Posttranslational Protein Modification in Archaea (original) (raw)
Abstract
One of the first hurdles to be negotiated in the postgenomic era involves the description of the entire protein content of the cell, the proteome. Such efforts are presently complicated by the various posttranslational modifications that proteins can experience, including glycosylation, lipid attachment, phosphorylation, methylation, disulfide bond formation, and proteolytic cleavage. Whereas these and other posttranslational protein modifications have been well characterized in Eucarya and Bacteria, posttranslational modification in Archaea has received far less attention. Although archaeal proteins can undergo posttranslational modifications reminiscent of what their eucaryal and bacterial counterparts experience, examination of archaeal posttranslational modification often reveals aspects not previously observed in the other two domains of life. In some cases, posttranslational modification allows a protein to survive the extreme conditions often encountered by Archaea. The various posttranslational modifications experienced by archaeal proteins, the molecular steps leading to these modifications, and the role played by posttranslational modification in Archaea form the focus of this review.
INTRODUCTION
With complete genome sequences appearing at an ever more rapid rate, attention is becoming increasingly directed towards describing the protein complement of a given organism, i.e., the proteome. Studies of proteins conducted both at the level of the individual polypeptide and cellwide have revealed that the repertoire of expressed proteins can expand beyond what is predicted by direct translation of the complement of open reading frames contained within a genome. For example, the proteome can assume additional levels of complexity with differential expression of individual polypeptides or members of protein families as a function of developmental stage or in response to environmental cues. The various permutations of protein-protein interactions possible further expand the complexity of the proteome. However, one of the most important and fundamental aspects of proteomic complexity comes from the various processing events that many proteins experience following their synthesis, i.e., posttranslational modification.
Proteins can be modified posttranslationally by covalent attachment of one or more of several classes of molecules, by the formation of intra- or intermolecular linkages, by proteolytic processing of the newly synthesized polypeptide chain, or by any combination of these events. By chemically linking various modifying groups either permanently or temporarily and by allowing for changes in the molecular composition of the modifying moieties, covalent modifications can endow proteins with properties that are very different from those that are predicted by the encoding genes. Examples of such covalent modifications include glycosylation, lipid attachment, phosphorylation, and methylation.
The covalent bonding of pairs of Cys residues to form disulfide bridges not only modulates the three-dimensional conformation of a polypeptide chain but can also be used to maintain proteins in multisubunit complexes. Controlled reduction and reoxidation of protein disulfide bonds is also employed in electron transfer reactions fundamental to many cellular processes. Proteolytic processing of newly synthesized polypeptide chains similarly allows the cell to control the folding and function of a protein. By removing specific targeting sequences or other stretches of amino acid residues, the cell is able to control where, when, and how a protein will act. As such, posttranslational modifications can significantly modulate the physicochemical and biological properties of a protein through effects on protein function, subcellular localization, oligomerization, folding, or turnover. The distribution of posttranslational modifications and their effects on protein chemistry and cell biology become even broader when one also considers the effects of additional, secondary posttranslational modification steps such as the addition of organic (e.g., flavins) or inorganic (e.g., metal groups) cofactors. Such modifications, however, lie beyond the scope of this review.
Long-known to be widespread in Eucarya and Bacteria, it is becoming clear that posttranslational modification of proteins also takes place in Archaea. Best known in their capacities as extremophiles, i.e., microorganisms able to thrive in the harshest environmental conditions on this planet, Archaea express proteins that enable them to succeed in such habitats. Indeed, archaeal proteins are able to remain properly folded and functional in the face of extremes of salinity, temperature, and other adverse physical conditions that would normally lead to protein denaturation, loss of solubility, and aggregation. Although posttranslational modifications may help archaeal proteins overcome the challenges presented by their surroundings, in most cases, the reason for posttranslational modification of a particular archaeal protein remains unclear. Table 1 lists the posttranslational modifications that archaeal proteins may experience.
TABLE 1.
Posttranslational modifications of archaeal proteins
| Posttranslational modification | Comment |
|---|---|
| Glycosylation | N-glycosylation, O-glycosylation |
| Lipid modification | Lipoproteins, isoprenylation, acylation, GPI anchoring |
| Phosphorylation | Phosphoaspartate, phosphohistidine, phosphoserine, phosphothreonine, phosphotyrosine |
| Disulfide bonds | Cytosolic proteins |
| Proteolytic processing | Signal sequence cleavage, intein excision, amino-terminal and carboxy-terminal maturation |
| Methylation | Methylarginine, methylaspartic acid, methylcysteine, methylglutamic acid, methylglutamine, methylhistidine, methyllysine |
| Acetylation | |
| Amino acid modification | Hypusination, thiolation |
Analysis of the various posttranslational modifications experienced by archaeal proteins has served to reveal not only novel protein modifications not previously observed in Eucarya or Bacteria but also variations of previously characterized posttranslational modifications. By and large, however, archaeal posttranslational modifications often resemble their eucaryal or bacterial counterparts. Hence, elucidating such similarities provides insight into evolutionary relationships across the three domains of life. Moreover, the mosaic profile of eucaryal, bacterial, and archaeal traits that describes posttranslational protein modification in Archaea also holds true when one examines the enzymes and mechanistic steps involved in archaeal protein modification processes. Here too, examination of archaeal systems has served to expand our understanding of natural pathways or to underscore the similarities between archaeal, eucaryal, and/or bacterial biology. Nonetheless, numerous aspects of archaeal posttranslational processing remain poorly described. In the following review, what is currently known of posttranslational protein modification in Archaea is considered.
PROTEIN GLYCOSYLATION
One of the more prevalent posttranslational modifications experienced by eucaryal proteins is glycosylation. Indeed, protein glycosylation, which begins in the lumen of the endoplasmic reticulum and continues in the Golgi apparatus, is thought to be experienced by more than half of all eucaryal proteins (12). Upon translocation into the endoplasmic reticulum, proteins can be N-glycosylated, when branched oligosaccharide trees of 14 subunits are initially added to selected Asn residues. O-glycosylation of Ser or Thr residues usually takes place in the Golgi. In Eucarya, the glycan moieties of glycosylated proteins fulfill a multitude of roles related to protein solubility, folding, stability and turnover, and subcellular localization as well as participating in numerous recognition events (46, 157, 333, 409, 448). Long believed to be an exclusively eucaryal trait, it is now clear that both Bacteria and Archaea are also capable of attaching glycan moieties to selected proteins (285, 292, 381, 425, 445, 456). A list of those archaeal strains reported to contain glycosylated proteins is provided in Table 2.
TABLE 2.
