Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation (original) (raw)
Background
The microRNAs (miRNAs) are an extensive class of small non-coding RNAs with mature transcripts of 18 to 25 nucleotides [1–12]. The enzymatic machinery involved in the biogenesis of miRNAs in plants and animals is well conserved. RNAse type III-like Dicer, together with Argonaute proteins, cleaves a miRNA hairpin precursor (70 to 75 nucleotides) to yield a stable, ~22 nucleotides miRNA from one arm of the hairpin [13]. Some miRNA genes are often organized into clusters [1–3, 14, 15], and in some cases clustered miRNAs are co-expressed [14, 15]. Coordinate transcription is the first of multiple levels for regulation of miRNA expression, including miRNA maturation, transport, and stability [15–17]. This gives plants and animals a great flexibility to modulate the levels of miRNAs in different cell types and tissues and at different stages of their development.
The biological functions of a handful of miRNAs have been identified in plants and animals [13, 18]. The most apparent difference between plant and animal miRNAs is the way they interact with target genes. Animal miRNAs are generally thought to recognize their messenger RNA (mRNA) targets by incomplete base-pairing, leading to translational inhibition of the target. Plant miRNAs apparently also inhibit translation [19], but many plant miRNAs are precisely complementary to their targets, and in these cases they act as guides to trigger transcript degradation through RNA interference (RNAi) [20–22]. In Caenorhabditis elegans, upregulation of lin-4 and let-7 RNAs during the first and fourth larval stage, respectively, triggers an ordered downregulation of target genes that is required for the sequential expression of stage-specific developmental programs [23–26]. lin-4 RNA binds to the 3' untranslated region (UTR) of lin-14 and lin-28 mRNAs and inhibits their translation [27, 28]. Similarly, let-7 RNA binds to 3' UTR of lin-41 and lin-57 mRNAs and inhibits their expression at a post-transcriptional level [29–31]. In Drosophila melanogaster, the mRNAs of the pro-apoptotic gene hid and the apoptotic effector gene drice possess binding sites for bantam and miR-14, respectively [32, 33]. The phenotypes of bantam and mir-14 loss-of-function suggest that these miRNAs regulate apoptosis by inhibiting translation of these apoptosis-promoting genes [32, 33].
In mammals, certain miRNAs have been implicated in the maintenance of pluripotent cell state during early embryogenesis [34], and roles have been suggested for other miRNAs in tissue-specific or organ-specific development [2]. However, hundreds of miRNAs have been cloned from mouse and human organs and cell lines [1–3, 9, 10], and numerous additional miRNAs have been predicted with computational algorithms [8]. To begin to explore the possible roles for these numerous miRNAs in mammalian development, we used northern analysis to characterize the expression of 119 previously reported miRNAs in adult organs from mouse and human. Of these, 30 miRNAs were specifically expressed or greatly enriched in a particular organ, suggesting an organ- or tissue-specific function. A small subset of brain-expressed miRNAs was induced in mouse and human embryonal carcinoma (EC) cells treated with all-_trans_-retinoic acid (RA). This induction coincided closely with the time of terminal neuronal differentiation. The evolutionary conserved expression of these miRNAs suggests that they may play a role in the specification and/or progression of neuronal lineage. The orthologous protein of C. elegans LIN-28 was repressed in these EC cells treated with RA. In addition, mammalian Lin-28 mRNAs contain conserved predicted binding sites in their 3' UTRs for neuron-expressed miR-125b (a lin-4 ortholog), let-7a, and miR-218. Thus, Lin-28 may also be subjected to miRNA regulation in mammalian cells.
Results and discussion
Diverse miRNA expression patterns in mammalian organs
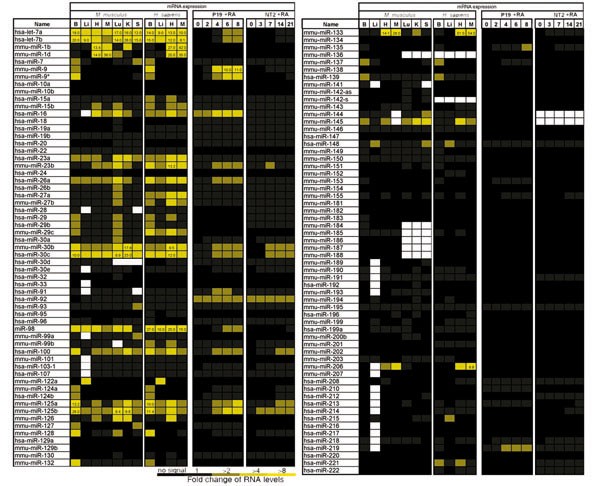
Using northern analysis, we characterized the expression pattern of 92 previously cloned miRNAs [1–3, 9, 10] and 27 computationally predicted miRNAs [8] in adult organs from mouse and human (Figure 1). This study confirms the expression of previously cloned miRNAs [1–3, 9, 10], and validates the existence of computationally predicted miRNAs [8]. Although a wide range of expression profiles was represented among these miRNAs, in general those miRNAs that had been efficiently cloned from a particular organ in previous studies [2, 3], were also detected in those same organ(s) by northern blot hybridization in this study. We detected 21 out of 27 computationally predicted miRNAs by northern blot hybridization. This result indicates that computational predictions are an appealing method for miRNA gene discovery [8], but one should be cautious not to take computer predictions as proof of existence until newly predicted miRNAs are validated experimentally. The inability to detect some of the cloned and predicted miRNAs might be due to low levels of expression, expression limited to a small number of cells or in a narrow window of time during development, or simply that these miRNAs are not expressed in organs examined in this study.
