Biomarkers for the acute respiratory distress syndrome: how to make the diagnosis more precise (original) (raw)
Review Article
Background
The acute respiratory distress syndrome (ARDS) is an acute inflammatory process of the lungs caused by direct or indirect insults to the alveolar-capillary membrane (1-5), and is associated with an overall mortality ranging from 35% to 50% (6). Pathologically, ARDS is characterized by diffuse alveolar damage (DAD) with injury to the epithelium and endothelium as well as interstitial and alveolar protein-rich edema, inflammatory cellular infiltration, atelectasis, capillary thrombosis, neovascularization, and pulmonary fibrosis (7). Clinically, this syndrome is defined by acute hypoxemia and bilateral pulmonary infiltrates, in the absence of congestive heart failure. As with other inflammatory process in the body, lung injury in ARDS is accompanied by several biochemical and cellular processes; some might initiate the syndrome, others might perpetuate it, and others could inactivate the inflammatory mediators (4).
ARDS cannot be diagnosed by a single laboratory test. Since no specific ARDS biomarker has yet been described, it is likely that the incidence of what we currently consider to be ARDS is overestimated, since patients with transient or persistent hypoxemic respiratory failure from other diseases accompanied with bilateral pulmonary infiltrates could be erroneously diagnosed as having ARDS (8). The lack of a specific biomarker for ARDS is arguably one of the main obstacles in the diagnosis and successful treatment of this syndrome (9). There are five major reasons to study and identify biomarkers in ARDS: (I) to predict the development of ARDS in high-risk patients; (II) to stratify disease severity into more accurate phenotypes or categories; (III) to provide new insights into its pathogenesis with the goal of developing novel therapeutics; (IV) to monitor response to treatment; and (V) to help in predicting outcome. Thus, the goal of having a biomarker is to provide clinicians with a better understanding of the importance of, and tools for, assessing risk and determining disease severity in ARDS patients.
Several investigators have questioned whether the concept of ARDS as a discrete entity is useful since only 50% of patients diagnosed as having ARDS have DAD lesions on pathological examination (10). It is critical that patients are defined as carefully as possible. Clinical studies on heterogeneous groups of patients with ARDS have marked differences in the underlying mechanism of lung injury, age, comorbidities, and/or number of extrapulmonary organ dysfunctions. Different diseases damage the lung parenchyma in diverse ways producing multiple signals from numerous injured cell types. For an adequate interpretation of the results of clinical trials, appropriate stratification of patients with ARDS should be related to two measures of disease severity: one that quantifies severity of lung damage and another one that quantifies the general physiologic response and associated comorbidities. It is plausible that a revised definition of ARDS based on biological criteria of DAD, rather than only on clinical variables, will identify a less heterogeneous population of ARDS patients (11).
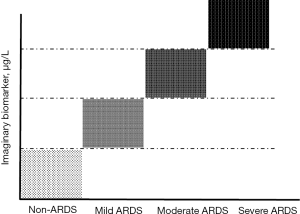
An ideal biological marker should provide information for identification of patients at risk for ARDS and with different ARDS phenotypes during the progression of lung injury (Figure 1). Ideally, such a marker should be 100% sensitive, 100% specific, easy to measure in any biological sample, be modified by management and treatment, and be cost-effective (12). It is anticipated that using biomarkers for defining or stratifying subsets of ARDS patients that could benefit from a given therapy will have a major effect on both clinical practice and the development of new diagnostic tools and drugs (9), potentially leading to better individual treatments. Before such biomarkers could be part of standard care, it is important to ensure that they are accurate, reliable, associated with severity, and predictive of individual patient responses (13). These features will help precision medicine to fulfill the potential for improving ARDS management and outcome (14,15) by facilitating the tailoring of treatment to the patient’s physiological and genetic characteristics.
Figure 1 Plot of (plasma, pulmonary edema fluid, exhaled air) levels of an imaginary specific biomarker for endothelial or epithelial lung injury in the ARDS. ARDS, acute respiratory distress syndrome.
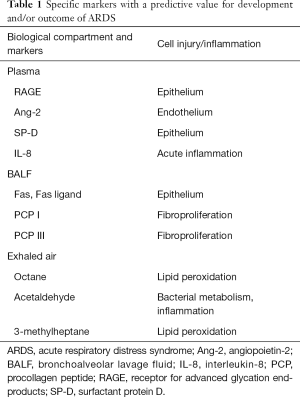
Investigators have identified several candidate biomarkers for ARDS that have been investigated in blood, pulmonary edema fluid, and exhaled air, but currently they are not reliable enough for clinical use or have not yet been validated (16-18). In this article, we will briefly review the present understanding on some appealing biomarkers that could be used for identifying patients with ARDS, for enrolling ARDS patients into clinical trials, or for better monitoring of patient’s management (Table 1). For a more detailed analysis we recommend a number of more in-depth reviews (16-18).