Archaeal species reported to contain glycoproteins
| Species | Evidence for glycosylationa | Reference(s) |
|---|---|---|
| Haloarcula japonica | G | 299 |
| Haloarcula marismortui | C | 136 |
| Halobacterium saccharovorum | E | 389 |
| Halobacterium salinarum | A, B | 246, 280 |
| Haloferax mediterranei | E | 232 |
| Haloferax volcanii | A, B, D | 98, 421 |
| Methanobacterium bryantii | D, F | 219 |
| Methanococcus deltae | F, H | 27 |
| Methanococcus mazei | D | 481 |
| Methanococcus voltae | A, B | 453 |
| Methanosaeta soehngenii | A | 340 |
| Methanospirillum hungatei | E, F | 406 |
| Methanothermus fervidus | A, B, E | 196, 204 |
| Methanothermus sociablis | A, B | 41 |
| Natrialba magadii | E | 197 |
| Pyrococcus furiosus | C, D, E | 44, 230, 231, 455, 464 |
| Sulfolobus acidocaldarius | A, B, E, G, H | 146, 147, 161, 258, 286 |
| Sulfolobus shibatae | E | 112 |
| Sulfolobus solfataricus | D, E | 106, 146, 262 |
| Staphylothermus marinus | C, F | 345 |
| Thermococcus litoralis | E | 44, 145 |
| Thermococcus stetteri | E | 184 |
| Thermoplasma acidophilum | A, B | 478 |
| Thermoplasma volcanium | E | 112 |
Glycosylated Archaeal Proteins
S-layer glycoproteins.
The surface (S)-layer glycoprotein of the halophilic archaeon Halobacterium salinarum was the first prokaryotic glycoprotein to be described in detail (246, 283). Subsequently, S-layer glycoproteins have been studied in numerous prokaryotes (292, 381-383). Serving as the main, if not sole, component of the protein layer surrounding many archaeal cells (101, 382, 383) (Fig. 1), S-layer glycoproteins remain among the best-characterized archaeal glycoproteins. Indeed, examination of the processes used for glycosylation of archaeal S-layer glycoproteins has not only served to enhance our understanding of prokaryotic cell surface biogenesis but has also provided insight into the general phenomenon of protein glycosylation in Archaea.
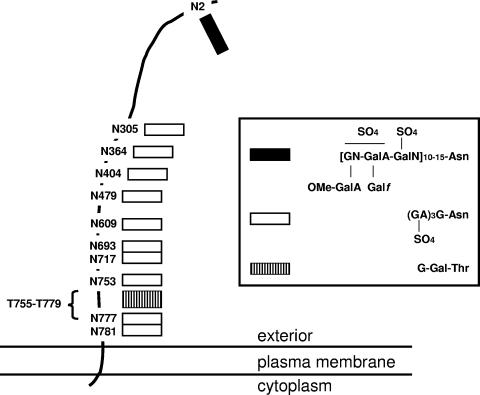
FIG. 1.
Schematic depiction of the glycosylation of the Halobacterium salinarum S-layer glycoprotein. The topology of the S-layer glycoprotein, the positions of the 11 Asn residues that undergo N-glycosylation, and the heavily O-glycosylated Thr-rich region between Thr-755 and Thr-779 are indicated (246). The inset shows the composition of the three oligosaccharide moieties bound to the protein (247). Abbreviations used: G, glucose; GA, glucaronic acid; Gal, galactose; GalA, galacturonic acid; Gal_f_, galactofuranose; GalN, _N_-acetylgalactosamine; GN, _N_-acetylglucosamine; OMe, _O_-methyl; SO4, sulfate. Approximately a third of the glucaronic acid residues may be replaced by iduronic acid.
While the glycosylated nature of S-layer proteins has been proposed in many archaeal species, experimental proof for this posttranslational modification is limited to the S-layer glycoproteins of Halobacterium salinarum (246), Haloferax volcanii (421), Haloarcula japonica (299), Methanothermus fervidus (204), Methanothermus sociablis (41), Sulfolobus spp. (146), and components of the S-layer of Staphylothermus marinus (345). Although the experimental evidence for glycosylation, ranging from chemical characterization of the bound glcyan moieties to glycol staining, is stronger in some cases than others, it has been calculated that these S-layer glycoproteins experience an overall degree of glycosylation of up to 15% (292, 382).
Like eucaryal glycoproteins, archaeal S-layer glycoproteins can undergo both N- and O-glycosylation. In contrast, bacterial S-layer glycoproteins contain only O-linked glycans (285, 445), although examples of N-glycosylation of other bacterial proteins have been shown (107, 425, 456). Analysis of the composition of the N-linked glycan moieties of archaeal S-layer glycoproteins has revealed the wide variety of saccharides available for protein glycosylation in Archaea, including galactofuranose, galactouronic acid, glucose, glucuronic acid, iduronic acid, mannose, _N_-acetylgalactosamine, _N_-acetylglucosamine, and rhamnose (204, 280, 335, 421, 456). In many cases, these sugar subunits may themselves be modified by methylation or sulfation. Such diversity in the range of saccharides used in archaeal S-layer glycoprotein N-glycosylation exceeds that seen in the bacterial and eucaryal N-glycosylation processes (425, 456).
(i) S-layer glycoproteins reveal unique aspects of archaeal protein glycosylation.
Despite the proposed evolution of the eucaryal N-glycosylation system from a precursor process in Archaea (46, 157), studies of archaeal S-layer glycoprotein glycosylation, and in particular glycosylation of the Halobacterium salinarum S-layer glycoprotein, have revealed differences in N-glycosylation in the two domains. Such differences are reflected, for example, in the failure thus far to detect antennary structures in Archaea similar to those employed in eucaryal protein N-glycosylation (46, 157, 235, 333, 409, 442), or in the identified amino acid sequence motifs recognized by the archaeal N-glycosylation machinery.
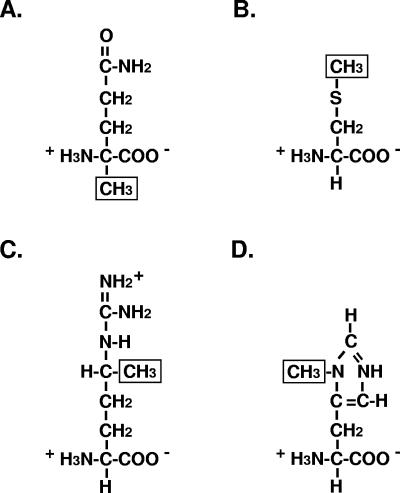
It was observed that replacement of the Ser residue of the Asn-2-Ala-3-Ser-4 sequence of the Halobacterium salinarum S-layer glycoprotein with Val, Leu, or Asn did not prevent N-glycosylation at the Asn-2 position (486). By contrast, the eucaryal system almost invariably recognizes the Asn-X-Ser/Thr sequence motif, where X is any residue apart from Pro (46, 157, 235, 333, 409, 442), although a rare exception of N-glycosylation at an Asn-Gly-Gly-Thr motif has been reported (211). The ability of Archaea to glycosylate proteins at Asn residues that are not part of the consensus Asn-X-Ser/Thr motif suggests that predictions of the glycosylation status of archaeal proteins may have overlooked similar or novel N-glycosylation sites. Moreover, the finding that the repeating sulfated pentasaccharide moiety attached at the Asn-2 position of the Halobacterium salinarum S-layer glycoprotein through an _N_-acetylgalactosamine link is chemically distinct from the sulfated polysaccharide unit attached via glucose subunits found at the other ten N-glycosylation sites in the S-layer glycoprotein (247) implies the existence of two different _N_-saccharyltransferases in this species. At present, it remains unclear how the cell would distinguish between the different N-glycosylation sites.