Figure 1
The miRNA expression in mammalian organs and carcinoma cells. Mouse and human organs analyzed by northern blotting were brain (B), liver (Li), heart (H), skeletal muscle (M), lung (Lu), kidney (K), and spleen (S). P19 and NT2/D1 were treated with RA at time 0, cells were harvested and total RNA extracted at the indicated times (days). The miRNA levels detected by northern blot hybridization are represented as fold change of radioactive signal over background, as described in Materials and methods. The intensity of the yellow scale corresponds to the intensity of miRNA expression; black indicates no signal was detected over background, either by quantification or by visual inspection of the phosphoimager image. White indicates cases that were not tested. The stringency of hybridization was not sufficient to distinguish single nucleotide mismatches; very similar expression profiles were observed for members of closely related miRNA families, and so these profiles may reflect a composite of expression profiles from each member.
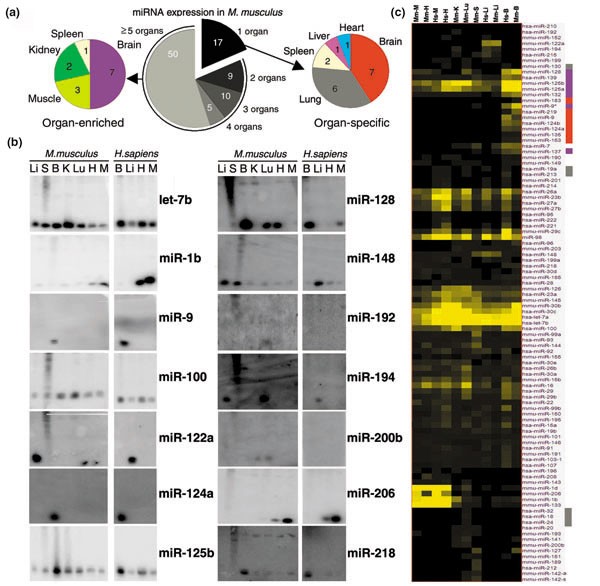
A total of 17 of the expressed miRNAs were detected exclusively in a particular mouse organ; these included: seven brain-specific miRNAs (miR-9, -124a, -124b, -135, -153, -183, -219), six lung-specific miRNAs (miR-18, -19a, -24, -32, -130, -213), two spleen-specific miRNAs (miR-189, -212), one liver-specific miRNA (miR-122a), and one heart-specific miRNA (miR-208) (Figure 2). All of the indicated mouse brain-, liver- and heart-specific miRNAs were also detected in the human counterpart organs (miRNA expression was not examined in human kidney, lung or spleen), with the exception of miR-183 in the human brain. Among the 75 miRNAs that were detected in two or more mouse organs, the levels of 14 of these were detected in a particular mouse organ at levels at least two-fold higher than in any other organ; these included: seven brain-enriched miRNAs (miR-9*, -125a, -125b, -128, -132, -137, -139), three skeletal muscle-enriched miRNAs (miR-1d, -133, -206), two kidney-enriched miRNAs (miR-30b, -30c), and one spleen-enriched miRNA (miR-99a) (Figure 2). All brain-enriched and skeletal muscle-enriched miRNAs had similar elevated levels in the human counterpart organs. The high conservation of expression of these organ-specific and organ-enriched miRNAs between mouse and human suggests that they may play a conserved role in the establishment and/or maintenance of a cell or tissue type of that particular organ.
Figure 2
The miRNA expression profiles in mammalian organs. (a) Distribution of all expressed miRNAs in mouse organs (center), miRNAs expressed only in a particular organ (right), and miRNA enriched in a particular organ at least two-fold over other organs (left). Note that mouse brain-specific miRNAs were detected at higher levels than mouse lung-specific miRNAs. Indeed, four of the seven brain-specific miRNA radioactive signals were detected at least two-fold over background. (b) Northern blots of representative miRNAs with similar expression patterns in organs of mouse and human. (c) Clustering of expressed miRNAs by their similar expression profiles in mouse (Mm) and human (Hs) brain (B), liver (Li), heart (H), skeletal muscle (M), lung (Lu), kidney (K), and spleen (S). Note that some mouse lung-specific miRNAs (miR-19a, miR -130, miR -213) are also expressed in other human organs. Colored bars to the right of miRNA names refer to the corresponding color-coded classes in the right hand pie chart in (a): brain-specific (red), brain-enriched (purple), or lung-specific (light grey).