Table 1 Specific markers with a predictive value for development and/or outcome of ARDS
Biomarkers in blood
Blood is the most common biological sample used to examine the presence of candidate biomarkers. Blood is easy to collect, to process, and to make serial measurements during disease progression. However, the use of blood as a sample source implies the measurement of circulating biomarkers, which may be due to other processes (coagulation, response to infection, inflammation) apart from ARDS, as well as the possible absence or lower levels of specific lung endothelial or epithelial injury markers. Serial serum levels of lipopolysaccharide binding protein (LBP), a crucial element in the response to infection, were measured in 180 septic patients to examine whether LBP levels differed among septic patients with different degrees of lung injury (19). The investigators found higher LBP levels in septic patients who developed ARDS, as well as in patients with more severe lung injury, suggesting that serial LBP serum levels may turn out to be a useful biomarker for identifying septic patients with the worst outcomes and with highest probability of developing ARDS.
“Omics” technologies hold great promise in providing precise characterization of diseases to more effectively predict the clinical course of a patient or to choose the most beneficial therapeutic approaches (20). Genomic science will allow investigators to understand the reasons why a protein fails to function, to develop a drug to improve its function, and to use genetic data to select the appropriate patients for a given medicine. Genetic approaches have been studied in patients with ARDS from several causes, both using a more classical approach of hypothesis-driven study of plausible genes or, more recently, by genome-wide analysis for identification of candidate genes (21-23). However, although some genetic variants have been postulated as potential markers, it is unlikely that an intrinsic characteristic of an individual could serve as a marker in such a heterogeneous disease process as ARDS, but they may identify those patients at highest risk for developing ARDS (or for not developing ARDS). Gene expression profiling, which can reflect the changes in gene expression under specific conditions, has been applied to acute lung injury and evolution of ARDS (24,25).
Metabolomics is a more recent field with a good potential for diagnosis and monitoring of several diseases. In a study using plasma of patients with sepsis-induced ARDS, elevated levels of several metabolites compared to healthy controls were reported (26). The main limitations of this study were the absence of described normal ranges, and the fact that metabolites may be affected by many processes and hence, systemic analysis may have less potential in defining or quantifying damage that is organ specific.
Classically, most clinical studies using blood have been performed in serum or in plasma by direct measurement of the concentrations of candidate proteins. In general, biomarkers of lung epithelial injury are expected to be more specific than inflammatory markers or even more specific than those related to endothelial injury. Markers reflecting the different phases of DAD and relevant signaling pathways for acute lung injury have been reported. Here we describe several clinical reports regarding some of the most promising biomarker proteins measurable in plasma or serum in patients with ARDS.
Receptor for advanced glycation end-products (RAGE)
RAGE is a promising biomarker of lung epithelium injury. RAGE is often known as a pattern recognition receptor. Soluble RAGE (sRAGE) is an isoform of the RAGE protein which lacks the signaling domain of the full-length receptor. This receptor is constitutively expressed in all cells at low levels, but it is highly expressed in the lung epithelium, primarily in alveolar type-I cells (27). Its activation modulates cell signaling and propagation of the inflammatory response (17). Calfee et al. (28) reported an increase in the RAGE plasma levels in patients with severe ARDS, as well as a correlation with mortality in ARDS patients ventilated with high tidal volume. Later studies found an association of sRAGE with severity (29) and outcome (30) in patients with ARDS. Other investigators (31) have reported higher levels of sRAGE in ARDS patients with or without sepsis when compared to patients that only had sepsis but not ARDS. These investigators also showed a correlation of RAGE with lung injury severity, but not with outcome (31). A recent meta-analysis suggested sRAGE as a biomarker strongly associated with diagnosis of ARDS in a high-risk population, but not associated with mortality (16). Several studies analyzing panels of biomarkers have pointed at RAGE as a valuable candidate for the diagnosis of ARDS (32,33).
Angiopoietin-2 (Ang-2)
Ang-2 is an endothelial growth factor produced by endothelial cells. After release, it binds to the tyrosine kinase receptor Tie 2, playing a role in endothelial junctional integrity, promoting vascular regression and cell death (34). The study of this regulator of vascular permeability has produced interesting results. Ang-2 levels have been found to be higher in ARDS patients than in patients with hydrostatic pulmonary edema (35). Higher levels have been also linked with occurrence of ARDS in critically ill patients (36-38) as well as with severity (36) and mortality (37,39). A clinical study demonstrated that, in infection-related ARDS patients, an increase of Ang-2 levels from day-0 to day-3 was associated with an increase in the risk of death when compared to patients with decreases in Ang-2 (40). A meta-analysis also found that Ang-2 was more relevant as a biomarker for ARDS mortality than for diagnosis (16). In panels of biomarkers, Ang-2 has been reported as a relevant marker for diagnosis and/or mortality (32). In addition, Ang-2 levels have been found to be an indicator of non-pulmonary ARDS (41).
