Adaptive protein evolution in Drosophila (original) (raw)
- Letter
- Published: 28 February 2002
Nature volume 415, pages 1022–1024 (2002)Cite this article
- 5489 Accesses
- 637 Citations
- 5 Altmetric
- Metrics details
Abstract
For over 30 years a central question in molecular evolution has been whether natural selection plays a substantial role in evolution at the DNA sequence level1,2. Evidence has accumulated over the last decade that adaptive evolution does occur at the protein level3,4, but it has remained unclear how prevalent adaptive evolution is. Here we present a simple method by which the number of adaptive substitutions can be estimated and apply it to data from Drosophila simulans and D. yakuba. We estimate that 45% of all amino-acid substitutions have been fixed by natural selection, and that on average one adaptive substitution occurs every 45 years in these species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
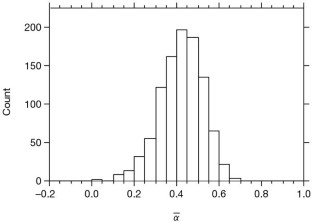
Figure 1: The distribution of 1,000 bootstrap values of \(\overline{α}\) for the divergence between Drosophila simulans and D. yakuba for genes in which _P_s > 5.
Similar content being viewed by others
References
- Gillespie, J. H. The Causes of Molecular Evolution (Oxford Univ. Press, Oxford, 1991).
Google Scholar - Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, 1983).
Book Google Scholar - Kreitman, M. & Akashi, H. Molecular evidence for natural selection. Annu. Rev. Ecol. Syst. 26, 403–422 (1995).
Article Google Scholar - Yang, Z. & Bielawski, J. P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503 (2000).
Article CAS Google Scholar - Charlesworth, B. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63, 213–227 (1994).
Article CAS Google Scholar - Fay, J., Wycoff, G. J. & Wu, C.-I. Positive and negative selection on the human genome. Genetics 158, 1227–1234 (2001).
CAS PubMed PubMed Central Google Scholar - McDonald, J. H. & Kreitman, M. Adaptive evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991).
Article ADS CAS Google Scholar - Charlesworth, B., Morgan, M. T. & Charlesworth, D. The effect of deleterious mutations on neutral molecular variation. Genetics 134, 1289–1303 (1993).
CAS PubMed PubMed Central Google Scholar - Maynard Smith, J. & Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 23, 23–35 (1974).
Article Google Scholar - Begun, D. J. & Aquadro, C. F. levels of naturally occuring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356, 519–520 (1992).
Article ADS CAS Google Scholar - Begun, D. The frequency distribution of nucleotide variation in Drosophila simulans. Mol. Biol. Evol. 18, 1343–1352 (2001).
Article CAS Google Scholar - Kliman, R. Recent selection on synonymous codon usage in Drosophila. J. Mol. Evol. 49, 343–351 (1999).
Article ADS CAS Google Scholar - Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000).
Article Google Scholar - Powell, J. R. & DeSalle, R. Drosophila molecular phylogenies and their uses. Evol. Biol. 28, 87–138 (1995).
Article CAS Google Scholar - Haldane, J. B. S. The cost of natural selection. J. Genet. 55, 511–524 (1957).
Article Google Scholar - Kimura, M. Evolutionary rate at the molecular level. Nature 217, 624–626 (1968).
Article ADS CAS Google Scholar - Thompson, J. D., Higgins, D. G. & Gibson, T. J. ClustalW—improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680 (1994).
Article CAS Google Scholar - Xia, X. Data Analysis in Molecular Biology and Evolution (Kluwer Academic, London, 2000).
Google Scholar - Rozas, J. & Rozas, R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175 (1999).
Article CAS Google Scholar - Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
CAS PubMed Google Scholar
Acknowledgements
We thank B. Charlesworth, C.-I. Wu, S. Otto, M. Whitlock, T. Johnson, P. Awadalla, J. Gillespie, G. McVean and P. Keightley for helpful discussions, and E. Moriyama for help with data collection. N.G.C.S. was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) and A.E.-W. is funded by the Royal Society and the BBSRC.
Author information
Author notes
- Nick G. C. Smith
Present address: Department of Evolutionary Biology, Evolutionary Biology Centre, Uppsala University, Norbyvägen 18D, SE-752 36, Uppsala, Sweden
Authors and Affiliations
- Centre for the Study of Evolution and School of Biological Sciences, University of Sussex, Brighton, BN1 9QG, UK
Nick G. C. Smith & Adam Eyre-Walker
Authors
- Nick G. C. Smith
- Adam Eyre-Walker
Corresponding author
Correspondence toAdam Eyre-Walker.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Smith, N., Eyre-Walker, A. Adaptive protein evolution in Drosophila.Nature 415, 1022–1024 (2002). https://doi.org/10.1038/4151022a
- Received: 15 May 2001
- Accepted: 19 December 2001
- Issue Date: 28 February 2002
- DOI: https://doi.org/10.1038/4151022a