To identify additional classes of miRNAs with similar expression profiles, all 119 miRNAs were hierarchically clustered on the basis of their expression profiles in our experiments, applying Pearson's distribution. From this analysis, at least six major groups of miRNAs with similar expression profiles were discerned (Figure 2c). A group of six miRNAs was expressed primarily in mouse spleen (miR-127, -142-a, -142-s, -151, -189, -212). A group of five miRNAs was expressed in mouse and human liver (miR-122a, -152, -194, -199, -215) with some scattered expression in other organs including lung and kidney. A group of seven miRNAs was expressed in mouse lung and kidney (miR-18, -20, -24, -32, -141, -193, -200b). Together, the last two groups might reflect a role of miRNAs in an epithelial cell type since liver, lung, and kidney are organs containing epithelial tissues. A group of 17 miRNAs was expressed in mouse and human brain (miR-7, -9, -9*, -124a, -124b, -125a, -125b, -128, -132, -135, -137, -139, -153, -149, -183, -190, -219) with scattered expression in other organs. This group included all seven brain-specific and seven brain-enriched miRNAs described above. The brain is a complex organ with several cell types, among which neurons and glial cells are predominant. Future work will determine the specific neuronal cells in which these miRNAs are expressed. A group of six miRNAs was expressed in mouse and human skeletal muscle and heart: miR-1b, -1d, -133 and -206 had elevated expression in heart and skeletal muscle with low expression in other organs and miR-143 and -208 were almost exclusively detected in heart and skeletal muscle. Skeletal muscle and heart derive from the mesodermal embryonic layer and possess similar striated muscle cells. Thus, these miRNAs could be developmentally expressed in muscle cells of both organs. A group of five miRNAs showed abundant expression across organs (let-7a, -7b, miR-30b, -30c). let-7 family members have been suggested to play a role in terminal differentiation during development in invertebrates and vertebrates [16]. Overall, about 50% of the expressed miRNAs were assigned to a specific or restricted set of organs. Other miRNAs had a more complex pattern of expression and these were not readily assigned to a particular group.
The majority of miRNAs presented here were previously cloned by Lagos-Quintana and colleagues [2, 3]. Our characterization of organ expression of miRNAs correlates with the organ(s) from which these miRNAS were originally cloned [2, 3]. miRNAs that were abundantly cloned in a particular organ, such as miR-135 and miR-153 from brain [2], were exclusively detected by northern analysis in mouse and human brain. However, we observed some discrepancies between our expression profiling by northern analysis and expression profiling by cloning [2, 3]. This is particularly so for certain less-abundant miRNAs - those that were represented in size-selected RNA libraries by three or fewer copies. We believe that northern analysis is a more statistically robust method for expression profiling of less abundant miRNAs than cloning frequency is. The panel of organs that we tested in our study did not include all the organs and tissues that miRNAs were cloned from [1–3, 9, 10], and thus some organ-specific or organ-enriched miRNAs may be expressed in additional organs or tissues.
Coordinate expression of closely linked miRNAs
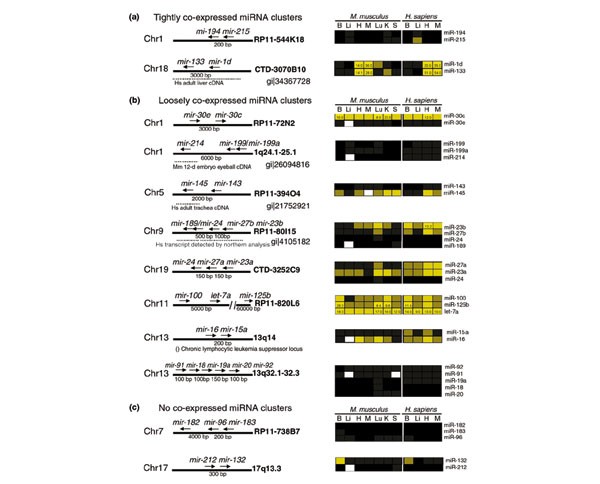
Closely linked miRNAs that are arranged in tandem often exhibit similar expression profiles, consistent with shared _cis_-regulatory elements, and possibly a polycistronic precursor transcript. We observed similar expression profiles for the pair of linked miRNAs, mir-194 and mir-215, and also for both members of the _mir-1d_~mir-133 cluster (Figure 3). The miR-1d~miR-133 hairpin precursors were found contained within the same human cDNA clone (gi|34367728), suggesting that miR-1d and miR-133 were expressed from the same di-cistronic transcript.
Figure 3
Genomic organization and correlated expression of clustered miRNAs. (a) In some miRNA clusters, all members of the cluster can be expressed in a highly correlated pattern; (b,c) for other clusters, the expression of different cluster members can vary in level or pattern. In some cases, entire clusters, or individual cluster members, have been cloned in human or mouse cDNA libraries, or detected by northern blot hybridization (dotted lines). Some miRNAs are associated with aberrant cytological locations (parenthesis). Chromosomal location of miRNAs is given by their cytological coordinates or BAC clone. Expression profiles are shown for mouse and human brain (B), liver (Li), heart (H), skeletal muscle (M), lung (Lu), kidney (K), and spleen (S).