Finally, the linkage of glycan moieties to the Halobacterium salinarum S-layer glycoprotein at selected Asn residues through either _N_-acetylgalactosamine or glucose subunits (335) is in contrast to the _N_-acetylglucosamine linkage largely employed in eucaryal N-glycosylation (46, 157, 235, 333, 409, 442). In the case of the eucaryal protein laminin, however, N-glycosylation involves a β-glucosyl-Asn protein linkage (385). It is of note that laminin is a component of the extracellular basement membrane, a structural layer surrounding mammalian cells in a manner reminiscent of the archaeal S-layer.
In addition to N-glycosylation, archaeal S-layer glycoproteins can also be modified by O-glycosylation of selected Ser or Thr residues. In both Halobacterium salinarum and Haloferax volcanii, Thr-rich regions adjacent to the membrane-spanning domain of the protein are decorated at numerous positions with galactose-glucose disaccharides (283, 421). Interestingly, a glycoprotein isolated from a eucaryal basement membrane contains a similar disaccharide (254). Presently, little is known of the steps involved in archaeal O-glycosylation or the relation of such steps to the parallel eucaryal or bacterial processes.
Flagellins.
In Archaea, cell motility mediated by flagella has been reported for representatives of the major phenotypic groups, i.e., the halophiles, the methanogens, the thermophiles, and the hyperthermophiles, largely based on microscopic investigation (20, 184, 436). Although fulfilling similar roles, archaeal flagella bear little resemblance to their better-characterized bacterial counterparts (7, 265) in terms of structure or assembly. Such differences become evident when one considers the flagellar filament in the two domains. Ultrastructural studies have shown that, unlike bacterial filaments, archaeal flagellar filaments are not hollow structures (72) and that the archaeal structures are generally thinner than their bacterial counterparts (79, 185, 190, 406).
Archaeal and bacterial flagella also differ at the level of flagellin, the major structural component of the flagellar filament. Whereas bacterial flagella are, for the most part, composed of a single type of flagellin, archaeal flagellar filaments are made up of several types of flagellins (with the possible exception of Sulfolobus solfataricus, where genome annotation efforts have reported the existence of only a single flagellin-encoding gene) (20, 184, 436). Indeed, archaeal and bacterial flagellins do not share sequence similarity (19). Moreover, many archaeal flagellins are glycosylated (184), a posttranslational modification that is considered rare for bacterial flagellins (95, 139, 291, 384, 435).
(i) Evidence for flagellin glycosylation.
Glycosylation has been reported for flagellins of numerous archaeal strains (112, 184, 196, 197, 389, 436), including Halobacterium salinarum (470), Methanococcus deltae (27), Methanococcus voltae (453), and Methanospirillum hungatei (406). In most of these examples, the evidence for glycosylation comes from studies employing glycan-detecting stains, such as thymol-sulfuric acid or periodic acid-Schiff reagent. Such techniques, however, may not always accurately reflect the glycosylated nature of a protein (222). Hence, additional evidence for glycosylation is desirable.
This has been achieved for the flagellins of Halobacterium salinarum and Methanococcus voltae, for which the chemical compositions of the covalently linked glycan moieties have been elucidated. The Halobacterium salinarum flagellin contains a sulfated glycoconjugate, N-linked through a glucose bridge and based on glucuronic or iduronic acid, similar to the glycan moiety found on the S-layer glycoprotein (420, 468). More recently, Methanococcus voltae flagellins have been shown to contain a novel N-linked trisaccharide (453), despite the fact that earlier glycoprotein staining-based studies had failed to detect flagellar glycosylation in this species (195). Analysis of trypsin-generated peptides derived from the Methanococcus voltae S-layer glycoprotein also revealed modification by the same novel trisaccharide (453), suggesting a common glycosylation process for the two proteins. Support for the glycosylation of Methanospirillum hungatei flagella beyond glycan staining was presented by chemical deglycosylation with trifluoromethansulfonic acid, a treatment that decreased molecular mass, as estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (406). The same was noted for Halobacterium salinarum flagellins upon similar treatment (247).
The glycosylated nature of Methanococcus deltae flagellins was indicated upon incubation of cultures with bacitracin, an antibiotic that interferes with protein glycosylation (see below) (247). Such treatment resulted in more rapid migration of the protein as reflected by SDS-PAGE analysis (27). Similar bacitracin treatment, however, had no effect on the glycosylation of Halobacterium salinarum flagellins, as gauged by migration in SDS-PAGE, although incubation with EDTA, thought to specifically inhibit an externally oriented Mg2+-dependent oligosaccharidetransferase (420), successfully modified flagellin migration. By contrast, treating cells with EDTA did lead to the appearance of Methanococcus deltae flagellins of lower apparent molecular weight (27). Together, these observations point to differences in the glycosylation machineries of the two species.
Other proteins.
While the bulk of attention on archaeal protein glycosylation has focused on S-layer glycoproteins and flagellins, other archaeal glycoproteins have been identified. Of those additional glycoproteins whose identities are known, the majority are membrane associated. In many instances, these are binding proteins involved in nutrient uptake (see below), such as the maltose/trehalose-binding proteins of Thermococcus litoralis, shown to react with glyco-stain (145) and of Pyrococcus furiosus, shown to contain glucose-containing glycan moieties by lectin binding and molecular analysis (231), or the Pyrococcus furiosus cellobiose-binding protein, which reacts with lectins and glyco-stain (230). Glyco-staining also indicated the glycosylated nature of Pyrococcus furiosus CipA and CipB, two ABC transporter binding proteins whose expression is up-regulated in response to cold shock in this hyperthermophile (464). Glycosylation of pyrolysin, a thermostable serine-protease also associated with Pyrococcus furiosus membranes, was proposed on the basis of sequence analysis that revealed the presence of numerous potential N-glycosylation sites and supported by glyco-staining of the protein (455).
