RNA versus protein toxicity in C9orf72 ALS/FTLD (original) (raw)
A (G4C2) n expansion with several hundred or thousand repeats in the first intron upstream of the C9orf72 coding region is the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), but the driver mechanism remains unclear [[7](/article/10.1007/s00401-018-1823-1#ref-CR7 "Edbauer D, Haass C (2016) An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr Opin Neurobiol 36:99–106. https://doi.org/10.1016/j.conb.2015.10.009
")\]. Three main pathomechanisms have been proposed, but their relative role is vigorously debated because they require partially opposing therapeutic strategies. Three reports in this issue take a provocative stance strongly arguing for either RNA or protein toxicity in _C9orf72_ pathogenesis (Fig. [1](/article/10.1007/s00401-018-1823-1#Fig1)).Fig. 1
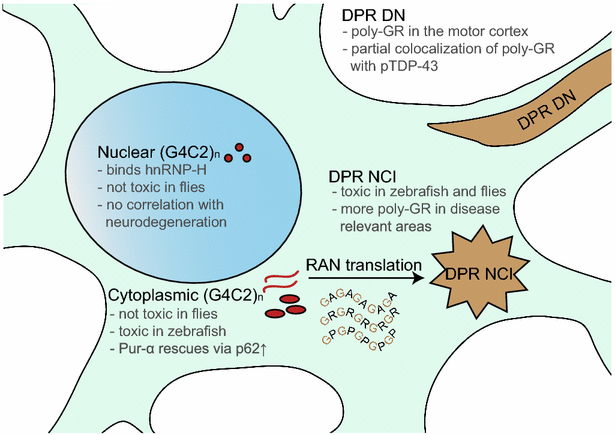
Novel insights into C9orf72 pathomechanisms. Three reports in this issue dissect RNA and protein toxicity mediated by the (G4C2) n repeat expansion. The (C4G2) n antisense transcript has similar effects in drosophila and zebrafish and is translated into additional rare DPR proteins poly-PR and poly-PA (not depicted)
Sense and antisense transcripts of the repeat accumulate in ubiquitous small nuclear, and occasionally cytoplasmic, RNA foci. Many (G4C2) n -binding proteins have been identified that are partially sequestered by the repeat RNA. Several of the trapped RNA-binding proteins are involved in alternative splicing and splicing abnormalities have been reported in C9orf72 patients [[22](/article/10.1007/s00401-018-1823-1#ref-CR22 "Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. https://doi.org/10.1038/nn.4065
"), [25](/article/10.1007/s00401-018-1823-1#ref-CR25 "Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A et al (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5:208ra149.
https://doi.org/10.1126/scitranslmed.3007529
")\]. However, a sophisticated study on 63 _C9orf72_ cases shows no correlation of sense and antisense foci with neurodegeneration or clinical parameters \[[6](/article/10.1007/s00401-018-1823-1#ref-CR6 "DeJesus-Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, Vasilevich A, Murray ME, Rousseau L, Weesner R et al (2017) In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol.
https://doi.org/10.1007/s00401-017-1725-7
")\], although antisense foci have been linked to TDP-43 pathology by others \[[5](/article/10.1007/s00401-018-1823-1#ref-CR5 "Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, Pickering-Brown S, Kirby J, Hautbergue GM, Shaw PJ (2015) Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol.
https://doi.org/10.1007/s00401-015-1429-9
")\].Repeat-associated non-ATG (RAN) translation of both sense and antisense repeat transcripts in all reading frames generates five co-aggregating dipeptide repeat (DPR) proteins: poly-GA/GP/GR from the sense transcript and poly-GP/PA/PR from the antisense transcript. The sense-strand derived DPRs are abundant throughout the neocortex, hippocampus, thalamus and cerebellum, but scarce in brain stem and spinal cord. Although the DPR proteins co-aggregate predominantly in cytoplasmic and less frequently in intranuclear inclusions in neurons, the individual proteins have very different biophysical properties. Cryoelectron tomography shows that poly-GA forms twisted ribbons that interfere with proteasome function [[9](/article/10.1007/s00401-018-1823-1#ref-CR9 "Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU et al (2018) In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. https://doi.org/10.1016/j.cell.2017.12.030
")\]. Poly-GR and -PR undergo liquid–liquid phase separation in vitro and interfere with the dynamics of the nucleolus and stress granules \[[13](/article/10.1007/s00401-018-1823-1#ref-CR13 "Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A et al (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167(774–788):e717.