We have also found several cases were closely linked miRNAs are not tightly coexpressed. Closely linked miRNAs such as mir-23b~mir-27b~mir-24/mir-189 exhibited more loosely correlated expression patterns (Figure 3). However, a transcript containing miR-23b~miR-27b~miR-24/miR-189 was detected by northern analysis in humans (gi|4105182), which suggests that these clustered miRNAs probably belong to the same transcriptional unit, but accumulate differentially in vivo as a result of differences in post-transcriptional processing or stability. Other cases of clustered miRNAs with different expression profiles, such as mir-132 and mir-212, could represent closely linked but independent transcription units.
miRNAs can be expressed from long transcripts
We found in GenBank [[35](/article/10.1186/gb-2004-5-3-r13#ref-CR35 "National Center for Biotechnology Information[ http://www.ncbi.nlm.nih.gov/
]")\] evidence that single, isolated miRNA genes can be expressed from long transcripts. miR-9/miR-9\* and miR-145 were cloned in cDNA libraries from human hypothalamus (gi|28436750) and trachea (gi|21752921), respectively. Both miR-191 and miR-214 were contained in long transcripts cloned in cDNA libraries from mouse neonate head (gi|26096865) and eyeball (gi|26094816), respectively. Where the miRNA transcriptional unit starts is an important consideration since regulatory regions for miRNA transcription may reside kilobases upstream of the predicted hairpin precursor. In other cases, a miRNA may be regulated by a cryptic or nearby promoter. For example, _mir-208_, which is expressed in the mouse and human heart, resides in an intron of human cardiac α-myosin heavy chain. Perhaps _mir-208_ is co-transcribed with α-myosin and then processed out.Some miRNAs are associated with chromosomal rearrangements
Some miRNAs reside in genomic regions that have been associated with a disease state. mir-15 and mir-16 reside on chromosome 13q14, a region associated with chronic lymphocytic leukemia (CLL) (Figure 3b). Calin et al. reported that miR-15 and miR-16 were expressed at reduced levels or unexpressed at all in patients with CLL [36]. Notably, mir-142s/mir-142as are located in the junction of the t(8;17) chromosomal rearrangement. This translocation fuses Bcl3 and Myc genes, thus resulting in an aggressive B-cell leukemia [2, 37]. mir-29 resides on chromosome 7q31. This FRA7G region is frequently altered in different tumor types including prostate, breast, and ovarian cancer [38]. It has also been hypothesized that FRA7G region contains a tumor suppressor gene, which is mutated during the development of these cancers or an oncogene that is amplified [38]. It will be interesting to see if miR-142s/miR-142as and miR-29 expression is altered in patients with a t(8;17) chromosomal rearrangement or mutated FRA7G region, respectively.
A subset of brain-expressed miRNAs is induced in retinoic acid-treated EC cells
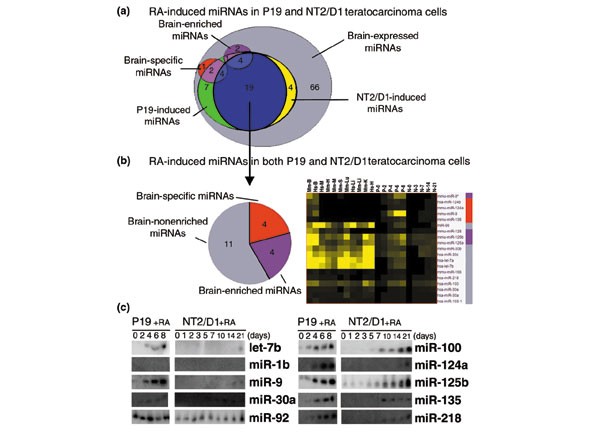
We observed a high percentage of brain-specific and brain-enriched miRNAs compared to miRNAs enriched or specific to other organs (14 out of 30 miRNAs in mouse), and an overall prevalence of miRNAs expressed in the brain (66 out of 90 miRNAs in mouse). This suggested that miRNA-based regulation of gene activity might be engaged during nervous system development and neuronal differentiation. To determine which of brain-expressed miRNAs were associated with neuronal differentiation, we examined their expression during neuronal differentiation of EC cells. Mouse P19 and human NTera-2 clone D1 (NT2/D1) are EC cells that differentiate into neurons upon RA-treatment [39–41]. P19 and NT2/D1 have similar programs of neuronal differentiation, though the differentiation of P19 cells progresses more rapidly than that of NT2/D1 cells [42, 43]. An early phase of commitment to neuronal fate occurs within two days after RA-treatment for P19 and NT2/D1 cells. A later phase of terminal differentiation takes place between days five to eight in P19 cells and is delayed for more than three weeks in NT2/D1 cells [42, 43]. We reasoned that miRNAs with evolutionarily conserved roles in neurons should be induced during neuronal differentiation of both mouse and human EC cells. P19 and NT2/D1 cells were treated with 1 μM and 10 μM of RA, respectively, to induce neuronal differentiation. Cells were collected at key days during differentiation and total RNA was analyzed by northern blotting (Figures 1, 4). Remarkably, 19 miRNAs were induced in both P19 and NT2/D1 cell lines (Figure 5). They included four of the brain-specific (miR-9, -124a, -124b, -135), four of the brain-enriched (miR-9*, -125a, -125b, -128), and 11 of the brain-nonenriched (let-7a, let-7b, miR-30a, -30b, -30c, -30e, -98, -100, 103-1, -156, -218). The expression of all 19 miRNAs was induced in P19 cells typically by day four after RA-treatment and progressively accumulated to higher levels thereafter. This time corresponds to a second wave of transcriptional activation of protein-encoding genes that are involved in terminal differentiation [44]. Thus, miRNA could collaborate with or fine-tune the regulation mediated by neurogenic proteins that specify neuronal identity. Since we did not directly examine expression of miRNAs in glial cells, we could not eliminate the possibility that RA-induced miRNAs were also expressed in glial cells.