Based on lectin binding, a series of glycosylated sugar-binding proteins, apparently containing mannose, glucose, galactose, and _N_-acetylglucosamine, was detected in Sulfolobus solfataricus membranes (106). Sulfolobus acidocaldarius cytochrome _b_558/566 was shown to be a highly glycosylated integral membrane protein, containing both O-linked mannose subunits and N-linked hexasaccharides (161). Analysis of the composition of the latter glycan moiety revealed the presence of glucose, mannose, and _N_-acetylglucosamine in addition to 6-sulfoquinovose (484). 6-Sulfoquinovose (or 6-deoxy-6-sulfoglucose) is a rare acidic sugar, commonly found in the glycolipids of chloroplasts and photosynthetic bacteria (177), but not previously found in a glycoprotein. The glycosylated character of a membrane-associated Sulfolobus solfataricus protein serine/threonine kinase was confirmed through precipitation of a protein with kinase activity using lectin-conjugated agarose beads and by the decreased apparent molecular mass of the protein and resistance to glyco-staining following treatment with chemical deglycosylation agents (262).
In addition to membrane proteins, secreted archaeal glycoproteins have also been detected. Lectin binding and chemical deglycosylation confirmed the glycosylated nature of the copper response extracellular proteins secreted by the copper-resistant methanogen Methanobacterium bryantii BKYH (219). Indeed, differential glycosylation is responsible for the appearance of multiple isoforms of the copper response protein. A secreted, inducible alkaline phosphatase purified from Haloarcula marismortui was shown to be glycosylated, in part through the use of radiolabeled glucosamine-containing growth medium (136). Quantitative analysis revealed that glycosylation accounted for 3% of the mass of the protein. Based on glyco-staining, a secreted enzyme possessing thermostable amylopullulanase activity, i.e., capable of hydrolyzing both α-1,6 linkages in pullulan and α-1,4 linkages in amylose and soluble starch, was detected in the growth media of both Pyrococcus furiosus and Thermococcus litoralis (44). Based on aberrant SDS-PAGE migration and sequencing data, it has been proposed that the partially secreted acid protease of Sulfolobus acidocaldarius, thermopsin, is also glycosylated (258).
In addition to these identified membrane and secretory glycoproteins, numerous other glycoproteins, uncharacterized apart from their glycosylated nature, have been reported. Using lectin-based purification techniques, a 152-kDa glycoprotein was isolated from Thermoplasma acidophilum membranes (478). Subsequent analysis of the glycan moiety of the protein revealed it to be a highly branched, mannose-based structure, N-linked to the polypeptide chain through an _N_-acetylglucosamine subunit. Several lectin-binding proteins have been observed in Methanococcus mazei S-6, with the levels of these glycoproteins related to the adoption of morphologically distinct forms by the cells (481). In Haloferax volcanii, membrane glycoproteins of 150, 98, 58, and 54 kDa, distinct from the S-layer glycoprotein, were identified in lectin-based studies (98). A second study of the same strain noted the presence of glycoproteins of 105, 56, and 52 kDa in whole-cell lysates (489). It remains to be seen whether any of the proteins identified in the two studies are the same and whether the smaller glycoproteins are derived from the heavier polypeptides.
Relying on glyco-staining, lectin-binding techniques, and treatments with inhibitors of glycosylation or deglycosylating agents, the membranes of both Sulfolobus acidcaldarius and Sulfolobus solfataricus were shown to contain unidentified glycoproteins distinct from the S-layer glycoprotein (147, 262). Glycoprotein staining was used to identify a series of glycosylated proteins in Pyrococcus furiosus membranes that are distinct from CipA and CipB and the expression of which is related to growth temperature (464).
Process of Protein N-Glycosylation in Archaea
In Eucarya, N-glycosylation begins on the cytoplasmic face of the endoplasmic reticulum membrane, where nucleotide-activated monosaccharides are sequentially added by membrane-embedded monosaccharyltransferases to the saturated polyisoprenol-based lipid carrier dolichol pyrophosphate. This generates the heptasccharide core of the glycan structure initially found on all eucaryal N-glycosylated proteins (46, 157, 235, 333, 409, 442). Once assembled, the glycan-charged lipid translocates (or “flips”) across the plane of the endoplasmic reticulum membrane bilayer so that the oligosaccharide is now oriented within the endoplasmic reticulum lumen. The translocation of the glycan-charged dolichol pyrophosphate across the membrane is catalyzed by an ATP-independent flippase (165), identified as the RTF1 protein in Saccharomyces cerevisiae (159), with homologues reported in other Eucarya (158). Additional sugar subunits are then added to the lipid-bound polysaccharide, transferred from flipped, lumen-facing dolichol phosphate glucose or mannose carriers (158). The completed oligosaccharide is next transferred to appropriate Asn residues of a nascent polypeptide chain entering the endoplasmic reticulum (46, 157, 235, 333, 409, 442). This is mediated by oligosaccharide transferase, a multisubunit complex associated with the translocon, the membrane protein complex responsible for protein translocation across the endoplasmic reticulum membrane (392).
If, as proposed (46, 157), the elaborate process responsible for protein N-glycosylation in Eucarya originated from a simpler archaeal system, then many of the fundamental steps and central components involved in eucaryal protein N-glycosylation should also be present in Archaea. As summarized in Table 3 and discussed in the following section, available evidence suggests that this is indeed the case.
TABLE 3.
N-glycosylation of proteins across the three domains of lifea
| Parameter | Eukarya | Archaea | Bacteria (Campylorbacter jejuni) |
|---|---|---|---|
| Site | ER (Golgi) | Plasma membrane | Plasma membrane |
| Saccharide donors | UDP-GlcNAc, GDP-Man, dolicholphosphate-Man/Glc | UDP-saccharide, GDP-Man?, dolicholphosphate-Man/Glc? | UDP-saccharide |
| Lipid carrier | Dolicholpyrophosphate | Dolicholphosphate, dolicholpyrophosphate | Undecaprenolpyrophosphate |
| Addition of saccharides following lipid flipping | Yes | No | No |
| Modification of lipid-bound oligosaccharide | No | Yes | Yes |
| Final oligosaccharide composition | GlcNAc2Man9Glc3 | Variable | GalNAc2(Glc)GalNAc3Bac |
| Protein glycosylation motif | Asp-X-Ser/Thr | Asp-X-Ser/Thr/Val/Leu/Asp | Asp-X-Ser/Thr |
| Linking sugar | GlcNAc | Variable | GalNAc |
| Oligosaccharide-transferring enzyme | Oligosaccharide transferase complex | STT3 (isoforms?), additional proteins? | Pg1B |
| Oligosaccharide modification following protein transfer | Yes | ? | No |
Dolichol carrier.