https://doi.org/10.1016/j.cell.2016.10.002
")\] and disturb nucleocytoplasmic transport \[[12](/article/10.1007/s00401-018-1823-1#ref-CR12 "Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1229.
https://doi.org/10.1038/nn.4085
")\]. The aggregate distribution of neither DPR species has been shown to correlate with TDP-43 pathology or neurodegeneration \[[16](/article/10.1007/s00401-018-1823-1#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879.
https://doi.org/10.1007/s00401-013-1181-y
"), [17](/article/10.1007/s00401-018-1823-1#ref-CR17 "Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D et al (2015) Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130:845–861.
https://doi.org/10.1007/s00401-015-1476-2
"), [27](/article/10.1007/s00401-018-1823-1#ref-CR27 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Degeneration GCfFL, Alliance BBB, Arzberger T et al (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130:537–555.
https://doi.org/10.1007/s00401-015-1450-z
")\].Finally, the repeat expansion interferes with transcription and/or splicing and leads to lower C9orf72 expression, a protein that has recently been linked to autophagy [[8](/article/10.1007/s00401-018-1823-1#ref-CR8 "Gao FB, Almeida S, Lopez-Gonzalez R (2017) Dysregulated molecular pathways in amyotrophic lateral sclerosis–frontotemporal dementia spectrum disorder. EMBO J 36:2931–2950. https://doi.org/10.15252/embj.201797568
")\]. Homozygous knockout of _C9orf72_ causes a variable (lupus-like) immune phenotype due to strong expression of _C9orf72_ in the myeloid lineage, but the mice show no overt neurodegeneration \[[1](/article/10.1007/s00401-018-1823-1#ref-CR1 "Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, Suzuki-Uematsu S, Ghosh S, Singh A, Merkle FT et al (2016) Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 8:347–393.
https://doi.org/10.1126/scitranslmed.aaf6038
"), [20](/article/10.1007/s00401-018-1823-1#ref-CR20 "O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J et al (2016) C9orf72 is required for proper macrophage and microglial function in mice. Science 351:1324–1329.
https://doi.org/10.1126/science.aaf1064
")\]. However, _C9orf72_ haploinsufficiency sensitizes patient-derived motoneurons to DPR toxicity and other stressors, suggesting it may contribute to neurodegeneration in _C9orf72_ ALS/FTD \[[28](/article/10.1007/s00401-018-1823-1#ref-CR28 "Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY et al (2018) Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med.
https://doi.org/10.1038/nm.4490
")\]. Thus, lowering _C9orf72_ expression to treat gain-of-function mechanisms may be detrimental.Swinnen et al. [[29](/article/10.1007/s00401-018-1823-1#ref-CR29 "Swinnen B, Bento-Abreu A, Gendron TF, Boeynaems S, Bogaert E, Nuyts R, Timmers M, Scheveneels W, Hersmus N, Wang J et al (2018) A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1796-5
")\] injected zebrafish embryos with sense and antisense _C9orf72_ repeat RNA or synthetic genes encoding individual DPR proteins. This method is more rapid than transgenesis and allows dosage studies; however, only developing embryos can be analyzed. They focus their analysis on the length and branching pattern of motor axons during outgrowth. Consistent with severe toxicity in other models, (GR)50 and (PR)50 expression resulted in shorter axons with abnormal branching pattern, while the other individual DPRs had no effect. Expression of sense or antisense RNA had a similar effect starting at 35 or 70 repeats, respectively. In all cases, toxicity was dose dependent. Strikingly, repeat RNA injection did not result in significant DPR expression via RAN-translation in zebrafish embryos. Poly-GR/PR was not detectable by dot blot, although poly-GR/PR could still be detected at non-toxic levels in embryos injected with synthetic DPR constructs. In contrast to the fly model \[[18](/article/10.1007/s00401-018-1823-1#ref-CR18 "Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194.
