6 | ||
| rs28399442,* % (N) | ||
| AA | 0 (0) | 0 (0) |
| AG | 5.6 (85) | 0.8 (2) |
| GG | 94.2 (1,433) | 99.2 (245) |
| # of A alleles, mean (SD) | 0.56 (0.23) | 0.01 (0.09) |
| CYP2A6 | ||
| Metabolism metric,† mean (SD) | 0.86 (0.07) | 0.87 (0.06) |
Definition of abbreviations: CO = mean exhaled carbon monoxide; CPD = number of cigarettes smoked per day; FTND = Fagerstrom Test for Nicotine Dependence score.
* Quality control measures zeroed the genotypes of single-nucleotide polymorphisms rs16969968, rs4950, and rs28399442 for N = 9, 5, and 3 European Americans, respectively.
† Metabolism metric available for N = 1,348 European Americans and N = 224 African Americans due to limited DNA.
Table 1.
Sample characteristics by self-reported ethnicity
| Characteristic | European American (N = 1,521) | African American (N = 247) |
|---|---|---|
| Sex, % (N) | ||
| Male | 42.9 (653) | 32.8 (81) |
| Female | 57.1 (868) | 67.2 (166) |
| Age, mean (SD), yr | 43.0 (11.5) | 45.2 (9.3) |
| CO, mean (SD), ppm | 26.5 (12.3) | 21.4 (9.5) |
| CPD, mean (SD) | 22.4 (9.0) | 18.6 (7.7) |
| Categorized baseline CPD, % (N) | ||
| ≤10 | 4.5 (69) | 10.1 (25) |
| 11–20 | 51.2 (777) | 63.2 (156) |
| 21– 30 | 30.4 (461) | 19.8 (49) |
| 31+ | 14.0 (212) | 6.9 (17) |
| FTND, mean (SD) | 5.4 (2.2) | 5.5 (2.0) |
| CHRNA5-CHRNA3-CHRNB4 | ||
| rs16969968,* % (N) | ||
| AA | 12.8 (193) | 0.4 (1) |
| AG | 48.7 (737) | 10.5 (26) |
| GG | 38.5 (582) | 89.1 (220) |
| # of A alleles, mean (SD) | 0.74 (0.67) | 0.11 (0.33) |
| CHRNA6-CHRNB3 | ||
| rs4950,* % (N) | ||
| AA | 63.0 (955) | 8.1 (20) |
| AG | 32.9 (498) | 41.3 (102) |
| GG | 4.2 (63) | 50.6 (125) |
| # of G alleles, mean (SD) | 0.41 (0.57) | 1.43 (0.64) |
| CYP2A6 | ||
| rs28399442,* % (N) | ||
| AA | 0 (0) | 0 (0) |
| AG | 5.6 (85) | 0.8 (2) |
| GG | 94.2 (1,433) | 99.2 (245) |
| # of A alleles, mean (SD) | 0.56 (0.23) | 0.01 (0.09) |
| CYP2A6 | ||
| Metabolism metric,† mean (SD) | 0.86 (0.07) | 0.87 (0.06) |
| Characteristic | European American (N = 1,521) | African American (N = 247) |
|---|---|---|
| Sex, % (N) | ||
| Male | 42.9 (653) | 32.8 (81) |
| Female | 57.1 (868) | 67.2 (166) |
| Age, mean (SD), yr | 43.0 (11.5) | 45.2 (9.3) |
| CO, mean (SD), ppm | 26.5 (12.3) | 21.4 (9.5) |
| CPD, mean (SD) | 22.4 (9.0) | 18.6 (7.7) |
| Categorized baseline CPD, % (N) | ||
| ≤10 | 4.5 (69) | 10.1 (25) |
| 11–20 | 51.2 (777) | 63.2 (156) |
| 21– 30 | 30.4 (461) | 19.8 (49) |
| 31+ | 14.0 (212) | 6.9 (17) |
| FTND, mean (SD) | 5.4 (2.2) | 5.5 (2.0) |
| CHRNA5-CHRNA3-CHRNB4 | ||
| rs16969968,* % (N) | ||
| AA | 12.8 (193) | 0.4 (1) |
| AG | 48.7 (737) | 10.5 (26) |
| GG | 38.5 (582) | 89.1 (220) |
| # of A alleles, mean (SD) | 0.74 (0.67) | 0.11 (0.33) |
| CHRNA6-CHRNB3 | ||
| rs4950,* % (N) | ||
| AA | 63.0 (955) | 8.1 (20) |
| AG | 32.9 (498) | 41.3 (102) |
| GG | 4.2 (63) | 50.6 (125) |
| # of G alleles, mean (SD) | 0.41 (0.57) | 1.43 (0.64) |
| CYP2A6 | ||
| rs28399442,* % (N) | ||
| AA | 0 (0) | 0 (0) |
| AG | 5.6 (85) | 0.8 (2) |
| GG | 94.2 (1,433) | 99.2 (245) |
| # of A alleles, mean (SD) | 0.56 (0.23) | 0.01 (0.09) |
| CYP2A6 | ||
| Metabolism metric,† mean (SD) | 0.86 (0.07) | 0.87 (0.06) |
Definition of abbreviations: CO = mean exhaled carbon monoxide; CPD = number of cigarettes smoked per day; FTND = Fagerstrom Test for Nicotine Dependence score.
* Quality control measures zeroed the genotypes of single-nucleotide polymorphisms rs16969968, rs4950, and rs28399442 for N = 9, 5, and 3 European Americans, respectively.
† Metabolism metric available for N = 1,348 European Americans and N = 224 African Americans due to limited DNA.
Participants completed baseline assessments of demographics, smoking history, and current number of cigarettes smoked per day. Number of cigarettes smoked per day at the baseline assessment was evaluated by the question “On average, about how many cigarettes do you currently smoke per day?” and categorized into four levels (≤10, 11–20, 21–30, and ≥31). Tobacco dependence was measured by the FTND, a score that ranges from 0 to 10 (21). Pack-years was defined as cigarettes smoked per day times length of time smoked. A baseline breath sample for CO analysis was collected before the initiation of smoking cessation pharmacotherapy and the quit attempt. Participants received no instructions to cut down or modify their smoking before sample collection, and the CO measures were taken without regard to the time of the last cigarette smoked. CO samples were collected using standard assessment methods for smoking cessation clinical trials and occurred after a check-in process that entailed a delay between smoking and CO collection to reduce distortion by immediate smoking. The distributions of the CO measure and cigarettes per day phenotype are provided in Figure 1 and Figure E1 in the online supplement for subjects of European and African descent.