Across evolution, isoprene-based lipids play essential roles in the glycosylation process by delivering their bound glycan cargo to selected protein targets (46, 362). In Archaea, glucose-, mannose-, _N_-acetylglucosamine-, and sulfated tetrasaccharyl-containing phospho- and pyrophosphopolyisoprene (containing 11 to 12 isoprene units) were first observed in Halobacterium salinarum by ion exchange and thin-layer chromatography (281). Later studies (248) confirmed that the lipid moiey of these compounds is C60 dodecaprenol. This is similar to the dolichol used in eucaryal protein N-glycosylation (46) but distinct from undecaprenol, which is composed of 11 unsaturated isoprene units and used by Bacteria for protein glycosylation and peptidoglycan synthesis (362, 425). Mass spectrometry and nuclear magnetic resonance-based approaches revealed the presence of _Eucarya_-like sugar carriers in Haloferax volcanii, including mannosyl-galactosyl-phosphodolichol, lesser quantities of a dihexosyl-phosphodolichol and a tetrasaccharyl-phosphodolichol containing mannose, galactose, and rhamnose, all linked to a dolichol containing 11 or 12 isoprene units (242).
(i) Antibiotics that affect dolichol processing interfere with archaeal protein glycosylation.
The use of various antibiotics and other compounds known to prevent protein glycosylation by interfering with the processing of dolichol carriers has provided insight into the role of this lipid in archaeal protein N-glycosylation. Tunicamycin hinders transfer of UDP-_N_-acetylglucosamine to polysaccharide-loaded dolichol carriers (105). Treatment with this antibiotic interferes with Sulfolobus acidocaldarius S-layer glycoprotein glycosylation (147). In contrast, tunicamycin has no effect on the biosynthesis of the Haloferax volcanii S-layer glycoprotein (99) and accordingly, the glycan moiety of the Haloferax volcanii S-layer glycoprotein does not include _N_-acetylglucosamine (242, 280). Bacitracin is another drug that interferes with protein glycosylation via an interruption of the recycling of pyrophosphate-containing dolichol species (420). Accordingly, in Halobacterium salinarum, bacitracin interferes with the attachment of the repeating sulfated pentasaccharide found at the Asn-2 position of the S-layer glycoprotein (284, 469), although not with the attachment of the sulfated polysaccharide found at the other N-glycosylation sites of the protein (469).
Bacitracin also inhibits glycosylation of flagellins in Methanococcus deltae (27) and slowed Sulfolobus acidocaldarius growth, possibly through interference with the protein N-glycosylation pathway (286). In contrast, bacitracin had no effect on the glycosylation of the S-layer glycoprotein or a second 98-kDa glycoprotein in Haloferax volcanii (99, 232). The failure of the antibiotic to prevent Haloferax volcanii glycoprotein biogenesis is likely related to the fact that, unlike Halobacterium salinarum, in which both monophosphate- and pyrophosphate-containing dolichol oligosaccharide carriers are present (247), only bacitracin-insensitive monophosphate-containing oligosaccharide-dolichol intermediates are found in Haloferax volcanii (242). Incorporation of glucose from UDP-glucose into Haloferax volcanii glycoproteins was, however, inhibited by amphomycin and two sugar nucleotide analogs, PP36 and PP55 (489), compounds reported to block transfer of nucleotide-conjugated sugars to phosphopolyisoprenols in Eucarya (201, 202, 336).
(ii) Analysis of dolichol-bound glcyans.
Evidence for the involvement of dolichol phosphate-linked oligosaccharides in archaeal protein N-glycosylation also comes from examination of the carrier-bound glycan moieties. The transfer of radiolabeled glucose from UDP-[3H]glucose to Haloferax volcanii glycoproteins proceeds through a glucose-containing phosphopolyisoprenol intermediate (489). The dolichol-linked sulfated polysaccharide moiety found in Halobacterium salinarum is identical to glycan moieties found on the S-layer glycoprotein and flagellin in this species (248, 470). On the other hand, the sulfated polysaccharide is methylated at the dolichol-linked stage, whereas no 3-_O_-methylglucose is detected in the protein-linked polysaccharide (249).
The importance of this transient methylation is illustrated by the detrimental effect of inhibiting _S_-adenosylmethionine-dependent methylation. Such treatment interfered with glycoprotein biosynthesis but did not affect either general protein biogenesis or the biosynthesis of sulfated phosphodolichol-bound oligosaccharides. It thus appears that methylation is an essential step in the biosynthesis of the sulfated oligosaccharide moiety prior to being transferred to its nascent polypeptide target. By contrast, the hexasaccharide moiety attached to the Methanothermus fervidus S-layer glycoprotein retains its methylation (204). It is not clear whether such methylation is involved in the translocation of the sulfated oligosaccharide phosphodolichol across the membrane or the subsequent transfer of the glycan moiety to the nascent polypeptide chain. In Eucarya, chemical modification of glycoprotein glycan moieties occurs only after the oligosaccharide has been transferred to the nascent polypeptide (449).
Enzymes of N-glycosylation.
Just as archaeal N-glycosylation relies on the dolichol carriers implicated in eucaryal protein glycosylation, Archaea also contain homologues of many of the enzymes involved in eucaryal N-glycosylation. These include those involved in oligosaccharide charging of the lipid carrier, translocation of the dolichol carrier across the membrane, and transfer of the oligosaccharide entity to the nascent polypeptide chain (Fig. 2).
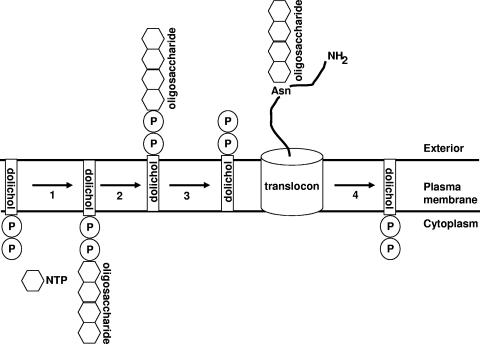
FIG. 2.
Schematic depiction of archaeal N-glycosylation. Step 1. A dolichol pyrophosphate (or monophosphate) species is glycosylated by transfer of saccharide subunits from nucleotide sugars (or possibly from lipid-bound sugar precursors). Step 2. Glycosylated phosphodolichol “flips” across the plasma membrane, likely in an enzyme-mediated process. Step 3. The oligosaccharide structure is transferred to selected Asn residues of a newly translocated polypeptide. The figure does not consider the relationship between protein translation and protein translocation or the relationship between protein translocation and protein glycosylation. Step 4. Following transfer of the oligosaccharide moiety to a protein target, the phosphorylated dolichol carrier is recycled to its original topology. See references 247, 420, and 468, the text, and Table 3 for additional information.
(i) Genomic studies.
Analysis of the NCBI protein database (www.ncbi.nlm.nih.gov) reveals the presence of genes encoding homologues of the staurosporine- and temperature-sensitive yeast protein 3 (Stt3p) (425), an essential protein thought to contain the active site of the multisubunit yeast oligosaccharide transferase complex (309, 493), in 18 archaeal strains. In Bacteria, such as Campylobacter jejuni, it is believed that the Stt3p homologue PglB is the only component needed for transfer of glycans to Asn residues during protein N-glycosylation (425).