Figure 4
The miRNA expression profiles in EC cells differentiating in response to RA. (a) Sets of miRNAs expressed in P19 and NT2/D1 cells treated with RA. All RA-induced miRNAs in P19 and NT2/D1 cells are also expressed in the brain (Figures 1, 2). Numbers in overlapping areas indicate the amount of miRNAs that belong to intersecting sets. (b) Clustering of the 19 miRNAs induced by RA-treatment in both P19 and NT2/D1 cells by their similar expression profile in mouse (Mm) and human (Hs) brain (B), liver (Li), heart (H), muscle (M), lung (Lu), kidney (K) and spleen (S). Colored bars to the right of miRNA names indicate miRNA expression classes in reference to the pie chart on the left: brain-specific (red), brain-enriched (purple), or brain-nonenriched (grey). (c) Northern blots of representative RA-induced miRNAs, constitutively expressed miRNAs (miR-92), and nonexpressed miRNAs (miR-1d) in P19 and NT2/D1 cells treated with RA.
Figure 5
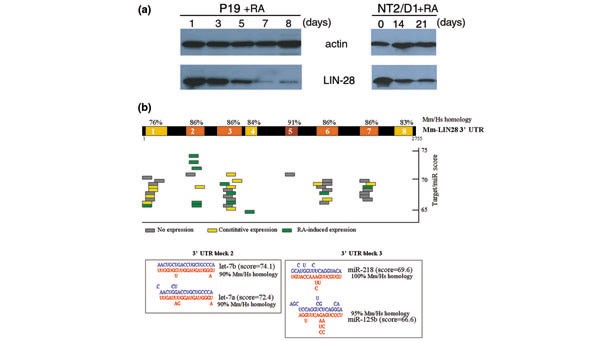
LIN-28: a potential miRNA target in P19 and NT2/D1 cells. (a) Western blot of LIN-28 protein in P19 and NT2/D1 cells treated with RA at day 0. Actin is shown as loading control for protein concentration. (b) Depiction of 3' UTR of mouse Lin-28 mRNA, colored blocks indicated strong sequence homology with human Lin-28 3' UTR. Colored bars indicate predicted miRNA sites on Lin-28 3' UTR conserved blocks. Insets of block 2 and block 3 show predicted RNA-RNA interaction of Lin-28 3' UTR and RA-induced let-7a, miR-125b, and miR-218.
Only a subset of the brain-expressed miRNAs were detected in neurons derived from RA-treated EC cells suggesting that other brain-expressed miRNAs are present primarily in nonneuronal cell types such as glia. Alternatively, these other brain-expressed miRNAs may be present in neuronal subtypes not represented in differentiated neurons derived from EC cells. In a recent study, Krichevsky et al. described a series of miRNAs that were developmentally regulated in rat brain during corticogenesis [17]. We observed that the same series of miRNAs was upregulated in EC cells upon RA-treatment. Krichevsky et al. also showed that miR-9/miR-9* were expressed at lower levels in the presenillin-1 knockout mouse, which had an abnormal forebrain development [17]. We observed a strong induction of miR-9/miR-9* expression in P19 cells. Together, these results suggest that miR-9/miR-9* may play an important role during neurogenesis in rodents.
The coordinate activation of miRNAs (miR-9/9*, -103-1, -124a, -124b, -128, -135, -156, -218) during neuronal differentiation represents a global action of RA in the genome, since these miRNAs constitute unlinked loci. This contrasts with the localized repression exerted by RA upon embryonic stem cell-specific miRNAs (miR-290 to miR-295) arranged in two tightly linked loci [34].