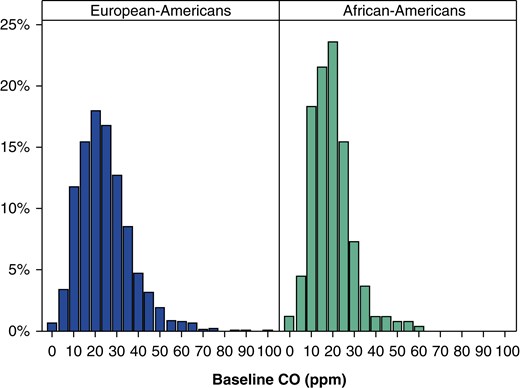
Figure 1.
Distribution of mean baseline exhaled carbon monoxide (CO) among current smokers stratified by ancestry group.
Genomewide genotyping was performed by the Center for Inherited Disease Research at Johns Hopkins University using the Illumina Omni2.5 microarray (www.illumina.com), and data cleaning was led by the GENEVA Coordinating Center at the University of Washington (25). All single-nucleotide polymorphisms (SNPs) analyzed conformed to Hardy-Weinberg equilibrium. Genotyping was successful in more than 99% of subjects. Population stratification was assessed and self-reported race confirmed by principal components analysis with HapMap samples as anchors (26).
A genome-wide association study (GWAS) of CO in European Americans was performed using a linear model with SNP genotype additively coded and sex, age, and study as covariates. Analyses were performed using PLINK v1.07 (27). Additional association analyses with CO focused on targeted regions that represent three key genetic loci known to be associated with smoking behavior at genomewide significance in European-ancestry populations (28, 29): the chromosome 15q25.1 locus, which includes the α5-α3-β4 nicotinic receptor subunit genes (CHRNA5-CHRNA3-CHRNB4); the chromosome 19q13.2 locus, which includes the primary gene responsible for nicotinic metabolism (CYP2A6); and the chromosome 8p11.21 locus, which includes the β3-α6 nicotinic receptor subunit genes (CHRNB3-CHRNA6). Analyses were performed using SAS (Cary, NC).
Because the CYP2A6 locus is heterogeneous and no single variant can act as a proxy for CYP2A6 activity, we performed additional genotyping and used a predictive model of CYP2A6 activity that provides a continuous estimate of nicotine metabolism based on CYP2A6 genotype (30, 31). This estimate is based on six SNPs: rs28399433 (TATA box *9), rs1137115 (V17V), rs28399435 (S29N *14), rs1801272 (L160H *2), rs28399442 (correlated with *12), rs148166815 (Y351H *38), and variation in CYP2A6 gene copy number. We computed a predicted nicotine metabolism metric from haplotypes using regression model parameters originally determined from an oral deuterated nicotine metabolism experiment in European Americans in which CYP2A6 haplotypes explained more than 70% of the variance in the conversion of nicotine to cotinine (cotinine/[nicotine + cotinine]) 30 minutes after oral administration. The metric was calculated by the equation:
where α = 0.56 and β1 or β2 = 1.00 for haplotypes *2, *4, *12, and *38 carriers; 0.65 for haplotype *9; 0.57 for haplotype *1A; and 0.42 for all other CYP2A6 haplotypes. This metabolism estimate was available for a subsample because of limited availability of DNA (n = 1,348 European descent subjects and 224 African descent subjects).
Results
Genome-wide association analysis of European ancestry subjects identifies the chromosome 15q25.1 region that includes the nicotinic receptor genes CHRNA5-CHRNA3-CHRNB4 as the locus most strongly associated with CO (Figures 2A and 2B), with several SNPs surpassing GWAS significance. Variants that demonstrate GWAS-significant association with CO include rs16969968 in CHRNA5 (β = 2.66; 95% confidence interval [CI], 1.74–3.58; P = 1.65 × 10−8), which causes an amino acid change in the receptor protein and alters receptor activity (32). The direction of effect is as expected given the previous association with heaviness of smoking. In European ancestry populations, this variant is highly correlated with rs1051730 in CHRNA3 (r 2 = 1) and rs8031948 in ADGPH1 (r 2 = 0.934), which, in addition to rs16969968, have been previously associated with smoking behavior, COPD, and lung cancer (4, 33–35). The SNP most significantly associated with CO, rs55958997 (β = 2.75; 95% CI, 1.86–3.64; P = 1.60 × 10−9), is also in high linkage disequilibrium with rs16969968 (r 2 = 0.82) in Europeans (36).
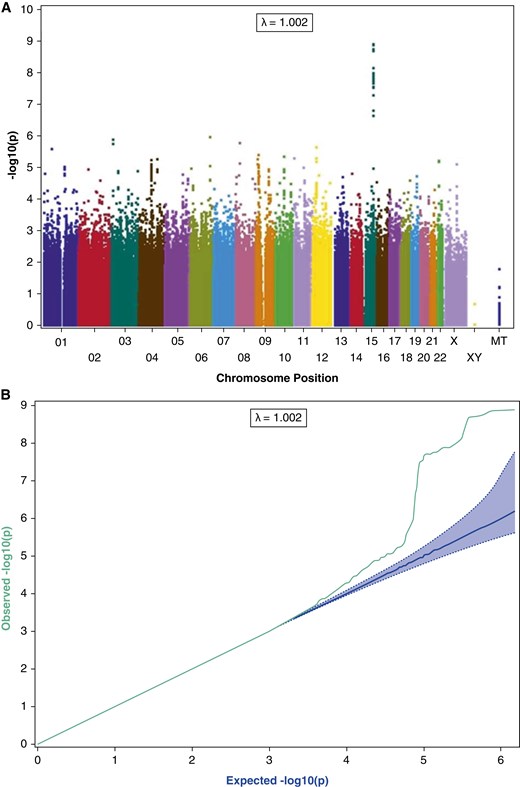
Figure 2.