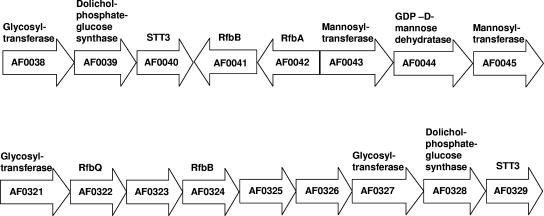
A close examination of the Archaeoglobus fulgidus genome sequence revealed genes encoding STT3-like proteins within two gene clusters encoding putative homologues of other enzymes involved in yeast protein glycosylation (Fig. 3) (46). One of these clusters contains three adjacent open reading frames (ORFs), one of which encodes a polypeptide that appears to contain a motif present in the yeast Alg1p and Alg2p glycosyltransferase proteins. In the yeast proteins, this motif is involved in the transfer of nucleotide sugars to the phosphodolichol carrier (46). The other two ORFs putatively encode a dolichyl-phosphoglucose synthase homologue and a homologue of Stt3p. Other ORFs in this cluster show high sequence similarity to RfbA and RfbB, components of a transporter family presumably involved in the flipping of bacterial O-antigen (467) and lipopolysaccharides (364) across the plasma membrane. While the functions of these putative gene products remain to be shown, it has been postulated that this Archaeoglobus fulgidus gene cluster encodes a functional unit involved in the assembly, translocation, and transfer of dolicholphosphate-linked oligosaccharides to protein targets (46). The second gene cluster in Archaeoglobus fulgidus includes ORFs also encoding putative glycosyltransferase, dolichyl-phosphoglucose synthase, and STT3 proteins, and lies near six ORFs bearing similarity to genes encoding proteins involved in bacterial lipopolysaccharide biosynthesis (46).
FIG. 3.
Schematic depiction of two Archaeoglobus fulgidus gene clusters putatively involved in protein glycosylation. Putative gene products are given above each ORF. For further details, see reference 46.
(ii) Biochemical studies.
In addition to such gene-based predictions, enzymatic activity has also been demonstrated for some archaeal glycosylation-related proteins. Biochemical characterization of Pyrococcus furiosus UDP-α-d-glucose pyrophosphorylase, responsible for UDP-glucose synthesis, represents the first analysis of an archaeal sugar nucleotidyltransferase (290). An _N_-acetylglucosamine transferase was also partially characterized from membranes of Halobacterium salinarum (281). Dolichylphosphate mannose synthase, which is able to transfer GDP-mannose to a dolichol phosphate carrier, was purified from Thermoplasma acidophilum (490). Amphomycin, an inhibitor of dolichylphosphate mannose synthases (202), blocked the activity of the enzyme (490). Using 5-azido-[32P]UDP-glucose in a photoaffinity approach, a single 45-kDa species was identified in Haloferax volcanii homogenates that is thought to correspond to dolichylphosphate glucose synthase (489).
Pyrophosphatases with their active site oriented towards the cell exterior have been purified from the membranes of two different Sulfolobus acidocaldarius strains (8, 286). The pyrophosphate-hydrolyzing activity of the enzymes, proposed to participate in the hydrolysis of dolicholpyrophosphate-linked oligosaccharides during protein glycosylation, was stimulated in the presence of Sulfolobus membrane lipids. Sequence analysis of one of these pyrophosphatases has led to the identification of putative homologues in the genome sequences of Sulfolobus tokodaii and Solfolobus solfataricus as well as in Methanobacterium thermoautotrophicum (294). This study also revealed the presence of a strongly conserved phosphatase tripartite sequence motif, Lys-XXXXX-Arg-Pro-X12-54-Pro-Ser-Gly-His-X31-54-Ser-Arg-XXXXX-His-XXX-Asp, also detected in Lpp1p and Dpp1p, Saccharomyces cerevisiae proteins showing hydrolytic activity towards dolichylphosphate, dolichylpyrophosphate, and other isoprenoid phosphates/pyrophosphates (116).
Subcellular localization of glycosylation.
Several lines of evidence suggest that archaeal glycosylation occurs at the outer cell surface, the topological equivalent of the luminal-facing leaflet of the endoplasmic reticulum membrane bilayer, the site of N-glycosylation in Eucarya (46, 157, 235, 333, 409, 442). Despite its inability to cross the plasma membrane of haloarchaea (284), bacitracin is nonetheless able to interfere with Halobacterium salinarum protein glycosylation by preventing transfer of sulfated oligosaccharides to the S-layer glycoprotein (284, 469). The external orientation of the archaeal glycosylation apparatus is further supported by the decoration of exogenously added, soluble cell-impermeable hexapeptides containing the Asn-based N-glycosylation motif with sulfated oligosaccharides by living Halobacterium salinarum cells (248). Other observations also favor the assignment of archaeal protein glycosylation to the cell's outer surface. These include the ecto-enzymatic nature of a Sulfolobus acidocaldarius pyrophosphatase (8, 286), the proposed specific inhibition of an externally oriented Mg2+-dependent oligosaccharidetransferase by EDTA, a non-cell-permeant reagent, and subsequent interference with Halobacterium salinarum flagellin glycosylation (420), as well as studies supporting the cotranslational mode of membrane protein insertion in Archaea (360).
Role of Protein Glycosylation in Archaea
Structural roles.
Given the seemingly routine glycosylation of archaeal proteins, one can ask what role is played by this posttranslational modification in Archaea. The observation that bacitracin treatment transformed rod-shaped Halobacterium salinarum cells into spheres led to the proposed structural function of archaeal protein glycosylation (282). In fitting with a role for the sulfated S-layer glycoprotein oligosaccharide chains in maintaining the rod shape of Halobacterium salinarum cells, it was noted that similarities exist in the overall structures of the S-layer glycoprotein and proteoglycans, components of the extracellular matrix of animal cells (30, 468). For example, iduronic acid, a major component of proteoglycans (296), is found in the glycans decorating the Halobacterium salinarum S-layer glycoprotein. Similarly, the O-glycosylation cluster situated near the membrane-spanning base of the Haloferax volcanii S-layer glycoprotein has also been assigned a structural support role in the formation of a periplasmic-like space (217). In Thermoplasma acidophilum, an organism that lacks a cell wall, it has been suggested that the glycan moieties attached to the major glycosylated membrane-bound protein species coating the cell surface act to either trap water molecules or allow the cell surface proteins to interact with each other. In either scenario, glycosylation would contribute to the rigidity of the cell surface (478).
Functional roles.