https://doi.org/10.1126/science.1256800
")\], injection of sense and antisense RNA interrupted by stop codons in all reading frames (“RNA-only”) still reduced axon length, but had very little effect on branching. To identify mediators of toxicity of the RNA repeats and potential drug targets, Swinnen et al. analyzed RNA-binding proteins known to interact with the (G4C2) _n_ repeat. Co-expression of Pur-alpha rescued RNA toxicity depending on the G-rich and PUR2 domain, although these domains are not required for (G4C2) _n_ \-binding, which argues for an indirect effect. Other well-characterized repeat-binding proteins, such as hnRNPH and hnRNPA1 did not rescue, which highlights the role of Pur-alpha in repeat RNA toxicity. This effect depends on a \~ 50% upregulation of p62 by Pur-alpha expression, which is unexpected because p62 is the prototypical component of proteinaceous inclusions.A caveat of this interesting model is the predominantly cytoplasmic localization of the injected repeat RNA, which is in contrast to the predominantly nuclear RNA foci seen in patients. In C9orf72 post-mortem tissue, nuclear RNA foci show little correlation with neurodegeneration and disease parameters, but cytoplasmic repeat RNA has not been investigated so far [[6](/article/10.1007/s00401-018-1823-1#ref-CR6 "DeJesus-Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, Vasilevich A, Murray ME, Rousseau L, Weesner R et al (2017) In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1725-7
")\].Consistent with previous analysis in zebrafish, repeat RNA is clearly toxic in zebrafish upon RNA injection [[14](/article/10.1007/s00401-018-1823-1#ref-CR14 "Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C et al (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5:1178–1186. https://doi.org/10.1016/j.celrep.2013.10.049
")\] and transgene driven overexpression \[[21](/article/10.1007/s00401-018-1823-1#ref-CR21 "Ohki Y, Wenninger-Weinzierl A, Hruscha A, Asakawa K, Kawakami K, Haass C, Edbauer D, Schmid B (2017) Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol Neurodegen 12:6.
https://doi.org/10.1186/s13024-016-0146-8
")\]. However, motor axons only seem to be affected upon RNA injection and are unaffected when derived from a transgene \[[21](/article/10.1007/s00401-018-1823-1#ref-CR21 "Ohki Y, Wenninger-Weinzierl A, Hruscha A, Asakawa K, Kawakami K, Haass C, Edbauer D, Schmid B (2017) Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol Neurodegen 12:6.
https://doi.org/10.1186/s13024-016-0146-8
")\] potentially due to different timing of expression during development or different structure of in vitro versus in vivo generated RNA species. Poly-GA/GP/GR were not detectable by immunoblotting in these models, which also argues for a significant component of RNA toxicity. It is unclear, why RAN-translation is so inefficient in zebrafish, but the early developmental stage could be a key factor, because DPR expression is also barely detectable in most iPSC-derived neurons.In the same issue, Moens et al. [[19](/article/10.1007/s00401-018-1823-1#ref-CR19 "Moens TG, Mizielinska S, Niccoli T, Mitchell JS, Thoeng A, Ridler CE, Gronke S, Esser J, Heslegrave A, Zetterberg H et al (2018) Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1798-3
")\] analyzed RNA toxicity in drosophila comparing different length of sense and antisense RNA-only repeats targeted either to the nucleus using intronic expression or targeted to the cytoplasm using a polyA tail. The inducible transgenic system is very elegant because it allows direct comparison of each line with or without expression of the _C9orf72_ repeat to exclude integration-specific effects and other genetic variabilities. Importantly, the expression is restricted to adult neurons and allows life-long phenotyping. Some lines contain over 1000 repeats confirmed by Southern blotting like most typical _C9orf72_ patients. PolyA-tailed RNA forms cytoplasmic foci (similar to the zebrafish model), while intronic expression results in nuclear RNA foci that are predominant in patients. The interspersed stop codons in all reading frames completely suppress DPR expression in most lines. The nuclear (G4C2) _n_ foci also partially colocalized with Glorund, the fly ortholog of hnRNP-H, an RNA-binding protein consistently reported to bind the _C9orf72_ repeat RNA. Quite strikingly, none of the RNA-only fly lines showed any toxic phenotype or survival deficits. In fact, two independent lines expressing (G4C2)100 in an intron even survived significantly longer upon transgene induction, although only by a few days. The 1000-repeat lines had much more abundant foci, but no apparent behavioral or developmental phenotype. Thus, RNA foci of even large sense and antisense repeat transcripts cause no overt phenotype in Drosophila in the absence of DPR expression, although they bind at least one RNA-binding protein like in humans.Therefore, in fly models, (G4C2) n toxicity is clearly predominantly driven by poly-GR toxicity [[18](/article/10.1007/s00401-018-1823-1#ref-CR18 "Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194. https://doi.org/10.1126/science.1256800
"), [30](/article/10.1007/s00401-018-1823-1#ref-CR30 "Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z et al (2015) Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a drosophila model of C9ORF72 FTD/ALS. Neuron 87:1207–1214.