Genomewide association scan among European Americans for genes associated with exhaled carbon monoxide (CO). (A) The –log10 of the P value is plotted for each single nucleotide polymorphism (SNP) in chromosomal order. The spacing between SNPs on the plot is based on physical map length. (B) Q-Q plot for genomewide association scan among European Americans for genes associated with exhaled CO.
In addition to chromosome 15 associations, variants in the CYP2A6 region are associated with CO, although not surpassing the genomewide significance level. The variant rs28399442 is strongly associated with CO (β = −5.45; 95% CI, −8.13 to −2.75; P = 6.84 × 10−5); rs28399442 is a perfect proxy for CYP2A6*12, one of several known loss-of-function alleles. The predicted CYP2A6 metabolism metric is also strongly associated with CO (β = 20.83; 95% CI, 11.35–30.31; P = 1.67 × 10−5). These results are consistent with faster metabolism being associated with heavier smoking and thus higher CO levels.
The results for these two loci are summarized in Table 2, and association results with these genetic variants with cigarettes per day and nicotine dependence are included for comparison. Current cigarettes smoked per day and current nicotine dependence (FTND) measures were entered with CO into genetic models to gauge the magnitude and independence of their effects. The variant rs16969968 remains strongly associated with CO after adjusting for cigarettes per day (β = 2.18; 95% CI, 1.32–3.04; P = 7.47 × 10−7) and for FTND (β = 2.28; 95% CI, 1.42–3.14; P = 1.76 × 10−7). Figure 3 illustrates the relation between rs16969968 genotype and CO within strata of self-reported smoking rate. The CYP2A6 metric also continues to be associated with CO after correction for these measures of smoking behavior (rs28399442, β = −4.10; 95% CI, −6.60 to −1.60; P = 1.3 × 10−3 adjusted for cigarettes per day and β = −4.12; 95% CI, −6.59 to −1.64; P = 1.1 × 10−3 adjusted for FTND). Overall, the variant rs16969968 explains 2.06% (95% CI, 0.88–3.69%) of the variance in CO and 0.47% (95% CI, 0.03–1.39%) of the variance in current cigarettes per day; the CYP2A6 metric explains 0.84% (95% CI, 0.15–2.07%) of the variance in the CO and 0.51% (95% CI, 0.03–1.54%) of the variance in current cigarettes per day.
Table 2.
Associations between measures of CHRNA5-CHRNA3-CHRNB4, CYP2A6, CHRNA6-CHRNB3, and smoking-related phenotypes among European American (N = 1,521) subjects
| CHRNA5-CHRNA3-CHRNB4 (rs16969968) | CYP2A6 (rs28399442) | CYP2A6 * (metabolism metric) | CHRNA6-CHRNB3 (rs4950) | |||||
|---|---|---|---|---|---|---|---|---|
| β† (95% CI) | P Value | β† (95%CI) | P Value | β‡ (95%CI) | P Value | β† (95%CI) | P Value | |
| CO* | 2.66 (1.74 to 3.58) | 1.65 × 10−8 | −5.65 (−8.30 to −2.99) | 3.2 × 10−5 | 20.83 (11.35 to 30.31) | 1.7 × 10−5 | −0.25 (−1.33 to 0.82) | 6.5 × 10−1 |
| CPD | 0.08 (0.02 to 0.13) | 7.7 × 10−3 | −0.27 (−0.44 to −0.11) | 9.8 × 10−4 | 1.08 (0.49 to 1.66) | 2.9 × 10−4 | −0.02 (−0.09 to 0.05) | 5.5 × 10−1 |
| FTND | 0.17 (0.01 to 0.33) | 4.2 × 10−2 | −0.62 (−1.09 to −0.15) | 9.3 × 10−3 | 1.44 (−0.23 to 3.10) | 9.0 × 10−2 | 0.01 (−0.18 to 0.20) | 9.1 × 10−1 |
| CHRNA5-CHRNA3-CHRNB4 (rs16969968) | CYP2A6 (rs28399442) | CYP2A6 * (metabolism metric) | CHRNA6-CHRNB3 (rs4950) | |||||
|---|---|---|---|---|---|---|---|---|
| β† (95% CI) | P Value | β† (95%CI) | P Value | β‡ (95%CI) | P Value | β† (95%CI) | P Value | |
| CO* | 2.66 (1.74 to 3.58) | 1.65 × 10−8 | −5.65 (−8.30 to −2.99) | 3.2 × 10−5 | 20.83 (11.35 to 30.31) | 1.7 × 10−5 | −0.25 (−1.33 to 0.82) | 6.5 × 10−1 |
| CPD | 0.08 (0.02 to 0.13) | 7.7 × 10−3 | −0.27 (−0.44 to −0.11) | 9.8 × 10−4 | 1.08 (0.49 to 1.66) | 2.9 × 10−4 | −0.02 (−0.09 to 0.05) | 5.5 × 10−1 |
| FTND | 0.17 (0.01 to 0.33) | 4.2 × 10−2 | −0.62 (−1.09 to −0.15) | 9.3 × 10−3 | 1.44 (−0.23 to 3.10) | 9.0 × 10−2 | 0.01 (−0.18 to 0.20) | 9.1 × 10−1 |
Definition of abbreviations: CI = confidence interval; CO = mean exhaled carbon monoxide in ppm; CPD = four-level measure of the number of cigarettes smoked per day (≤10 CPD = 0, 11–20 CPD = 1, 21–30 CPD = 2, and ≥31 CPD = 3); FTND = Fagerstrom Test for Nicotine Dependence score (0–10 scale).
Analyses adjusted for sex, age, population-stratification principal components, and study.
* Metabolism metric available for N = 1,348 European Americans.
† Estimated effect of the number of copies of the minor allele.
‡ Parameter estimate is expressed as increase of 0.1 unit in estimated CYP2A6 activity (scale 0.0–1.0).
Table 2.