LIN-28 could be a potential target for miRNA regulation in embryonal carcinomas
The mir-125 and let-7 family members were among the brain-expressed miRNAs induced during neuronal differentiation. In C. elegans, lin-4 (the ortholog of mir-125) and let-7 are engaged in a genetic pathway that controls temporal cell fates during larval development [25, 26]. Upregulation of lin-4 RNA during the first larval stage (L1) triggers the progression from early (L1) to late (L2-L3) fate programs [24]. lin-4 RNA binds to the 3' UTR of target mRNAs, including lin-28, and inhibits their translation [27, 45]. LIN-28 is expressed at early stages in neuronal, hypodermal, and intestinal lineages before it is developmentally downregulated by lin-4 RNA. In mammals, LIN-28 protein is downregulated as embryonic development progresses, consistent with a possible conserved role in developmental timing [46]. Interestingly, LIN-28 is present in undifferentiated P19 and NT2/D1 cells, but it is not detected in neuronal or liver tissues [46]. To test if downregulation of LIN-28 could occur in conjunction with neuronal differentiation, western analysis of total protein from lysates of P19 and NT2/D1 cells was performed at various times after RA-treatment. LIN-28 levels began to decrease on about day five after RA-treatment, and almost disappeared by day eight, when cells reached their final state of neuronal differentiation. A similar result was observed in NT2/D1 cells; LIN-28 levels decreased slightly, typically by day 14, and were still detectable at day 21. The kinetics of LIN-28 downregulation in P19 and NT2/D1 cells during RA-induced differentiation parallels the upregulation of miR-125b, let-7a, and miR-218 in these EC cells. The 3' UTRs of the mouse and human Lin-28 mRNAs contain conserved sequences complementary to miR-125b, let-7a, and miR-218 (Figure 5) [46], consistent with a role for these miRNAs in the downregulation of LIN-28 in NT2/D1 and P19 cells. Yet, we did not detect downregulation, in differentiating P19 cells, of a luciferase reporter gene fused to the mouse Lin-28 3' UTR and driven by a cytomegalovirus (CMV) or thymidine kinase (TK) promoter (data not shown). We also noticed a decrease in endogenous Lin-28 mRNA levels in differentiated neurons derived from P19 cells (data not shown), suggesting that repression of the native Lin-28 gene in this system may be largely transcriptional. These results therefore suggest that, unlike C. elegans larval development, where lin-28 downregulation is primarily caused by translational repression by lin-4 RNA, the decline of Lin-28 during neuronal differentiation of P19 cells involves primarily transcriptional repression. However, the phylogenetic conservation of miRNA sites suggests that translational repression of Lin-28 by let-7a, miR-125b, and/or miR-218 may contribute secondarily to Lin-28 downregulation in the mammalian system.
Myc and Cyclin D1 are well-characterized cases of genes repressed by RA-treatment at the levels of transcription and translation in differentiating neurons [47, 48]. We subjected the conserved blocks between mouse and human Myc and Cyclin D1 3' UTRs to the same computer algorithm applied to Lin-28 3' UTR to identify potential binding sites for RA-induced miRNAs. Potential sites for let-7 and miR-30 family members were found on Myc and Cyclin D1 3' UTRs (data not shown). These findings suggest that translational repression by miRNAs, in combination with transcriptional repression and mRNA turnover, could contribute to the downregulation of growth and cell cycle regulators during neuronal differentiation.
Conclusions
Hundreds of miRNAs have recently been discovered in mammalian genomes. To begin to explore the potential roles of these miRNAs in mammalian development and physiology, we characterized the expression profiles of 119 miRNAs in mouse and human organs (Figure 1). We found that many of these miRNAs exhibit phylogenetically conserved organ-enriched or organ-specific expression. In both mouse and human examined organs, seven miRNAs were expressed specifically in a particular organ and ten miRNAs were greatly enriched in a particular organ over all others (Figure 2). These data associate miRNAs to particular organs and suggest their participation in conserved developmental or physiological pathways within these spatial domains in mouse and human. Interestingly, 13 of these 17 miRNAs were enriched or specifically expressed in brain of both mouse and human. Of these, eight brain miRNAs were also induced during differentiation of neurons derived from human and mouse EC cells treated with RA (Figure 4). These data strongly suggest a conserved role for these miRNAs in the specification and/or progression of neuronal fate in mammals. At least one gene whose protein product decreases during neuronal differentiation, Lin-28, contains potential sites in its 3' UTR for complementary base-pairing by the RA-induced miRNAs let-7a, miR-125b, and miR-218 (Figure 5). However, not only the protein levels but also the RNA levels of Lin-28 decreased upon RA treatment suggesting that other factors must also mediate this process. Unlike C. elegans lin-28 whose translational inhibition by lin-4 RNA is the primary mechanism for its downregulation, miRNAs may provide an auxiliary or reinforcing layer of regulation to completely shut down Lin-28 expression in EC cells. miRNA loss-of-function experiments in these EC cells could prove useful in understanding their regulatory contribution during the process of neuronal differentiation. In the case of targets whose expression is primarily controlled on the translational level, miRNAs could play a primary regulatory role; in other cases, such as mammalian Lin-28, where the primary mode of regulation is transcriptional, translational repression by miRNAs could play an auxiliary, or supplementary regulatory role.
Materials and methods
Cell culture and retinoic acid treatment
P19 cells were cultured in αMEM with 7.5% fetal calf serum and 2.5% fetal bovine serum supplemented with streptomycin and penicillin; NT2/D1 cells were cultured in high glucose DME with 10% fetal bovine serum supplemented with penicillin, streptomycin, and glutamine under humidified 5% CO2. P19 cells were treated with 1 μM RA solution as described in [40]; NT2/D1 were treated with 10 μM RA solution as described in [49]. Control cells were treated with an equal amount of the vehicle dimethyl-sulfoxide.