Surfactant protein D (SP-D)
SP-D is a biomarker of lung epithelial injury. This glycoprotein is mainly produced by type-II cells, playing a crucial role in maintaining the integrity of the alveolar-capillary interface. In addition to reducing the surface tension at the alveoli, SP-D also has a role in innate immunity, acting as an inflammatory molecule and having anti-microbial functions (42). Several studies have found an association with elevated plasma levels of SP-D and diagnosis and/or worse clinical outcome of ARDS. SP-D seems to be a good diagnostic indicator of ARDS in septic patients (43). An increase of SP-D plasma levels has been found after 48 h in patients with ARDS; the increase was smaller in those patients ventilated with a lung-protective ventilation approach (44). This same study showed increased levels of SP-D in non-survivors. Eisner et al. (45) showed an association between higher plasma levels of SP-D and a higher risk of death; they found a relationship between higher SP-D levels and worse clinical outcome, in terms of fewer ventilation- and organ failure-free days. They also demonstrated that a lower tidal volume strategy attenuated the rise in plasma levels of SP-D (45). Although several authors have reported no relevant association of SP-D with development of ARDS (38,46), a meta-analysis included SP-D in the list of clinically relevant biomarkers, although it appears not to be strongly associated with the diagnosis of ARDS (16). Nevertheless, SP-D has been included in several studies of biomarker panels for diagnosis and mortality prediction (33,47,48).
Interleukin-8 (IL-8)
During the inflammatory phase of DAD, immune cells of the lungs express inflammatory mediators, which in turn activate the entry of circulating inflammatory cells into the interstitium and alveolar spaces (17,49). IL-8 is a proinflammatory cytokine with a role in regulating neutrophils and monocytes chemotaxis in the lung (49). There is some evidence that IL-8 concentrations have predictive value in high-risk patients for developing ARDS, although early acute lung injury may have been present in some of those patients. Donnelly et al. (50) measured IL-8 in plasma and bronchoalveolar lavage fluid (BALF) of patients at risk for ARDS. They found that IL-8 levels in BALF on initial hospital presentation were higher in patients who subsequently progressed to ARDS, although there were no differences of the mean plasma IL-8 levels between both groups. Higher IL-8 levels have been found in non-surviving ARDS patients (51,52), and were associated with a decrease in ventilator- and organ failure-free days (52). Another report found an association of IL-8 at day-1 in ARDS patients, but failed to observe an association of IL-8 levels with mortality (53). As in the case of SP-D, although IL-8 was included in a meta-analysis of biomarkers for ARDS development and outcome (16), it was not considered among the most strongly associated markers. IL-8 has been also studied as part of biomarker panels to diagnose ARDS and predict mortality (32,33,47,48).
Biomarker panels
Since no specific clinical or biological marker has been described that predicts ARDS, several studies performed by a same group (32,33,47,48) examined a combination of markers of endothelial and epithelial lung injury, inflammation, and coagulation in ARDS patients from several trials. In an initial study in trauma patients, Fremont et al. (32) showed that a combination of biomarkers and clinical predictors was better for predicting mortality or stratifying ARDS patients than clinical predictors or biomarkers alone; however, sensitivity and specificity was low. In their model, RAGE, ANG-2, SP-D and IL-8, were among the selected biomarkers.
BALF
The most fundamental early physiological characteristic of ARDS is an increase in protein permeability across the endothelial and epithelial membrane of the lung with flooding of the interstitium and alveolar spaces with a protein-rich edema fluid (9). Measurement of protein concentration in pulmonary edema fluid obtained by sampling the distal airspaces provided the first direct evidence in patients to support the conclusion that ARDS resulted from increased lung microvascular and epithelial permeability (54).
BALF has been the second most common source of samples for searching for ARDS biomarkers. The main advantage of using BALF is that it is the closest sample to the site of injury (with the exception of lung tissue sampled by biopsy), reflecting the local lung environment. BALF contains locally produced proteins as well as immune cells involved in the processes under study. However, BALF sampling requires a flexible fiberoptic bronchoscopy. BALF contains soluble proteins, lipids, and adherent cells in the air spaces. Although the concentration of putative biomarkers of lung injury can be assessed sequentially in the same patient, this approach is limited because of the invasive procedure of this technique and because the quantitative assessment could be difficult to interpret due to a variable dilution of the samples. The assessment of the cellular profile in the distal airspaces is probably more accurate than in edema fluid samples.