Associations between measures of CHRNA5-CHRNA3-CHRNB4, CYP2A6, CHRNA6-CHRNB3, and smoking-related phenotypes among European American (N = 1,521) subjects
| CHRNA5-CHRNA3-CHRNB4 (rs16969968) | CYP2A6 (rs28399442) | CYP2A6 * (metabolism metric) | CHRNA6-CHRNB3 (rs4950) | |||||
|---|---|---|---|---|---|---|---|---|
| β† (95% CI) | P Value | β† (95%CI) | P Value | β‡ (95%CI) | P Value | β† (95%CI) | P Value | |
| CO* | 2.66 (1.74 to 3.58) | 1.65 × 10−8 | −5.65 (−8.30 to −2.99) | 3.2 × 10−5 | 20.83 (11.35 to 30.31) | 1.7 × 10−5 | −0.25 (−1.33 to 0.82) | 6.5 × 10−1 |
| CPD | 0.08 (0.02 to 0.13) | 7.7 × 10−3 | −0.27 (−0.44 to −0.11) | 9.8 × 10−4 | 1.08 (0.49 to 1.66) | 2.9 × 10−4 | −0.02 (−0.09 to 0.05) | 5.5 × 10−1 |
| FTND | 0.17 (0.01 to 0.33) | 4.2 × 10−2 | −0.62 (−1.09 to −0.15) | 9.3 × 10−3 | 1.44 (−0.23 to 3.10) | 9.0 × 10−2 | 0.01 (−0.18 to 0.20) | 9.1 × 10−1 |
| CHRNA5-CHRNA3-CHRNB4 (rs16969968) | CYP2A6 (rs28399442) | CYP2A6 * (metabolism metric) | CHRNA6-CHRNB3 (rs4950) | |||||
|---|---|---|---|---|---|---|---|---|
| β† (95% CI) | P Value | β† (95%CI) | P Value | β‡ (95%CI) | P Value | β† (95%CI) | P Value | |
| CO* | 2.66 (1.74 to 3.58) | 1.65 × 10−8 | −5.65 (−8.30 to −2.99) | 3.2 × 10−5 | 20.83 (11.35 to 30.31) | 1.7 × 10−5 | −0.25 (−1.33 to 0.82) | 6.5 × 10−1 |
| CPD | 0.08 (0.02 to 0.13) | 7.7 × 10−3 | −0.27 (−0.44 to −0.11) | 9.8 × 10−4 | 1.08 (0.49 to 1.66) | 2.9 × 10−4 | −0.02 (−0.09 to 0.05) | 5.5 × 10−1 |
| FTND | 0.17 (0.01 to 0.33) | 4.2 × 10−2 | −0.62 (−1.09 to −0.15) | 9.3 × 10−3 | 1.44 (−0.23 to 3.10) | 9.0 × 10−2 | 0.01 (−0.18 to 0.20) | 9.1 × 10−1 |
Definition of abbreviations: CI = confidence interval; CO = mean exhaled carbon monoxide in ppm; CPD = four-level measure of the number of cigarettes smoked per day (≤10 CPD = 0, 11–20 CPD = 1, 21–30 CPD = 2, and ≥31 CPD = 3); FTND = Fagerstrom Test for Nicotine Dependence score (0–10 scale).
Analyses adjusted for sex, age, population-stratification principal components, and study.
* Metabolism metric available for N = 1,348 European Americans.
† Estimated effect of the number of copies of the minor allele.
‡ Parameter estimate is expressed as increase of 0.1 unit in estimated CYP2A6 activity (scale 0.0–1.0).
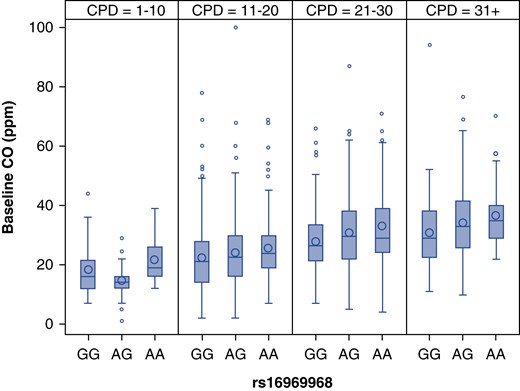
Figure 3.
Mean baseline exhaled carbon monoxide (CO) among current smokers divided by categorical cigarettes per day (CPD) and rs16969968 genotype among European Americans. The boxplot represents the interquartile range divided by a line indicating the median; whisker lines extend to values within 1.5× the interquartile range; further outliers are marked with circles.
In contrast to the CHRNA5-CHRNA3-CHRNB4 and CYP2A6 results, the region spanning CHRNB3-CHRNA6 on chromosome 8 does not have any genotyped variants that show association with CO, cigarettes per day, or nicotine dependence. For example, rs4950, a variant strongly associated with nicotine dependence at a genomewide significance level in previous studies (28, 37), is not associated with CO (β = −0.25; 95% CI, −1.33 to 0.83; P = 0.647).
We separately examined the associations of these targeted loci with CO in the African American sample. This sample is modest in size, and the expected power to detect association is reduced. The variant rs16969968 in CHRNA5-CHRNA3-CHRNB4 is uncommon in African Americans (minor allele frequency = 5.7%). The point estimate for the β coefficient is in the same direction as the effect seen in the European ancestry subjects, but this finding is not statistically significant (β = 0.91; 95% CI, −2.79 to 4.60; P = 0.63). The CYP2A6 variant rs28399442 is rare in African Americans (minor allele frequency = 0.4%), and no evidence of association is seen. The predicted CYP2A6 metric was developed in European ancestry subjects and was not expected to capture CYP2A6 metabolism well in other ancestral groups, and it does not predict CO in the African American subjects.
To further understand the relation between current cigarette smoking and CO, we examined the correlation between cigarettes per day and CO. The correlation between these smoking measures is modest in European-ancestry subjects (r = 0.36; 95% CI, 0.31–0.40; P = 0.0001). The correlation is markedly lower in African American subjects (r = 0.14; 95% CI, 0.02–0.26; P = 0.003), and there is a statistically different correlation between CO and cigarettes per day in European and African American subjects (z = 3.43; P = 6.07 × 10−4). Figure 4 illustrates these different relations between number of cigarettes smoked and CO level in the two populations.