The glycosylation of archaeal proteins has also been implicated in protein assembly and function. In archaeal flagellins, glycosylation is associated with proper flagellar assembly, since upon bacitracin-mediated interference with flagellin glycosylation, a loss of Methanococcus deltae flagellation was observed microscopically (196). In a mutant Halobacterium salinarum strain in which underglycosylated flagellins are overproduced, increased levels of flagella were detected in the growth medium, suggesting proper flagellin glycosylation to be important for correct flagellar incorporation into the plasma membrane (470). This explanation is, however, inconsistent with the apparent nonglycosylated nature of other archaeal flagellins (184) or the glycosylation of Methanospirillum hungatei flagellins, which only occurs in low-phosphate media (406). Similarly, evidence against glycosylation's playing a role in protein function comes from bacterial expression of archaeal binding proteins. Normally glycosylated in their native hosts, nonglycosylated heterologously expressed versions of these proteins were also capable of substrate binding (170, 230, 231). Nevertheless, glycosylation could play a role in stabilization against proteolysis or could affect the interaction of binding proteins with the cell membrane or envelope (4).
Glycosylation as an environmental adaptation.
Coping with the often harsh environmental conditions encountered by Archaea serves as the basis for yet another hypothesized role for archaeal protein glycosylation. In a comparison of the glycosylation profiles of S-layer glycoproteins from the moderate halophile Haloferax volcanii and the extreme halophile Halobacterium salinarum, it was noted that the latter experiences a higher degree of glycosylation than the former (280). Moreover, the glycan moieties of the extreme halophile were enriched in sulfated glucuronic acid subunits as opposed to the neutral sugars found in the moderate halophile. These properties endow the Halobacterium salinarum S-layer glycoprotein with a drastically increased surface charge density relative to its Haloferax volcanii counterpart.
The enhanced negative surface charges are thought to contribute to the stability of haloarchaeal proteins in the face of molar salt concentrations (266). Accordingly, the Halobacterium salinarum S-layer glycoprotein also contains 20% more acidic amino acid residues than does the corresponding protein in Haloferax volcanii (246, 421). The enhanced negative surface charge associated with protein glycosylation and the resulting protection that this would afford in the face of acidic conditions have been offered as the role of Sulfolobus acidocaldarius cytochrome b 558/566 glycosylation (161, 484). It has also been suggested that a significant amount of the protein surface is shielded from the ∼pH 2 environment by the high degree of glycosylation (484). Finally, glycosylation has also been implicated in the stabilization of thermophilic archaeal glycoproteins (4, 258, 455).
LIPID MODIFICATION
Lipid modification, defined herein as the permanent or temporary covalent attachment of lipid-based groups at various positions within a polypeptide chain, is a common modification experienced by both eucaryal and bacterial proteins. An examination of known lipid modifications reveals that a wide variety of lipid moieties can be directly or indirectly linked to a protein at any of numerous attachment sites through the use of any of several linkages (414). For instance, lipid modification can involve myristoyl or palmitoyl acyl groups (358), isoprenyl polymers of various lengths (393), or aminoglycan-linked phospholipids (103). These can be added at the amino terminus, the carboxy terminus, or at internal residues via ester, thioester, thioether, or amide bonds, or through mediating elements, such as the phosphopantethene group of the acyl carrier protein (267).
Lipid modification of proteins is largely a posttranslational event (115). It serves a variety of roles, the most obvious being to enhance the membrane affinity of the modified protein. Accordingly, amino-terminal acylation leads to the localization of numerous proteins to the outer membrane of gram-negative Bacteria (156, 379), as exemplified by Braun's lipoprotein in Escherichia coli (40). Similarly, otherwise soluble eucaryal proteins also become membrane associated upon the covalent attachment of one or more lipid moieties (102, 153, 194, 462). Lipid modification can also modulate protein-protein interactions in Eucarya, as shown by the effects of myristylation or prenylation upon trimeric G protein subunit affinity (124, 178, 462), and in viruses, exemplified by the involvement of myristylation of the capsid proteins of human immunodeficiency virus type 1 and picornavirus in virion particle assembly and secretion (65, 142).
Lipid modifications of eucaryal proteins has also been implicated in a variety of other cellular events. These include signal transduction (287), embryogenesis and pattern formation (271), protein trafficking through the secretory pathway (297), and evasion of the immune response by infectious parasites (369, 461). Yet another role for lipid modification is exemplified by the bacterial toxin hemolysin A, which requires fatty acid acylation on an internal Lys residue for its activation (414).
Given the ubiquitous distribution and numerous functions of lipid modifications in eucaryal and bacterial proteins, it is not surprising that lipid-modified proteins have also been identified in Archaea.
Membrane Lipids of Archaea
One of the defining traits of Archaea that distinguish them from Eucarya and Bacteria is the chemical composition of their membrane phospholipids (206, 208). First, unlike eucaryal and bacterial phospholipids which are built on a glycerol-3-phosphate backbone, archaeal phospholipids are based on the opposite stereoisomer, glycerol-1-phosphate. Second, archaeal phospholipids contain polyisoprenyl side chains rather than the acyl groups employed by eucaryal and bacterial phospholipids. Third, archaeal phospholipids rely on ether bonds to link the isoprenyl side chains to the glycerol-1-phosphate backbone. In Eucarya and Bacteria, ester bonds link acyl side chains to the glycerol-3-phosphate backbone. Of these three traits, the use of glycerol-1-phosphate is considered the most defining, since examples of ether-linked lipids have been observed in Eucarya and Bacteria (172, 328) and non-ester-linked phospholipid fatty acids and genes encoding components involved in the metabolism of fatty acids have been reported in Archaea (127, 342). Indeed, free fatty acids have been observed in the lipid phase of Methanosphaera stadtmanae and Pyrococcus furious (51, 191). Finally, archaeal phospholipids are generally organized into the bilayer structure that is also present in eucaryal and bacterial cells, although tetraether lipid-based monolayers can be found in thermophilic and hyperthermophilic Archaea (92, 226).
Whereas phospholipids and other polar lipids (phosphoglycolipids, glycolipids, and sulfolipids) account for the vast majority of archaeal membrane lipids, archaeal membranes also contain acetone-soluble nonpolar lipid species, primarily neutral squalenes and other isoprenoid-based polymers (206, 207, 334, 439, 440). In halophilic Archaea, in which membrane lipid composition has been most studied, pigmented carotenoids, in particular bacterioruberins, are major components of the nonpolar lipid pool (243, 438). These have been implicated in affording protection from UV-induced damage (390). In addition, many halophilic Archaea also contain retinal as part of bacteriorhodopsin, the purple retinal-containing protein complex that functions as a light-driven proton pump (244).