Several approaches have been used to identify or detect candidate biomarkers in BALF. Proteomic studies have been useful to characterize the protein expression in BALF, showing some known, but also some unknown candidates (55). The “omics” studies are becoming more useful as technology improves. The metabolome of the lung may provide information on the status of the lung environment that could be helpful for the discovery of ARDS biomarkers and for the identification of plausible drug targets. Evans et al. (56) compared metabolites in BALF samples from ARDS patients using a liquid chromatography-mass spectroscopy platform for untargeted metabolomics. Their study revealed networks associated with several metabolism pathways. These metabolomic observations need to be validated in studies with larger sample sizes and including other groups of patients to test the discrimination between ARDS and other pulmonary disease processes. As in the case for studies using blood samples, most of the studies of ARDS biomarkers in BALF have been performed by direct measurement of the levels of specific candidate proteins. We present below findings related to some representative biomarkers in BALF in ARDS patients.
Fas and Fas ligand
Apoptosis of epithelial cells occurs in ARDS (57). The Fas and Fas ligand system is the best-studied mechanism regulating epithelial cell death. Soluble Fas and Fas ligand are increased in lung edema fluid in early ARDS, and elevated levels of Fas and Fas ligand in lung tissue and edema fluid correlate with worse outcomes (58). Also, mRNA for Fas and Fas ligand were upregulated in BALF during the initial phase of sepsis-induced ARDS but not in sepsis without ARDS (59). Although these reports suggest that epithelial apoptosis occurs very early in ARDS and Fas and Fas ligand might be specific markers for ARDS, this system is not unique to lung epithelial cells.
Procollagen peptides (PCP)
The consecutive phases of DAD (exudative, proliferative, and fibrotic) are a gross oversimplification (6). While the development of pulmonary fibrosis predicts the need for prolonged ventilator support and a fatal outcome, fibrosis is evident histologically within the first week of ARDS onset in some patients. PCP III, a precursor of and a marker of collagen synthesis, is increased in the BALF at the time of initiating mechanical ventilation in ARDS patients, suggesting that the fibroproliferative response plays an active role in the progression of ARDS (60). PCP III increased in the BALF from patients on day-3 of ARDS and is an independent risk factor for mortality (61). Meduri et al. (62) reported high PCP I and PCP III levels in ARDS patients and showed that treatment with corticosteroids caused a sustained reduction in plasma and BALF levels of both peptides. However, the benefits for late, non-resolving ARDS were not confirmed in a trial by the ARDS Network (63). Whether targeting the pathway of those peptides could lead to a novel drug therapy for modulating or attenuating the fibrotic response requires further research.
Non-targeted proteomics
While most studies have targeted a pre-determined biomarker, several investigators have examined proteomic changes in BALF samples from ARDS patients using a non-targeted approach (64-66). These non-targeted proteomic studies compared the protein levels in the BALF or undiluted pulmonary edema fluid in ARDS patients and in healthy controls. The proteomic analysis of BALF can provide insights into ARDS pathology and response to therapy. Ideally, BALF proteomics could be useful for the molecular profiling of DAD. For example, Bhargava et al. (66) examined BALF from patients with different severities of lung injury, and identified 792 proteins, from which 161 were differentially expressed in survivors and non survivors in the early phase of ARDS. Pathways related to activation of immune response, wound healing and coagulation were predominant in survivors, while pathways of collagen synthesis and carbohydrate catabolism were more abundant in non-survivors. Although these findings should be considered exploratory due to the small sample size of the cohorts, many of these pathways are not specific to the lung or lung disease.
Exhaled breath
Alternative technological advances are focused on the use of samples easily available by noninvasive procedures and representing as best as possible the lung environment during the different phases of ARDS. The most innovative approach is, probably, metabolomics of exhaled gas. Exhaled breath contains a large number of metabolites from pulmonary and systemic processes. This approach has a number of potential major advantages: simple, cheap, continuous, and non-invasive. Thus, breath analysis can be performed repeatedly during the course of the disease process. The initial studies used exhaled breath condensates (EBCs), in which soluble metabolites were measured. A relevant advance was the possibility of separately collecting alveolar EBC, avoiding products from microbial colonization of the upper airways (67). In addition, EBC also contains small concentrations of proteins, which can be used for monitoring ARDS. For example, a study comparing EBC from ventilated patients with respiratory failure (most of them with ARDS) and from healthy volunteers, identified the presence of cytokeratins in EBC from patients, but not from controls (68).
Methods for measuring volatile organic compounds (VOCs) were later developed to analyze volatile metabolites of exhaled breath [for a review on exhaled breath in diagnosis, phenotyping and monitoring of respiratory diseases, see Boots _et al._ (69)]. Several studies have been published using analysis of VOCs by gas-chromatography and mass-spectrometry in ventilated patients (18,70), and three potential VOCs have been identified as potential markers of ARDS: octane, acetaldehyde and 3-methylheptane. These VOCs discriminated ARDS from controls, improving the discrimination when combining with the use of lung injury prediction score (18). Octane is an end-product of lipid peroxidation, one of the processes caused by oxidative stress. Acetaldehyde is produced by bacteria and leukocytes. In general, bacterial colonization of the airways occurs frequently in mechanically ventilated patients and neutrophil infiltration is a hallmark of ARDS. The 3-methylheptane is produced through lipid peroxidation, similar to octane (18). Thus, the exhaled gas seems to be mainly influenced by the oxidative stress in the lungs and affected by the inflammatory and infectious responses.