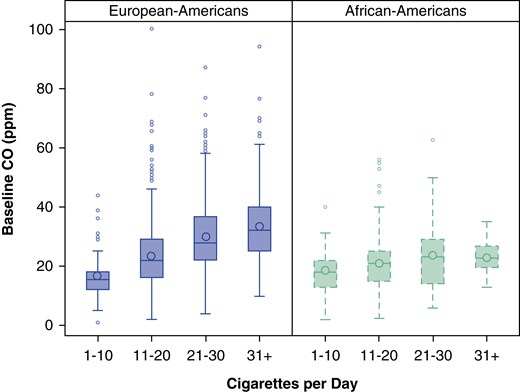
Figure 4.
Mean baseline exhaled carbon monoxide (CO) among current smokers divided by categorical cigarettes per day and ancestry group. The boxplot represents the interquartile range divided by a line indicating the median; whisker lines extend to values within 1.5× the interquartile range; further outliers are marked with circles.
Subsequent GWAS of pack-years did not identify any significant genetic associations in European or African American subjects (see Figures E2 and E3).
Discussion
This investigation undertakes the first GWAS of exhaled CO in current smokers. Specifically, this study demonstrates a genomewide significant association between the biomarker CO and the CHRNA5-CHRNA3-CHRNB4 locus in a modestly sized sample of heavy smoking European-ancestry subjects (β = 2.66; 95% CI, 1.74–3.58; P = 1.65 × 10−8). Importantly, the variant rs16969968 in CHRNA5 explains a larger portion of the variance in the CO phenotype (2.06%) than the smoking behavior measure of cigarettes per day (0.47%). This association of CHRNA5 with CO remains strongly associated after adjustment for current cigarettes smoked per day (β = 2.18; 95% CI, 1.32–3.04; P = 7.47 × 10−7). Genetic variation in CYP2A6, the gene primarily responsible for nicotine metabolism, is also strongly associated with CO, an association that remains after controlling for number of cigarettes smoked.
The locus CHRNA5-CHRNA3-CHRNB4 has been associated with COPD and lung cancer even after controlling for self-reported smoking rate; the current results suggest that this association may be due to the insensitivity of self-reported smoking as an index of cigarette smoke exposure. We find that self-reported smoking amount accounts for only 13% of variance in CO among European Americans. Furthermore, compared with number of cigarettes smoked per day, exhaled CO is more highly correlated with levels of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (20), a key biomarker of carcinogen exposure. Thus, using cigarettes per day and self-reported nicotine dependence as measures for smoking-related toxin and carcinogen exposure do not fully capture the health risks associated with smoking.
This evidence builds on prior findings that showed a strong association between the CHRNA5-CHRNA3-CHRNB4 locus and another smoking biomarker, cotinine, a metabolite of nicotine (7, 38, 39). Each biomarker has its advantages. Exhaled CO is easier and less expensive to collect, and it may more directly reflect the toxic effects of smoking. However, CO also has a shorter half-life than cotinine and therefore provides a less stable measure over time and may provide a poor indicator of exposure among light or occasional smokers. On the other hand, cotinine’s longer half-life reflects smoking behavior over a longer period of time, but its levels can be strongly influenced by variation in nicotine and cotinine metabolism unrelated to differences in smoke exposure (40). The variance in these two biomarkers, relative to cigarettes per day, explained by the key variant in CHRNA5-CHRNA3-CHRNB4 appears to be similar. We found rs16969968-rs1051730 explains 2.1% of the variance in CO compared with 0.5% of the variance in cigarettes per day, and a prior study in European smokers found this same locus explained 4.3% of the variance in cotinine versus 0.9% of the variance in cigarettes per day (38).
In contrast to the associations of CO with the CHRNA5 and CYP2A6 loci, another locus that demonstrated genomewide association with smoking behavior in large metaanalyses (28, 29), CHRNA6-CHRNB3 on chromosome 8, shows no evidence for association with CO in this sample. At present it is unclear why the CHRNA6-CHRNB3 region fails to demonstrate any association with CO. The lack of association with the CHRNA6-CHRNB3 locus may merely reflect its weaker association with nicotine dependence, consistent with its relatively modest association in GWAS of cigarettes per day in other studies. However, it is also possible that these genetic loci influence smoking behaviors via separate mechanisms that differentially impact levels of CO versus number of cigarettes smoked per day (e.g., influencing responses to nicotine in the initiation of smoking behaviors versus promoting more intense smoking of individual cigarettes and increased tolerance to smoking’s aversive effects) (41, 42). The existence of separable mechanisms influencing levels of CO versus amount smoked is consistent with findings that the CHRNA6-CHRNB3 locus shows little contribution to the genetic risk of COPD and lung cancer in the large-scale studies (43, 44).
Our findings have implications for the estimation of exposure to cigarette-related toxins. Current social and regulatory environments are leading to changes in smoking behavior. Because of smoking constraints (smoking restriction policies and increased cost), smokers are reducing the number of cigarettes they smoke (45). Among daily smokers, the proportion who smoked 30 or more cigarettes per day declined significantly from 12.5% in 2005 to 9.1% in 2011, whereas the proportion of those who smoked 1 to 9 cigarettes per day increased significantly from 16.4 to 22.0%. However, smoking fewer cigarettes may not reduce harm if smokers compensate by inhaling more intensively. At low levels of smoking, the number of cigarettes smoked per day may underestimate tobacco toxicant exposure (20, 46). Thus, biomarkers may increase in importance as the distribution of cigarettes smoked per day becomes truncated and smoking intensity becomes the primary driver of smoking-related toxin exposure. In addition, new smoking behaviors are developing, including the use of electronic cigarettes, which will likely change a person’s exposure to smoking-related toxins. Finally, our results suggest that prior studies that have evaluated genetic associations with smoking heaviness most likely have systematically underestimated the magnitude of the genetic contribution to smoking behavior. Indeed, in this study the genetic effect of variation in CHRNA5 (rs16969968) on CO is more than two times greater than its effect on number of cigarettes smoked per day (effect size defined by β/SE: 5.68 vs. 2.66).
Importantly, our results indicate that number of self-reported current cigarettes smoked per day is an especially poor proxy for tobacco smoke exposure among African Americans. We observe a very low correlation between number of cigarettes smoked per day and CO measures in African Americans relative to European Americans, confirming prior reports (47, 48). This observation may help to explain findings that African Americans are at higher risk for lung cancer than other racial groups despite lower cigarette consumption (49).