https://doi.org/10.1016/j.neuron.2015.09.015
")\]. A possible explanation for the lack of RNA toxicity in this fly model is the pronounced formation of RNA foci which is in contrast to the more diffusely distributed cytoplasmic repeat RNA in zebrafish. Moens et al. and Swinnen et al. are the first models for antisense RNA toxicity. Unfortunately, some important details about the antisense transcript in patients are still unclear, such as the exact start site, splicing events, capping and poly-adenylationation. Therefore, we cannot tell which of the models reflects the patient situation best.Saberi et al. [[24](/article/10.1007/s00401-018-1823-1#ref-CR24 "Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M et al (2017) Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1793-8
")\] analyzed _C9orf72_ haploinsufficiency and poly-GR toxicity through nucleocytoplasmic transport in a small cohort of _C9orf72_ ALS cases with short post-mortem intervals. The authors focused on areas with clear clinical relevance (motor cortex, frontal cortex and the anterior horn of the spinal cord) and compared them to areas with no clear clinical relevance for ALS/FTLD (parietal cortex, occipital cortex and posterior horn of the spinal cord). To address the _C9orf72_ loss of function component, they thoroughly validated a commercial antibody in mouse knockout tissue because raising good antibodies has been notoriously difficult. The reduction of C9orf72 protein in both frontal and occipital cortex extends previous findings \[[31](/article/10.1007/s00401-018-1823-1#ref-CR31 "Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ (2014) Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35:1779.
https://doi.org/10.1016/j.neurobiolaging.2014.01.016
(e1775–1779 e1713)
")\] and supports a haploinsufficiency component, while arguing against its primary role.Using home-made rabbit polyclonal DPR antibodies, they observed inclusions pathology largely consistent with previous observations. Strikingly, they found much higher levels of poly-GR but not of other DPR species in frontal and motor cortex compared to parietal and occipital cortex and detected dendritic poly-GR inclusions only in the motor cortex. 85% of these inclusions colocalized with phosphorylated TDP-43 (pTDP-43), although only 4% of dendritic pTDP-43 inclusions colocalized with poly-GR.
Both poly-GR and RNA toxicity has been linked to impaired nucleocytoplasmic transport in various model systems. However, the detailed analysis by Saberi et al. shows normal distribution of RanGAP and Lamin B1 in motor cortex or anterior horn of the spinal cord. In particular, neurons with poly-GR inclusions show similar RanGAP distribution as neighboring cells. The nuclear shape was slightly abnormal in both C9orf72 and non-hereditary ALS cases. This is consistent with a parallel study showing that nuclear pore defects are not unique for C9orf72 cases but are also directly connected to the TDP-43 pathology in ALS [[4](/article/10.1007/s00401-018-1823-1#ref-CR4 "Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin-Asp PG, Chen YH, Duong DM et al (2018) TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci. https://doi.org/10.1038/s41593-017-0047-3
")\].Thus, Saberi et al. support a causal role of poly-GR in C9orf72 pathogenesis, while questioning a key pathway proposed to mediate its toxicity. Unfortunately, the cohort is rather small (n = 4–5) and does not contain pure FTLD cases. Also, the degree of DPR pathology in the frontal cortex has not been correlated with the degree of neurodegeneration or the presence of FTD-like symptoms. Therefore, the study cannot explain why some C9orf72 patients develop pure ALS, FTLD or a mixture of both.