The same group of investigators have described the use of an electronic nose (eNose) based on cross-reactive sensor arrays, to analyze the exhaled breath of mechanically ventilated patients (71). This system was able to differentiate patients with moderate/severe ARDS from patients with pulmonary edema and pneumonia based on the exhaled breath profiles (71). The authors stated that the advantage of this approach was that the system is noninvasive, portable, and the data are available in real time. In addition, this methodology does not rely on single biomarkers but is integrative (72). It is worth mentioning that libraries of exhaled breath VOCs are already being built (69). These methodologies for analysis of metabolites in EBCs and VOCs still need to be replicated by other groups, or using larger samples sizes, and also characterization of standards in diverse conditions (as an example, the use of mechanical ventilation alters the metabolites produced). But it seems that exhaled breath is a very promising source of samples for analysis of biomarkers or biomarker profiles, with the exciting possibility of obtaining rapid point-of-care tests.
Biomakers in ARDS: lost in translation
Despite major advances and developments in the last five decades with respect to the definition and management of ARDS, our understanding of the mechanistic underpinnings of ARDS is still in its infancy. As discussed in this brief review, there is no a single biomarker that identifies patients with DAD, or that predicts outcome or that specifically identifies a pathological ARDS pathway (73). Potentially useful biomarker tests in ARDS have not been yet adopted into clinical practice.
We should test the hypothesis for the identification and validation of biomarkers with a clear question: what role must the biomarker play? Before biomarkers become part of standard of care, such tests must be proven to be accurate and reliable for ARDS diagnosis, or associated with the ARDS outcome of interest, and/or lead to improved patient outcomes as compared with standard treatment (74). Ideally, a diagnostic marker should add to clinical findings by increasing their specificity. However, this demand is difficult to satisfy due to the wide heterogeneity of the diseases causing ARDS, and by the variety of other coincident processes. In addition, different management regimes may contribute to patient heterogeneity, both in the face of clear evidence (for example, poor adherence to lung protective ventilation) and where evidence is lacking.
A plausible goal in the next few years would be to identify a reliable biomarker ARDS profile that could help to stratify and individualize therapy for improving outcome (75). Biomarkers for molecular-targeted therapies can aid in selecting effective therapies for subsets of ARDS patients and avoiding ineffective or harmful treatments. A clinically relevant biomarker assists by enriching patient populations in clinical trials with better responders, thereby reducing the size of the trial sample required to detect significant efficacy. In addition, the link between a clinical biomarker and the therapy for ARDS would create new opportunities for innovation and to the entry of other therapies. By allowing clinicians to rule out ARDS, it could prevent exposing patients to unnecessary therapies. As biomarkers are identified, a stratified medicine patient population might emerge as a clinically distinct ARDS-like syndrome, as postulated by Schuster (11) and Villar et al. (12). This formulation would help precision medicine to fulfill the potential for improving ARDS management and outcome.
Conclusions
The absence of a validated biomarker to define, diagnose, monitor responsiveness to therapy or predict prognosis of ARDS has limited progress in the field. In the era of personalized medicine, discovery and validation of a biomarker or panel of biomarkers would not only help identify ARDS patients and/or lung injury severity, but also, more importantly, direct therapy. Many candidate biomarkers have been investigated, but at the time of writing this review, a single, clear biomarker that is specific for ARDS has proven difficult to find. Since it is difficult to find a single biomarker that distinguishes ARDS patients from those patients who do not have ARDS, it is anticipated that given the complex and heterogeneous pathophysiology of ARDS, combinations of two or three biomarkers reflecting different aspects of DAD (as epithelial and endothelial injury, or inflammation) is more likely to be identified. It is plausible that the best prediction approach will likely combine clinical predictors with several biomarkers (76). Certainly, further work with newer and more sophisticated technological methods is required to identify specific phenotypes of ARDS patients.