These results should be interpreted in the context of the limitations of this study. The University of Wisconsin Center for Tobacco Research and Intervention smoking cessation trials include only subjects who actively sought treatment for smoking cessation (22–24). Treatment seeking is associated with high levels of tobacco dependence (50), higher number of cigarettes smoked, and perhaps higher smoking intensity as well (51). The unique features of this sample may have influenced the genetic associations observed.
In summary, the biomarker of current combustive cigarette exposure, exhaled CO, captures important aspects of smoking behavior including number of puffs per cigarette and depth of inhalation, as well as number of cigarettes smoked per day, all of which relate to smoking-related health risks. Genetic variation in CHRNA5 and CYP2A6 influence smoking behaviors, which in turn contribute to a person’s CO level. Furthermore, the relation between current cigarettes smoked per day and CO is markedly weaker in African Americans than in European Americans in our sample, perhaps providing further insight into the heightened lung cancer risk observed in African Americans compared with European Americans who smoke similar numbers of cigarettes. These findings point to the value of biomarkers such as CO to improve the measurement of current smoking behavior and toxin exposure relevant to smoking-related disease endpoints.
Acknowledgment
The authors thank the Wisconsin State Laboratory of Hygiene, which provided considerable technical assistance in the form of DNA extraction. Research was also aided by the Wisconsin Partnership Program. Medication was provided to patients at no cost under a research agreement with GlaxoSmithKline. The authors appreciate the contributions of Sherri Fisher in the development of this paper.
Footnotes
Supported by the National Institute of Mental Health grant T32MH014677 (A.J.B.); the National Cancer Institute grants P01CA089392 (L.J.B.) and K05CA139871 (T.B.B.); the National Institute on Drug Abuse grant K08DA030398 (L.-S.C.), R01DA026911 (N.L.S.), and K02DA021237 (L.J.B.); and the National Human Genome Research Institute grant U01HG004422 (L.J.B.). Funding support for genotyping, performed at the Center for Inherited Disease Research (CIDR), was provided by 1 X01 HG005274–01. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. Assistance with genotype cleaning and general study coordination was provided by the Gene Environment Association Studies (GENEVA) Coordinating Center grant U01HG004446. Funding support for collection of datasets and samples was provided by the Collaborative Genetic Study of Nicotine Dependence grant P01CA089392 and the University of Wisconsin Transdisciplinary Tobacco Use Research Center grants P50DA019706 and P50CA084724.
Author Contributions: A.J.B. contributed to generation, analysis, and interpretation of data and writing of the manuscript. S.M.H. contributed to data analysis and writing of the manuscript. L.J.B. and T.B.B. contributed to study design, generation of data, analysis, interpretation of data, and writing of the manuscript. M.E.P. contributed to data generation and writing of the manuscript. L.F. contributed to data analysis. M.M. and C.C.L. contributed to data generation. D.H., N.L.S., E.O.J., and L.-S.C. contributed to data interpretation and writing of the manuscript. A.G. contributed to data interpretation.
This article has an online supplement, which is accessible from this issue’s table of contents.
Author disclosures are available with the text of this article.
References
Anand
P
,
Kunnumakkara
AB
,
Sundaram
C
,
Harikumar
KB
,
Tharakan
ST
,
Lai
OS
,
Sung
B
,
Aggarwal
BB
.
Cancer is a preventable disease that requires major lifestyle changes
.
Pharm Res
2008
;
25
:
2097
–
2116
.
Lips
EH
,
Gaborieau
V
,
McKay
JD
,
Chabrier
A
,
Hung
RJ
,
Boffetta
P
,
Hashibe
M
,
Zaridze
D
,
Szeszenia-Dabrowska
N
,
Lissowska
J
, et al.
Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals
.
Int J Epidemiol
2010
;
39
:
563
–
577
.
Wilk
JB
,
Shrine
NR
,
Loehr
LR
,
Zhao
JH
,
Manichaikul
A
,
Lopez
LM
,
Smith
AV
,
Heckbert
SR
,
Smolonska
J
,
Tang
W
, et al.
Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction
.
Am J Respir Crit Care Med
2012
;
186
:
622
–
632
.
VanderWeele
TJ
,
Asomaning
K
,
Tchetgen Tchetgen
EJ
,
Han
Y
,
Spitz
MR
,
Shete
S
,
Wu
X
,
Gaborieau
V
,
Wang
Y
,
McLaughlin
J
, et al.
Genetic variants on 15q25.1, smoking, and lung cancer: an assessment of mediation and interaction
.
Am J Epidemiol
2012
;
175
:
1013
–
1020
.
Siedlinski
M
,
Tingley
D
,
Lipman
PJ
,
Cho
MH
,
Litonjua
AA
,
Sparrow
D
,
Bakke
P
,
Gulsvik
A
,
Lomas
DA
,
Anderson
W
, et al. ;
COPDGene and ECLIPSE Investigators
.
Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility
.
Hum Genet
2013
;
132
:
431
–
441
.
Munafò
MR
,
Timofeeva
MN
,
Morris
RW
,
Prieto-Merino
D
,
Sattar
N
,
Brennan
P
,
Johnstone
EC
,
Relton
C
,
Johnson
PC
,
Walther
D
, et al. ;
EPIC Study Group
.
Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure
.
J Natl Cancer Inst
2012
;
104
:
740
–
748
.
Thorgeirsson
TE
,
Geller
F
,
Sulem
P
,
Rafnar
T
,
Wiste
A
,
Magnusson
KP
,
Manolescu
A
,
Thorleifsson
G
,
Stefansson
H
,
Ingason
A
, et al.
A variant associated with nicotine dependence, lung cancer and peripheral arterial disease
.
Nature
2008
;
452
:
638
–
642
.
Spitz
MR
,
Amos
CI
,
Bierut
LJ
,
Caporaso
NE
.
Cotinine conundrum—a step forward but questions remain
.
J Natl Cancer Inst
2012
;
104
:
720
–
722
.
Benowitz
NL
,
Hall
SM
,
Herning
RI
,
Jacob
P
III,
Jones
RT
,
Osman
AL
.
Smokers of low-yield cigarettes do not consume less nicotine
.
N Engl J Med
1983
;
309
:
139
–
142
.