The different frequencies of both perinuclear and dendritic poly-GR inclusions in disease-related regions vs. disease-unrelated regions in this cohort are striking. In a previous study, we had reported similar levels of poly-GR inclusions in those areas [[27](/article/10.1007/s00401-018-1823-1#ref-CR27 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Degeneration GCfFL, Alliance BBB, Arzberger T et al (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130:537–555. https://doi.org/10.1007/s00401-015-1450-z
")\]. Furthermore, the findings of Saberi et al. are in contrast to absent or very low neuritic poly-GR pathology detected in the motor cortex with two different monoclonal poly-GR antibodies \[[17](/article/10.1007/s00401-018-1823-1#ref-CR17 "Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D et al (2015) Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130:845–861.
https://doi.org/10.1007/s00401-015-1476-2
"), [27](/article/10.1007/s00401-018-1823-1#ref-CR27 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Degeneration GCfFL, Alliance BBB, Arzberger T et al (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130:537–555.
https://doi.org/10.1007/s00401-015-1450-z
")\]. Re-evaluation of our own stainings confirmed our previous results that neuritic poly-GR pathology was generally rare with the highest frequency in hippocampal areas, but was not restricted to the motor cortex as observed by Saberi et al.For poly-GR the specificity of the antibodies used in the different studies is decisive, because poly-GR shows complex post-translational modifications. Arginine methylation gives rise to four different species that likely occur in a complex mixture even within one molecule: non-methylated, mono-methylated, symmetrically di-methylated and asymmetrically di-methylated. De-imination of arginine to citrulline is another conceivable modification. Elucidating the exact specificity of the new antibody for the different poly-GR modification and replication in a larger and more diverse cohort will be crucial to draw mechanistic conclusions.
What does that mean for developing a therapy for C9orf72 ALS/FTD? Not surprisingly, the papers conclude with opposing suggestions to either focus on the RNA or DPRs. Using the strength of the different available models wisely, is likely the best strategy to design and validate therapeutic approaches. Thanks to Moens et al., it is now clear that the C9orf72 repeat in the current fly models predominantly invokes poly-GR/PR toxicity. It will be interesting to test long repeat models not interupted by stop codons to study the effects of DPR proteins of physiological length. In zebrafish embryos, RNA-mediated toxicity seems to dominate, but forced expression of DPRs can boost toxicity. BAC-transgenic and AAV-based mouse models recapitulate RNA foci, DPR inclusions, (nuclear) TDP-43 pathology and neurodegeneration and argue for combination of RNA and DPR toxicity [[2](/article/10.1007/s00401-018-1823-1#ref-CR2 "Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME et al (2015) Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348:1151–1154. https://doi.org/10.1126/science.aaa9344
"), [15](/article/10.1007/s00401-018-1823-1#ref-CR15 "Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP (2016) C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90:521–534.
https://doi.org/10.1016/j.neuron.2016.04.005
")\], because at least the current DPR-only mice show less severe phenotypes and only faint hints of TDP-43 pathology \[[26](/article/10.1007/s00401-018-1823-1#ref-CR26 "Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J et al (2017) Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol.
https://doi.org/10.1007/s00401-017-1711-0
")\]. However, strain-specific effects such as a different genetic background seem to affect the phenotype of both _C9orf72_ BAC and knockout mice \[[10](/article/10.1007/s00401-018-1823-1#ref-CR10 "Hayes LR, Rothstein JD (2016) C9ORF72-ALS/FTD: transgenic mice make a come-BAC. Neuron 90:427–431.
https://doi.org/10.1016/j.neuron.2016.04.026
")\]. It is remarkable that only 6 years after the discovery of the C9orf72 mutation, the current mouse models are already closer to human pathology than Alzheimer mouse models. Human iPSC-derived neurons are widely popular, but DPR expression is very low in most differentiation protocols and robust TDP-43 pathology has not been reported. Thus, iPSC models may be most useful to study RNA toxicity and the consequences of reduced _C9orf72_ protein expression \[[28](/article/10.1007/s00401-018-1823-1#ref-CR28 "Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY et al (2018) Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med.