Acknowledgements
Funding: This work was supported in part by Instituto de Salud Carlos III, Madrid, Spain (CB06/06/1088 and PI16/0049 to J Villar, PI15/01942 to JA Lorente, PI14/00844 to C Flores), and by Asociación Científica Pulmón y Ventilación Mecánica, Las Palmas de Gran Canaria, Spain (to J Villar).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ashbaugh DG, Bigelow DB, Petty TL, et al. Acute respiratory distress in adults. Lancet 1967;2:319-23. [Crossref] [PubMed]
- Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818-24. [Crossref] [PubMed]
- Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA 2012;307:2526-33. [PubMed]
- Villar J. What is the acute respiratory distress syndrome? Respir Care 2011;56:1539-45. [Crossref] [PubMed]
- Villar J, Pérez-Méndez L, Blanco J, et al. A universal definition of ARDS: the PaO2/FiO2 ratio under a standard ventilatory setting –a prospective, multicenter, validation study. Intensive Care Med 2013;39:583-92. [Crossref] [PubMed]
- Villar J, Blanco J, Kacmarek RM. Current incidence and outcome of the acute respiratory distress syndrome. Curr Opin Crit Care 2016;22:1-6. [Crossref] [PubMed]
- Tomashefski JF Jr. Pulmonary pathology of acute respiratory distress syndrome. Clin Chest Med 2000;21:435-66. [Crossref] [PubMed]
- Guérin C, Thompson T, Brower R. The ten diseases that look like ARDS. Intensive Care Med 2015;41:1099-102. [Crossref] [PubMed]
- Villar J, Slutsky AS. GOLDEN anniversary of the acute respiratory distress syndrome: still much work to do! Curr Opin Crit Care 2017;23:4-9. [Crossref] [PubMed]
- de Hemptinne Q, Remmelink M, Brimioulle S, et al. ARDS: a clinicopathological confrontation. Chest 2009;135:944-9. [Crossref] [PubMed]
- Schuster DP. The search for “objective” criteria of ARDS. Intensive Care Med 2007;33:400-2. [Crossref] [PubMed]
- Villar J, Blanco J, Kacmarek RM. Acute respiratory distress syndrome definition: do we need a change? Curr Opin Crit Care 2011;17:13-17. [Crossref] [PubMed]
- Sweeney RM, McAuley D. Acute respiratory distress syndrome. Lancet 2016;388:2416-30. [Crossref] [PubMed]
- Jameson JL, Longo DL. Precision medicine – personalized, problematic and promising. N Engl J Med 2015;372:2229-34. [Crossref] [PubMed]
- Rubin R. A precision medicine approach to clinical trials. JAMA 2016;316:1953-5. [Crossref] [PubMed]
- Terpstra ML, Aman J, van Nieuw Amerongen GP, et al. Plasma biomarkers for acute respiratory distress síndrome: a systematic review and meta-analysis. Crit Care Med 2014;42:691-700. [Crossref] [PubMed]
- Blondonnet R, Constantin JM, Sapin V, et al. A Pathophysiologic Approach to Biomarkers in Acute Respiratory Distress Syndrome. Dis Markers 2016;2016:3501373. [Crossref] [PubMed]
- Bos LD, Weda H, Wang Y, et al. Exhaled breath metabolomics as a non-invasive diagnostic tool for acute respiratory distress syndrome. Eur Respir J 2014;44:188-197. [Crossref] [PubMed]
- Villar J, Pérez-Méndez L, Espinosa E, et al. Serum lipopolysaccharide binding protein levels predict severity of lung injury and mortality in patients with severe sepsis. PLoS One 2009;4:e6818. [Crossref] [PubMed]
- McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials. Nature 2013;502:317-20. [Crossref] [PubMed]
- Meyer NJ, Christie JD. Genetic heterogeneity and risk of acute respiratory distress syndrome. Semin Respir Crit Care Med 2013;34:459-74. [Crossref] [PubMed]
- Acosta-Herrera M, Pino-Yanes M, Pérez-Méndez L, et al. Assessing the quality of studies supporting genetic susceptibility and outcomes of ARDS. Front Genet 2014;5:20. [Crossref] [PubMed]
- Horhat FG, Gundogdu F, David LV, et al. Early Evaluation and Monitoring of Critical Patients with Acute Respiratory Distress Syndrome (ARDS) Using Specific Genetic Polymorphisms. Biochem Genet 2017;55:204-11. [Crossref] [PubMed]
- Howrylak JA, Dolinay T, Lucht L, et al. Discovery of the gene signature for acute lung injury in patients with sepsis. Physiol Genomics 2009;37:133-9. [Crossref] [PubMed]
- Wang Z, Beach D, Su L, et al. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am J Respir Cell Mol Biol 2008;38:724-32. [Crossref] [PubMed]
- Stringer KA, Serkova NJ, Karnovsky A, et al. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am J Physiol Lung Cell Mol Physiol 2011;300:L4-L11. [Crossref] [PubMed]
- Shirasawa M, Fujiwara N, Hirabayashi S, et al. Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells 2004;9:165-74. [Crossref] [PubMed]
- Calfee CS, Ware LB, Eisner MD, et al. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax 2008;63:1083-9. [Crossref] [PubMed]