Northern analysis
Frozen dissected organs from six-week-old C57BL/6 mice were a gift from Nathan Watson (Norris Cotton Cancer Center at Dartmouth-Hitchcock Medical Center). Total RNA was extracted from pooled organs of two to four mice or from cell lines with Trizol Reagent following the vendor's recommendations. Total RNA, 5 or 10 μg, was routinely loaded onto 12% urea-polyacrylamide gels as described in [50]. Total RNA from human organs was acquired from the Biochain Institute. Membranes were stripped by boiling in 0.1% SDS and re-hybridized up to five times without observing decreases in quality or intensity of radioactive signals.
StarFire oligonucleotide probes
We used StarFire probes, radioactively labeled with [α-32P]-dATP 6000 Ci/mmol following vendor's recommendations. The complete list of StarFire probes used in this study is enclosed in the additional data file.
Data clustering
Radioactivity contained in northern blot bands was quantified with the ImageQuant software package, and was expressed as a ratio of signal in the band of hybridization to signal in the background adjacent to the band. Hybridization/background ratios of ≤ 1.1 were not reported as positive signals, except when the band was readily detectable by eye (Figures 2, 5). Northern hybridization data were clustered with GeneTraffic 6.2 software package, applying Pearson's hierarchical clustering.
miRNA::target mRNA predictions
Potential RNA::RNA interactions between miRNAs and 3' UTR sequences were assessed by testing 20-nts segments of 3' UTR sequence for their predicted capacity to form RNA secondary structure with a candidate miRNA. Each miRNA sequence was appended to each 20-nucleotide 3' UTR segment by a poly-(N)10 linker, and analyzed for predicted RNA secondary structure using the RNAfold program of the Vienna RNA package [[51](/article/10.1186/gb-2004-5-3-r13#ref-CR51 "Vienna RNA package[ http://www.tbi.univie.ac.at/~ivo/RNA/
]")\]. The full length of a 3' UTR sequence was queried by extracting successive 20-nucleotide test segments, tiling along in two-nucleotide steps. RNA folding output generated by ViennaRNA was analyzed using the following software filters to evaluate the strength of the predicted target::miRNA interaction: at least two out of the three first and last bases of the miRNA had to be bound to the target sequence; at least 65% of the miRNA sequence had to be bound to the target sequence; the free energy (ΔG) of the binding had to be greater than -7 Kcal/mole; and the target::miRNA interaction site had to lie within regions of at least 76% identity between mouse and human target 3' UTRs.The scoring scheme was modeled based on the experimentally confirmed interaction between lin-4 RNA::lin-28 3' UTR in C. elegans. Those predicted miRNA::UTR structures that passed the above filters were further assigned a numerical score based on the following parameters: total number of miRNA bases bound to a target sequence - points for binding were weighed, having a less impact on the score of a mismatch in the central region of the miRNA, which tends to bulge out in documented miRNA/mRNA interactions [23, 45] (denoted (a)); number of consecutive miRNA bases bound to target sequence (denoted (b)); overall ΔG of RNA-RNA interaction (denoted (c)); and additional emphasis for miRNA with the first three and last three bases bound to target sequence (denoted (d)). This is the formula of the overall score = 8a + 2b + 84c + 6d.
Additional data file
The complete list of StarFire probes used in this study is provided in Additional data file 1, available with the complete version of this article, online.
References
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T: Identification of novel genes coding for small expressed RNAs.Science 2001, **294:**853–858.
Article Google Scholar - Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T: Identification of tissue-specific microRNAs from mouse.Curr Biol 2002, **12:**735–739.
Article Google Scholar - Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T: New microRNAs from mouse and human.RNA 2003, **9:**175–179.
Article Google Scholar - Lai EC, Tomancak P, Williams RW, Rubin GM: Computational identification of Drosophila microRNA genes.Genome Biol 2003, **4:**R42.
Article Google Scholar - Lau NC, Lim LP, Weinstein EG, Bartel DP: An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans .Science 2001, **294:**858–862.
Article Google Scholar - Lee RC, Ambros V: An extensive class of small RNAs in Caenorhabditis elegans .Science 2001, **294:**862–864.
Article Google Scholar - Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP: The microRNAs of Caenorhabditis elegans .Genes Dev 2003, **17:**991–1008.
Article Google Scholar - Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP: Vertebrate microRNA genes.Science 2003, **299:**1540.
Article Google Scholar - Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G: miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs.Genes Dev 2002, **16:**720–728.
Article Google Scholar - Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G: Numerous microRNPs in neuronal cells containing novel microRNAs.RNA 2003, **9:**180–186.
Article Google Scholar - Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP: MicroRNAs in plants.Genes Dev 2002, **16:**1616–1626.
Article Google Scholar - Llave C, Kasschau KD, Rector MA, Carrington JC: Endogenous and silencing-associated small RNAs in plants.Plant Cell 2002, **14:**1605–1619.
Article Google Scholar - Ke XS, Liu CM, Liu DP, Liang CC: MicroRNAs: key participants in gene regulatory networks.Curr Opin Chem Biol 2003, **7:**516–523.
Article Google Scholar - Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V: Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity.Dev Biol 2003, **259:**9–18.
Article Google Scholar - Lee Y, Jeon K, Lee JT, Kim S, Kim VN: MicroRNA maturation: stepwise processing and subcellular localization.EMBO J 2002, **21:**4663–4670.
Article Google Scholar - Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al.: Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA.Nature 2000, **408:**86–89.