https://doi.org/10.1038/nm.4490
")\]. Human neuropathology clearly remains the gold standard. A comprehensive quantitative analysis of proteins interacting the repeat RNA in nuclear and cytoplasmic RNA foci throughout the brain could be rewarding.Until we know more about the pathomechanisms, the safest bet is to target both the repeat RNA and the DPRs, for example, by antisense oligonucleotides [[11](/article/10.1007/s00401-018-1823-1#ref-CR11 "Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D et al (2016) Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90:535–550. https://doi.org/10.1016/j.neuron.2016.04.006
")\] that have been recently approved for the therapy of spinal muscular atrophy after a very successful clinical trials \[[3](/article/10.1007/s00401-018-1823-1#ref-CR3 "Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, Norris DA, Bennett CF, Bishop KM (2016) Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology 86:890–897.
https://doi.org/10.1212/WNL.0000000000002445
")\]. However, the long prodromal stage of _C9orf72_ disease might require very early treatment \[[23](/article/10.1007/s00401-018-1823-1#ref-CR23 "Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, van Minkelen R, Rombouts SA, Cardoso MJ, Clegg S et al (2015) Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the genetic frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol 14:253–262.
https://doi.org/10.1016/S1474-4422(14)70324-2
")\], which is less feasible with the current invasive delivery method. Dissecting the mechanistic cascade of _C9orf72_ disease will require continuous comparison of animal models with human pathology. Ultimately only an effective drug that targets only a subset of the proposed _C9orf72_ mechanisms (e.g., immunotherapy) will reveal the crucial component for _C9orf72_ ALS/FTLD.References
- Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, Suzuki-Uematsu S, Ghosh S, Singh A, Merkle FT et al (2016) Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 8:347–393. https://doi.org/10.1126/scitranslmed.aaf6038
Article Google Scholar - Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME et al (2015) Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348:1151–1154. https://doi.org/10.1126/science.aaa9344
Article CAS PubMed PubMed Central Google Scholar - Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, Norris DA, Bennett CF, Bishop KM (2016) Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology 86:890–897. https://doi.org/10.1212/WNL.0000000000002445
Article CAS PubMed PubMed Central Google Scholar - Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin-Asp PG, Chen YH, Duong DM et al (2018) TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci. https://doi.org/10.1038/s41593-017-0047-3
PubMed Google Scholar - Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, Pickering-Brown S, Kirby J, Hautbergue GM, Shaw PJ (2015) Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. https://doi.org/10.1007/s00401-015-1429-9
PubMed PubMed Central Google Scholar - DeJesus-Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, Vasilevich A, Murray ME, Rousseau L, Weesner R et al (2017) In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1725-7
PubMed PubMed Central Google Scholar - Edbauer D, Haass C (2016) An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr Opin Neurobiol 36:99–106. https://doi.org/10.1016/j.conb.2015.10.009
Article CAS PubMed Google Scholar - Gao FB, Almeida S, Lopez-Gonzalez R (2017) Dysregulated molecular pathways in amyotrophic lateral sclerosis–frontotemporal dementia spectrum disorder. EMBO J 36:2931–2950. https://doi.org/10.15252/embj.201797568
Article CAS PubMed Google Scholar - Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU et al (2018) In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. https://doi.org/10.1016/j.cell.2017.12.030
Google Scholar - Hayes LR, Rothstein JD (2016) C9ORF72-ALS/FTD: transgenic mice make a come-BAC. Neuron 90:427–431. https://doi.org/10.1016/j.neuron.2016.04.026
Article CAS PubMed Google Scholar - Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D et al (2016) Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90:535–550. https://doi.org/10.1016/j.neuron.2016.04.006
Article CAS PubMed PubMed Central Google Scholar - Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1229. https://doi.org/10.1038/nn.4085
Article CAS PubMed PubMed Central Google Scholar - Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A et al (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167(774–788):e717. https://doi.org/10.1016/j.cell.2016.10.002
Google Scholar - Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C et al (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5:1178–1186. https://doi.org/10.1016/j.celrep.2013.10.049
Article CAS PubMed PubMed Central Google Scholar - Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP (2016) C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90:521–534. https://doi.org/10.1016/j.neuron.2016.04.005