- Mauri T, Masson S, Pradella A, et al. Elevated plasma and alveolar levels of soluble receptor for advanced glycation end-products are associated with severity of lung dysfunction in ARDS patients. Tohoku J Exp Med 2010;222:105-12. [Crossref] [PubMed]
- Nakamura T, Sato E, Fujiwara N, et al. Increased levels of soluble receptor for advanced glycation end products (sRAGE) and high mobility group box 1 (HMGB1) are associated with death in patients with acute respiratory distress syndrome. Clin Biochem 2011;44:601-4. [Crossref] [PubMed]
- Jabaudon M, Futier E, Roszyk L, et al. Soluble form of the receptor for advanced glycation end products is a marker of acute lung injury but not of severe sepsis in critically ill patients. Crit Care Med 2011;39:480-8. [Crossref] [PubMed]
- Fremont RD, Koyama T, Calfee CS, et al. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma 2010;68:1121-7. [Crossref] [PubMed]
- Ware LB, Koyama T, Zhao Z, et al. Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit Care 2013;17:R253. [Crossref] [PubMed]
- Scholz A, Plate KH, Reiss Y. Angiopoietin-2: a multifaceted cytokine that functions in both angiogenesis and inflammation. Ann N Y Acad Sci 2015;1347:45-51. [Crossref] [PubMed]
- Bhandari V, Choo-Wing R, Lee CG, et al. Hyperoxia causes angiopoietin 2-mediated acute lung injury and necrotic cell death. Nat Med 2006;12:1286-93. [Crossref] [PubMed]
- van der Heijden M, van Nieuw Amerongen GP, Koolwijk P, et al. Angiopoietin-2, permeability oedema, occurrence and severity of ALI/ARDS in septic and non-septic critically ill patients. Thorax 2008;63:903-9. [Crossref] [PubMed]
- Wada T, Jesmin S, Gando S, et al. The role of angiogenic factors and their soluble receptors in acute lung injury (ALI)/ acute respiratory distress syndrome (ARDS) associated with critical illness. J Inflamm (Lond) 2013;10:6. [Crossref] [PubMed]
- Agrawal A, Matthay MA, Kangelaris KN, et al. Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am J Respir Crit Care Med 2013;187:736-42. [Crossref] [PubMed]
- Gallagher DC, Parikh SM, Balonov K, et al. Circulating angiopoietin 2 correlates with mortality in a surgical population with acute lung injury/adult respiratory distress syndrome. Shock 2008;29:656-61. [PubMed]
- Calfee CS, Gallagher D, Abbott J, et al. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med 2012;40:1731-7. [Crossref] [PubMed]
- Calfee CS, Janz DR, Bernard GR, et al. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest 2015;147:1539-48. [Crossref] [PubMed]
- Sorensen GL, Husby S, Holmskov U. Surfactant protein A and surfactant protein D variation in pulmonary disease. Immunobiology 2007;212:381-416. [Crossref] [PubMed]
- Endo S, Sato N, Nakae H, et al. Surfactant protein A and D (SP-A, AP-D) levels in patients with septic ARDS. Res Commun Mol Pathol Pharmacol 2002;111:245-51. [PubMed]
- Determann RM, Royakkers AA, Haitsma JJ, et al. Plasma levels of surfactant protein D and KL-6 for evaluation of lung injury in critically ill mechanically ventilated patients. BMC Pulm Med 2010;10:6. [Crossref] [PubMed]
- Eisner MD, Parsons P, Matthay MA, et al. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax 2003;58:983-8. [Crossref] [PubMed]
- Cheng IW, Ware LB, Greene KE, et al. Prognostic value of surfactant proteins A and D in patients with acute lung injury. Crit Care Med 2003;31:20-7. [Crossref] [PubMed]
- Ware LB, Koyama T, Billheimer DD, et al. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest 2010;137:288-96. [Crossref] [PubMed]
- Calfee CS, Ware LB, Glidden DV, et al. Use of risk reclassification with multiple biomarkers improves mortality prediction in acute lung injury. Crit Care Med 2011;39:711-7. [Crossref] [PubMed]
- Han S, Mallampalli RK. The acute respiratory distress syndrome: from mechanism to translation. J Immunol 2015;194:855-60. [Crossref] [PubMed]
- Donnelly SC, Strieter RM, Kunkel SL, et al. Interleukin-8 and development of adult respiratory distress syndrome in at-risk patients. Lancet 1993;341:643-7. [Crossref] [PubMed]
- McClintock D, Zhuo H, Wickersham N, et al. Biomarkers of inflammation, coagulation and fibrinolysis predict mortality in acute lung injury. Crit Care 2008;12:R41. [Crossref] [PubMed]
- Parsons PE, Eisner MD, Thompson BT, et al. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med 2005;33:1-6. [Crossref] [PubMed]
- Cartin-Ceba R, Hubmayr RD, Qin R, et al. Predictive value of plasma biomarkers for mortality and organ failure development in patients with acute respiratory distress syndrome. J Crit Care 2015;30:219.e1-7. [Crossref] [PubMed]