Benowitz
NL
,
Jacob
P
III,
Yu
L
,
Talcott
R
,
Hall
S
,
Jones
RT
.
Reduced tar, nicotine, and carbon monoxide exposure while smoking ultralow- but not low-yield cigarettes
.
JAMA
1986
;
256
:
241
–
246
.
Herning
RI
,
Jones
RT
,
Bachman
J
,
Mines
AH
.
Puff volume increases when low-nicotine cigarettes are smoked
.
Br Med J (Clin Res Ed)
1981
;
283
:
187
–
189
.
Etter
JF
,
Vu Duc
T
,
Perneger
TV
.
Saliva cotinine levels in smokers and nonsmokers
.
Am J Epidemiol
2000
;
151
:
251
–
258
.
Krall
EA
,
Valadian
I
,
Dwyer
JT
,
Gardner
J
.
Accuracy of recalled smoking data
.
Am J Public Health
1989
;
79
:
200
–
202
.
Vesey
CJ
,
Saloojee
Y
,
Cole
PV
,
Russell
MA
.
Blood carboxyhaemoglobin, plasma thiocyanate, and cigarette consumption: implications for epidemiological studies in smokers
.
Br Med J (Clin Res Ed)
1982
;
284
:
1516
–
1518
.
Benowitz
NL
,
Jacob
P
III,
Kozlowski
LT
,
Yu
L
.
Influence of smoking fewer cigarettes on exposure to tar, nicotine, and carbon monoxide
.
N Engl J Med
1986
;
315
:
1310
–
1313
.
Robinson
JC
,
Forbes
WF
.
The role of carbon monoxide in cigarette smoking. I. Carbon monoxide yield from cigarettes
.
Arch Environ Health
1975
;
30
:
425
–
434
.
Zacny
JP
,
Stitzer
ML
,
Brown
FJ
,
Yingling
JE
,
Griffiths
RR
.
Human cigarette smoking: effects of puff and inhalation parameters on smoke exposure
.
J Pharmacol Exp Ther
1987
;
240
:
554
–
564
.
Marrone
GF
,
Shakleya
DM
,
Scheidweiler
KB
,
Singleton
EG
,
Huestis
MA
,
Heishman
SJ
.
Relative performance of common biochemical indicators in detecting cigarette smoking
.
Addiction
2011
;
106
:
1325
–
1334
.
Joseph
AM
,
Hecht
SS
,
Murphy
SE
,
Carmella
SG
,
Le
CT
,
Zhang
Y
,
Han
S
,
Hatsukami
DK
.
Relationships between cigarette consumption and biomarkers of tobacco toxin exposure
.
Cancer Epidemiol Biomarkers Prev
2005
;
14
:
2963
–
2968
.
Heatherton
TF
,
Kozlowski
LT
,
Frecker
RC
,
Fagerström
KO
.
The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire
.
Br J Addict
1991
;
86
:
1119
–
1127
.
McCarthy
DE
,
Piasecki
TM
,
Lawrence
DL
,
Jorenby
DE
,
Shiffman
S
,
Fiore
MC
,
Baker
TB
.
A randomized controlled clinical trial of bupropion SR and individual smoking cessation counseling
.
Nicotine Tob Res
2008
;
10
:
717
–
729
.
Piper
ME
,
Federman
EB
,
McCarthy
DE
,
Bolt
DM
,
Smith
SS
,
Fiore
MC
,
Baker
TB
.
Efficacy of bupropion alone and in combination with nicotine gum
.
Nicotine Tob Res
2007
;
9
:
947
–
954
.
Piper
ME
,
Smith
SS
,
Schlam
TR
,
Fiore
MC
,
Jorenby
DE
,
Fraser
D
,
Baker
TB
.
A randomized placebo-controlled clinical trial of 5 smoking cessation pharmacotherapies
.
Arch Gen Psychiatry
2009
;
66
:
1253
–
1262
. [Published erratum appears in Arch Gen Psychiatry 2010;67:77.]
Laurie
CC
,
Doheny
KF
,
Mirel
DB
,
Pugh
EW
,
Bierut
LJ
,
Bhangale
T
,
Boehm
F
,
Caporaso
NE
,
Cornelis
MC
,
Edenberg
HJ
, et al. ;
GENEVA Investigators
.
Quality control and quality assurance in genotypic data for genome-wide association studies
.
Genet Epidemiol
2010
;
34
:
591
–
602
.
Patterson
N
,
Price
AL
,
Reich
D
.
Population structure and eigenanalysis
.
PLoS Genet
2006
;
2
:
e190
.
Purcell
S
,
Neale
B
,
Todd-Brown
K
,
Thomas
L
,
Ferreira
MA
,
Bender
D
,
Maller
J
,
Sklar
P
,
de Bakker
PI
,
Daly
MJ
, et al.
PLINK: a tool set for whole-genome association and population-based linkage analyses
.
Am J Hum Genet
2007
;
81
:
559
–
575
.
Thorgeirsson
TE
,
Gudbjartsson
DF
,
Surakka
I
,
Vink
JM
,
Amin
N
,
Geller
F
,
Sulem
P
,
Rafnar
T
,
Esko
T
,
Walter
S
, et al. ;
ENGAGE Consortium
.
Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior
.
Nat Genet
2010
;
42
:
448
–
453
.
Tobacco and Genetics Consortium
.
Genome-wide meta-analyses identify multiple loci associated with smoking behavior
.
Nat Genet
2010
;
42
:
441
–
447
.
Bloom
AJ
,
Harari
O
,
Martinez
M
,
Madden
PA
,
Martin
NG
,
Montgomery
GW
,
Rice
JP
,
Murphy
SE
,
Bierut
LJ
,
Goate
A
.
Use of a predictive model derived from in vivo endophenotype measurements to demonstrate associations with a complex locus, CYP2A6
.
Hum Mol Genet
2012
;
21
:
3050
–
3062
.
Bloom
J
,
Hinrichs
AL
,
Wang
JC
,
von Weymarn
LB
,
Kharasch
ED
,
Bierut
LJ
,
Goate
A
,
Murphy
SE
.
The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans
.
Pharmacogenet Genomics
2011
;
21
:
403
–
416
.