Article CAS PubMed Google Scholar - Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. https://doi.org/10.1007/s00401-013-1181-y
Article CAS PubMed Google Scholar - Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D et al (2015) Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130:845–861. https://doi.org/10.1007/s00401-015-1476-2
Article CAS PubMed Google Scholar - Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194. https://doi.org/10.1126/science.1256800
Article CAS PubMed PubMed Central Google Scholar - Moens TG, Mizielinska S, Niccoli T, Mitchell JS, Thoeng A, Ridler CE, Gronke S, Esser J, Heslegrave A, Zetterberg H et al (2018) Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1798-3
PubMed Google Scholar - O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J et al (2016) C9orf72 is required for proper macrophage and microglial function in mice. Science 351:1324–1329. https://doi.org/10.1126/science.aaf1064
Article PubMed PubMed Central Google Scholar - Ohki Y, Wenninger-Weinzierl A, Hruscha A, Asakawa K, Kawakami K, Haass C, Edbauer D, Schmid B (2017) Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol Neurodegen 12:6. https://doi.org/10.1186/s13024-016-0146-8
Article Google Scholar - Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. https://doi.org/10.1038/nn.4065
Article CAS PubMed PubMed Central Google Scholar - Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, van Minkelen R, Rombouts SA, Cardoso MJ, Clegg S et al (2015) Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the genetic frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol 14:253–262. https://doi.org/10.1016/S1474-4422(14)70324-2
Article PubMed Google Scholar - Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M et al (2017) Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1793-8
Google Scholar - Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A et al (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5:208ra149. https://doi.org/10.1126/scitranslmed.3007529
Article PubMed PubMed Central Google Scholar - Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J et al (2017) Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1711-0
PubMed PubMed Central Google Scholar - Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Degeneration GCfFL, Alliance BBB, Arzberger T et al (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130:537–555. https://doi.org/10.1007/s00401-015-1450-z
Article CAS PubMed PubMed Central Google Scholar - Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY et al (2018) Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. https://doi.org/10.1038/nm.4490
Google Scholar - Swinnen B, Bento-Abreu A, Gendron TF, Boeynaems S, Bogaert E, Nuyts R, Timmers M, Scheveneels W, Hersmus N, Wang J et al (2018) A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. https://doi.org/10.1007/s00401-017-1796-5
PubMed Google Scholar - Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z et al (2015) Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a drosophila model of C9ORF72 FTD/ALS. Neuron 87:1207–1214. https://doi.org/10.1016/j.neuron.2015.09.015
Article CAS PubMed PubMed Central Google Scholar - Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ (2014) Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35:1779. https://doi.org/10.1016/j.neurobiolaging.2014.01.016 (e1775–1779 e1713)
Article PubMed PubMed Central Google Scholar
Acknowledgements
We apologize for not citing all relevant original literature due to space constraints. D.E. received funding from the European Community’s Health Seventh Framework Programme under Grant agreement 617198 [DPR-MODELS].
Author information
Authors and Affiliations
- German Center for Neurodegenerative Diseases (DZNE), Munich, Feodor-Lynen-Str. 17, 81377, Munich, Germany
Thomas Arzberger, Martin H. Schludi, Carina Lehmer, Bettina Schmid & Dieter Edbauer - Center for Neuropathology and Prion Research, Ludwig-Maximilians-University Munich, Feodor-Lynen-Str. 23, 81377, Munich, Germany
Thomas Arzberger - Department of Psychiatry and Psychotherapy, Ludwig-Maximilians-University Munich, Nußbaumstraße 7, 80336, Munich, Germany
Thomas Arzberger - Munich Cluster of Systems Neurology (SyNergy), Feodor-Lynen-Str. 17, 81377, Munich, Germany
Martin H. Schludi, Carina Lehmer, Bettina Schmid & Dieter Edbauer
Authors
- Thomas Arzberger
- Martin H. Schludi
- Carina Lehmer
- Bettina Schmid
- Dieter Edbauer
Corresponding author
Correspondence toDieter Edbauer.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Arzberger, T., Schludi, M.H., Lehmer, C. et al. RNA versus protein toxicity in C9orf72 ALS/FTLD.Acta Neuropathol 135, 475–479 (2018). https://doi.org/10.1007/s00401-018-1823-1
- Received: 10 February 2018
- Accepted: 10 February 2018
- Published: 15 February 2018
- Issue Date: March 2018
- DOI: https://doi.org/10.1007/s00401-018-1823-1