- Fein A, Grossman RF, Jones JG, et al. The value of edema fluid protein measurement in patients with pulmonary edema. Am J Med 1979;67:32-8. [Crossref] [PubMed]
- Janz DR, Ware LB. Biomarkers of ALI/ARDS: pathogenesis, discovery, and relevance to clinical trials. Semin Respir Crit Care Med 2013;34:537-48. [Crossref] [PubMed]
- Evans CR, Karnovsky A, Kovach MA, et al. Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J Proteome Res 2014;13:640-9. [Crossref] [PubMed]
- Galani V, Tatsaki E, Bai M, et al. The role of apoptosis in the pathophysiology of acute respiratory distress síndrome (ARDS): an up-to-date cell specific review. Pathol Res Pract 2010;206:145-50. [Crossref] [PubMed]
- Albertine KH, Soulier MF, Wang Z, et al. Fas and fas ligand are up-regulated in pulmonary edema fluid and lung tissue of patients with acute lung injury and the acute respiratory distress syndrome. Am J Pathol 2002;161:1783-96. [Crossref] [PubMed]
- Hashimoto S, Kobayashi A, Kooguchi K, et al. Upregulation of two death pathways of perforin/granzyme and FasL/Fas in septic acute respiratory distress syndrome. Am J Respir Crit Care Med 2000;161:237-43. [Crossref] [PubMed]
- Chesnutt AN, Matthay MA, Tibayan FA, et al. Early detection of type III procollagen peptide in acute lung injury. Pathogenetic and prognostic significance. Am J Respir Crit Care Med 1997;156:840-5. [Crossref] [PubMed]
- Clark JG, Milberg JA, Steinberg KP, et al. Type III procollagen peptide in the adult respiratory distress syndrome. Association of increased peptide levels in bronchoalveolar lavage fluid with increased risk of death. Ann Intern Med 1995;122:17-23. [Crossref] [PubMed]
- Meduri GU, Tolley EA, Chinn A, et al. Procollagen types I and III aminoterminal propeptide levels during acute respiratory distress syndrome and in response to methylprednisolone treatment. Am J Respir Crit Care Med 1998;158:1432-41. [Crossref] [PubMed]
- Steinberg KP, Hudson LD, Goodman RB, et al. Efficacy and safety of corticosteroids for persistent acute respiratory distress syndrome. N Engl J Med 2006;354:1671-84. [Crossref] [PubMed]
- de Torre C, Ying SX, Munson PJ, et al. Proteomic analysis of inflammatory biomarkers in bronchoalveolar lavage. Proteomics 2006;6:3949-57. [Crossref] [PubMed]
- Chang DW, Hayashi S, Gharib SA, et al. Proteomic and computational analysis of bronchoalveolar proteins during the course of the acute respiratory distress syndrome. Am J Respir Crit Care Med 2008;178:701-9. [Crossref] [PubMed]
- Bhargava M, Becker TL, Viken KJ, et al. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One 2014;9:e109713. [Crossref] [PubMed]
- Vaschetto R, Corradi M, Goldoni M, et al. Sampling and analyzing alveolar exhaled breath condensate in mechanically ventilated patients: a feasibility study. J Breath Res 2015;9:047106. [Crossref] [PubMed]
- Gessner C, Dihazi H, Brettschneider S, et al. Presence of cytokeratins in exhaled breath condensate of mechanical ventilated patients. Respir Med 2008;102:299-306. [Crossref] [PubMed]
- Boots AW, Bos LD, van der Schee MP, et al. Exhaled molecular fingerprinting in diagnosis and monitoring: validating volatile promises. Trends Mol Med 2015;21:633-44. [Crossref] [PubMed]
- Bos LD, Wang Y, Weda H, et al. A simple breath sampling method in intubated and mechanically ventilated critically ill patients. Respir Physiol Neurobiol 2014;191:67-74. [Crossref] [PubMed]
- Bos LD, Schultz MJ, Sterk PJ. Exhaled breath profiling for diagnosing acute respiratory distress syndrome. BMC Pulm Med 2014;14:72. [Crossref] [PubMed]
- Bos LD, Sterk PJ, Schultz MJ. Measuring metabolomics in acute lung injury: choosing the correct compartment? Am J Respir Crit Care Med 2012;185:789. [Crossref] [PubMed]
- Proudfoot AG, Hind M, Griffiths MJ. Biomarkers of acute lung injury: worth their salt? BMC Med 2011;9:132. [Crossref] [PubMed]
- Lyman GH, Moses HL. Biomarker tests for molecularly targeted therapies- the key to unlocking precision medicine. N Engl J Med 2016;375:4-6. [Crossref] [PubMed]
- Trusheim MR, Berndt ER, Douglas FL. Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nat Rev Drug Discov 2007;6:287-93. [Crossref] [PubMed]
- Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014;2:611-20. [Crossref] [PubMed]
Cite this article as: García-Laorden MI, Lorente JA, Flores C, Slutsky AS, Villar J. Biomarkers for the acute respiratory distress syndrome: how to make the diagnosis more precise. Ann Transl Med 2017;5(14):283. doi: 10.21037/atm.2017.06.49