Article Google Scholar - Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS: A microRNA array reveals extensive regulation of microRNAs during brain development.RNA 2003, **9:**1274–1281.
Article Google Scholar - Carrington JC, Ambros V: Role of microRNAs in plant and animal development.Science 2003, **301:**336–338.
Article Google Scholar - Chen X: A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development.Science 2003, 101126/science.1088060.
Google Scholar - Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D: Control of leaf morphogenesis by microRNAs.Nature 2003, **425:**257–263.
Article Google Scholar - Tang G, Reinhart BJ, Bartel DP, Zamore PD: A biochemical framework for RNA silencing in plants.Genes Dev 2003, **17:**49–63.
Article Google Scholar - Xie Z, Kasschau KD, Carrington JC: Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation.Curr Biol 2003, **13:**784–789.
Article Google Scholar - Lee RC, Feinbaum RL, Ambros V: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14.Cell 1993, **75:**843–854.
Article Google Scholar - Feinbaum R, Ambros V: The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans .Dev Biol 1999, **210:**87–95.
Article Google Scholar - Pasquinelli AE, Ruvkun G: Control of developmental timing by microRNAs and their targets.Annu Rev Cell Dev Biol 2002, **18:**495–513.
Article Google Scholar - Rougvie AE: Control of developmental timing in animals.Nat Rev Genet 2001, **2:**690–701.
Article Google Scholar - Olsen PH, Ambros V: The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation.Dev Biol 1999, **216:**671–680.
Article Google Scholar - Seggerson K, Tang L, Moss EG: Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation.Dev Biol 2002, **243:**215–225.
Article Google Scholar - Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G: The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor.Mol Cell 2000, **5:**659–669.
Article Google Scholar - Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE: The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs.Dev Cell 2003, **4:**625–637.
Article Google Scholar - Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ: The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target.Dev Cell 2003, **4:**639–650.
Article Google Scholar - Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM: bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila .Cell 2003, **113:**25–36.
Article Google Scholar - Xu P, Vernooy SY, Guo M, Hay BA: The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism.Curr Biol 2003, **13:**790–795.
Article Google Scholar - Houbaviy HB, Murray MF, Sharp PA: Embryonic stem cell-specific MicroRNAs.Dev Cell 2003, **5:**351–358.
Article Google Scholar - National Center for Biotechnology Information[http://www.ncbi.nlm.nih.gov/]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al.: Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia.Proc Natl Acad Sci USA 2002, **99:**15524–15529.
Article Google Scholar - Gauwerky CE, Huebner K, Isobe M, Nowell PC, Croce CM: Activation of MYC in a masked t(8;17) translocation results in an aggressive B-cell leukemia.Proc Natl Acad Sci USA 1989, **86:**8867–8871.
Article Google Scholar - Huang H, Qian J, Proffit J, Wilber K, Jenkins R, Smith DI: FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites.Oncogene 1998, **16:**2311–2319.
Article Google Scholar - Jones-Villeneuve EM, Rudnicki MA, Harris JF, McBurney MW: Retinoic acid-induced neural differentiation of embryonal carcinoma cells.Mol Cell Biol 1983, **3:**2271–2279.
Article Google Scholar - Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI: Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells.J Cell Biol 1982, **94:**253–262.
Article Google Scholar - Andrews PW: Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro .Dev Biol 1984, **103:**285–293.
Article Google Scholar - Bain G, Ray WJ, Yao M, Gottlieb DI: From embryonal carcinoma cells to neurons: the P19 pathway.Bioessays 1994, **16:**343–348.
Article Google Scholar - Andrews PW: Teratocarcinomas and human embryology: pluripotent human EC cell lines. Review article.APMIS 1998, **106:**158–167.
Article Google Scholar - Wei Y, Harris T, Childs G: Global gene expression patterns during neural differentiation of P19 embryonic carcinoma cells.Differentiation 2002, **70:**204–219.
Article Google Scholar - Moss EG, Lee RC, Ambros V: The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA.Cell 1997, **88:**637–646.
Article Google Scholar - Moss EG, Tang L: Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites.Dev Biol 2003, **258:**432–442.
Article Google Scholar - Miller WH Jr, Moy D, Li A, Grippo JF, Dmitrovsky E: Retinoic acid induces down-regulation of several growth factors and proto-oncogenes in a human embryonal cancer cell line.Oncogene 1990, **5:**511–517.
Google Scholar - Spinella MJ, Freemantle SJ, Sekula D, Chang JH, Christie AJ, Dmitrovsky E: Retinoic acid promotes ubiquitination and proteolysis of cyclin D1 during induced tumor cell differentiation.J Biol Chem 1999, **274:**22013–22018.
Article Google Scholar - Freemantle SJ, Kerley JS, Olsen SL, Gross RH, Spinella MJ: Developmentally-related candidate retinoic acid target genes regulated early during neuronal differentiation of human embryonal carcinoma.Oncogene 2002, **21:**2880–2889.
Article Google Scholar - Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V: The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster .Dev Biol 2002, **244:**170–179.
Article Google Scholar - Vienna RNA package[http://www.tbi.univie.ac.at/~ivo/RNA/]