Bierut
LJ
,
Stitzel
JA
,
Wang
JC
,
Hinrichs
AL
,
Grucza
RA
,
Xuei
X
,
Saccone
NL
,
Saccone
SF
,
Bertelsen
S
,
Fox
L
, et al.
Variants in nicotinic receptors and risk for nicotine dependence
.
Am J Psychiatry
2008
;
165
:
1163
–
1171
.
Amos
CI
,
Wu
X
,
Broderick
P
,
Gorlov
IP
,
Gu
J
,
Eisen
T
,
Dong
Q
,
Zhang
Q
,
Gu
X
,
Vijayakrishnan
J
, et al.
Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1
.
Nat Genet
2008
;
40
:
616
–
622
.
Hung
RJ
,
McKay
JD
,
Gaborieau
V
,
Boffetta
P
,
Hashibe
M
,
Zaridze
D
,
Mukeria
A
,
Szeszenia-Dabrowska
N
,
Lissowska
J
,
Rudnai
P
, et al.
A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25
.
Nature
2008
;
452
:
633
–
637
.
Saccone
NL
,
Culverhouse
RC
,
Schwantes-An
TH
,
Cannon
DS
,
Chen
X
,
Cichon
S
,
Giegling
I
,
Han
S
,
Han
Y
,
Keskitalo-Vuokko
K
, et al.
Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD
.
PLoS Genet
2010
;
6
:
e1001053
.
Johnson
AD
,
Handsaker
RE
,
Pulit
SL
,
Nizzari
MM
,
O’Donnell
CJ
,
de Bakker
PI
.
SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap
.
Bioinformatics
2008
;
24
:
2938
–
2939
.
Rice
JP
,
Hartz
SM
,
Agrawal
A
,
Almasy
L
,
Bennett
S
,
Breslau
N
,
Bucholz
KK
,
Doheny
KF
,
Edenberg
HJ
,
Goate
AM
, et al. ;
GENEVA Consortium
.
CHRNB3 is more strongly associated with Fagerström test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results
.
Addiction
2012
;
107
:
2019
–
2028
.
Keskitalo
K
,
Broms
U
,
Heliövaara
M
,
Ripatti
S
,
Surakka
I
,
Perola
M
,
Pitkäniemi
J
,
Peltonen
L
,
Aromaa
A
,
Kaprio
J
.
Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15
.
Hum Mol Genet
2009
;
18
:
4007
–
4012
.
Le Marchand
L
,
Derby
KS
,
Murphy
SE
,
Hecht
SS
,
Hatsukami
D
,
Carmella
SG
,
Tiirikainen
M
,
Wang
H
.
Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine
.
Cancer Res
2008
;
68
:
9137
–
9140
.
Astrup
P
.
Some physiological and pathological effects of moderate carbon monoxide exposure
.
BMJ
1972
;
4
:
447
–
452
.
Fowler
CD
,
Lu
Q
,
Johnson
PM
,
Marks
MJ
,
Kenny
PJ
.
Habenular α5 nicotinic receptor subunit signalling controls nicotine intake
.
Nature
2011
;
471
:
597
–
601
.
Ehringer
MA
,
McQueen
MB
,
Hoft
NR
,
Saccone
NL
,
Stitzel
JA
,
Wang
JC
,
Bierut
LJ
.
Association of CHRN genes with “dizziness” to tobacco
.
Am J Med Genet B Neuropsychiatr Genet
2010
;
153B
:
600
–
609
.
Timofeeva
MN
,
Hung
RJ
,
Rafnar
T
,
Christiani
DC
,
Field
JK
,
Bickeböller
H
,
Risch
A
,
McKay
JD
,
Wang
Y
,
Dai
J
, et al. ;
Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team
.
Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls
.
Hum Mol Genet
2012
;
21
:
4980
–
4995
.
Siedlinski
M
,
Cho
MH
,
Bakke
P
,
Gulsvik
A
,
Lomas
DA
,
Anderson
W
,
Kong
X
,
Rennard
SI
,
Beaty
TH
,
Hokanson
JE
, et al. ;
COPDGene Investigators
;
ECLIPSE Investigators
.
Genome-wide association study of smoking behaviours in patients with COPD
.
Thorax
2011
;
66
:
894
–
902
.
Centers for Disease Control and Prevention (CDC)
.
Current cigarette smoking among adults - United States, 2011
.
MMWR Morb Mortal Wkly Rep
2012
;
61
:
889
–
894
.
Joseph
AM
,
Hecht
SS
,
Murphy
SE
,
Lando
H
,
Carmella
SG
,
Gross
M
,
Bliss
R
,
Le
CT
,
Hatsukami
DK
.
Smoking reduction fails to improve clinical and biological markers of cardiac disease: a randomized controlled trial
.
Nicotine Tob Res
2008
;
10
:
471
–
481
.
Benowitz
NL
,
Dains
KM
,
Dempsey
D
,
Wilson
M
,
Jacob
P
.
Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure
.
Nicotine Tob Res
2011
;
13
:
772
–
783
.
Mustonen
TK
,
Spencer
SM
,
Hoskinson
RA
,
Sachs
DP
,
Garvey
AJ
.
The influence of gender, race, and menthol content on tobacco exposure measures
.
Nicotine Tob Res
2005
;
7
:
581
–
590
.
Haiman
CA
,
Stram
DO
,
Wilkens
LR
,
Pike
MC
,
Kolonel
LN
,
Henderson
BE
,
Le Marchand
L
.
Ethnic and racial differences in the smoking-related risk of lung cancer
.
N Engl J Med
2006
;
354
:
333
–
342
.
Fiore
MC
,
Novotny
TE
,
Pierce
JP
,
Giovino
GA
,
Hatziandreu
EJ
,
Newcomb
PA
,
Surawicz
TS
,
Davis
RM
.
Methods used to quit smoking in the United States. Do cessation programs help?
JAMA
1990
;
263
:
2760
–
2765
.
Perkins
KA
,
Karelitz
JL
,
Giedgowd
GE
,
Conklin
CA
.
The reliability of puff topography and subjective responses during ad lib smoking of a single cigarette
.
Nicotine Tob Res
2012
;
14
:
490
–
494
.
Copyright © 2014 by the American Thoracic Society
Supplementary data
Citations
Views
Altmetric
See also
Citing articles via
More from Oxford Academic