Lipid-Modified Archaeal Proteins
In Archaea, lipid-modified proteins have been reported from a wide range of species. In many cases, modification involves uncharacterized lipid entities, whereas in others, direct proof for the presence of attached lipid groups remains lacking. Table 4 summarizes the various lipid-based modifications shown or presumed to exist in Archaea, while Fig. 4 offers a schematic presentation of representative archaeal lipid-modified proteins.
TABLE 4.
Lipid modifications observed and proposed in Archaea
| Modification | Species | Observed or predicteda | Reference(s) |
|---|---|---|---|
| N-terminally linked lipid (lipoprotein) | Archaeoglobus fulgidus | Predicted | 4 |
| Halobacterium salinarum | Predicted | 228 | |
| Halobacterium sp. strain NRC-1 | Predicted | 4 | |
| Methanococcus jannaschii | Predicted | 4 | |
| Methanosarcina acetivorans | Predicted | 4 | |
| Methanosarcina mazei | Predicted | 4 | |
| Natronobacterium pharaonis | Predicted | 274 | |
| Pyrococcus abyssii | Predicted | 4 | |
| Pyrococcus furiosus | Predicted | 4 | |
| Pyrococcus horikoshii | Predicted | 4 | |
| Thermococcus litoralis | Predicted | 170 | |
| Isoprenylation | Halobacterium cutirubrum | Observed | 376 |
| Halobacterium salinarum | Observed | 218, 376 | |
| Haloferax volcanii | Observed | 233 | |
| Acylation | Halobacterium cutirubrum | Observed | 350 |
| Methanobacterium thermoautotrophicum | Observed | 350 | |
| GPI anchor | Sulfolobus acidocaldarius | Observed | 224 |
| Methanosarcina barkeri | Predicted | 310 |
FIG. 4.
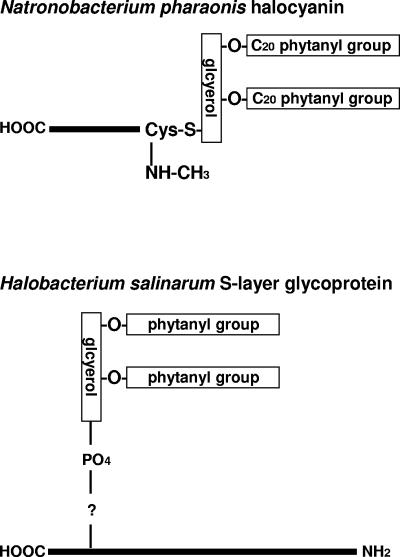
Schematic depiction of representative archaeal lipid-modified proteins. Shown are Natronobacterium pharaonis halocyanin and Halobacterium salinarum S-layer glycoprotein. The lipid modification and acetylation of the amino-terminal Cys of Natronobacterium pharaonis halocyanin have not been experimentally proven, nor has the linkage or exact position of the diphytanylglycerylphosphate group found within the Thr-rich carboxy-terminal region of the Halobacterium salinarum S-layer glycoprotein. See text for details.
Lipoproteins.
In the haloalkaliphile Natronobacterium pharaonis, halocyanin, a small blue copper protein, was proposed to undergo amino-terminal lipid modification based on the presence of the so-called lipobox sequence motif near the start of predicted amino acid sequence (274). In Bacteria, the Leu-Ala-Gly-Cys lipobox sequence motif (156) lies at the end of the signal sequence, the short N-terminal extension not found in the mature, lipid-modified protein (see below). At the membrane, the bacterial lipobox motif is sequentially recognized and processed by three enzymes. The sulfydryl group of the Cys residue is first modified with a diacylglyceride by prolipoprotein diacylglyceryl transferase, after which the upstream Gly-Cys bond is cleaved by signal peptidase II. The newly exposed N-terminal Cys residue of the protein then undergoes additional acylation by apolipoprotein _N_-acyltransferase to yield the mature, lipid-modified lipoprotein (379). Direct proof for such modification of halocyanin has not been provided since the amino-terminal sequence of the protein could not be determined, possibly due to modification of the amino-terminal residue. Support for lipid modification of Natronobacterium pharaonis halocyanin, however, extends beyond the presence of the lipobox motif. Halocyanin is predicted to contain a β-turn after the lipobox, a structural feature that is characteristic of bacterial lipoproteins (130). Furthermore, mass spectroscopic analysis of halocyanin was consistent with the presence of two C20 phytanyl groups ether linked to a glyceryl group (274).
In gram-positive bacteria, it is accepted that substrate-binding proteins, components of multisubunit ABC transporters responsible for cellular uptake of substrates, are lipoproteins (131, 422, 430). The same may well be true in Archaea. The trehalose/maltose-binding protein of the hyperthermophile Thermococcus litoralis contains a lipobox-like sequence motif and requires detergent for its solubilization (170). Similar motifs have been identified in other ABC sugar transporter binding proteins identified in Archaea, suggesting that amino-terminal lipid modification of binding proteins takes place in other species (4, 228).
Lipid modification is not, however, the sole mode of membrane association for archaeal sugar-binding proteins. For example, a membrane-spanning domain is predicted to anchor the glucose-binding protein of Sulfolobus solfataricus (3). It should be noted, however, that binding proteins in this organism differ from those in other Archaea in terms of amino-terminal sequence and subsequent posttranslational processing (see below). In Halobacterium salinarum, BasB and CosB, the first examples of binding proteins involved in chemotaxis in Archaea, are also thought to be lipoproteins due to their membrane localization and bearing of the lipobox sequence motif (228). Indeed, sequence analysis of putative substrate-binding proteins in Halobacterium salinarum, be they involved in nutrient uptake or chemotaxis, suggests that all are lipoproteins (228). Finally, in the case of Pyrococcus species peptide-binding proteins, a conserved Gly-Cys motif reminiscent of the lipobox sequence located near the carboxy terminus may also be a target for lipid modification (4).
Despite the proposed presence of lipoproteins in Archaea, no archaeal homologue of signal peptidase II, one of the enzymes involved in lipoprotein precursor maturation, has been observed. Whether this is because there is no such enzyme in Archaea or because its sequence differs beyond recognition from that of its bacterial homologues, possibly in adaptation to the ether-based phospholipids of the archaeal membrane, remains unknown.
Isoprenylated proteins.
Growth of Halobacterium cutirubrum, Halobacterium salinarum, and Haloferax volcanii in the presence of radiolabeled mevalonate, a precursor of the isoprene building block used to synthesize archaeal lipids (38, 398), led to the appearance of several proteins radiolabeled through the covalent attachment of a lipid entity (233, 376). Subsequent chemical analysis of the modifying lipid moiety in Halobacterium salinarum revealed a novel diphytanylglycerol methyl unit, linked to Cys residues of the modified proteins by a thioetheric bond (376). Further analysis of isoprenoid-modified proteins in Halobacterium salinarum using other radiolabeled isoprenyl derivatives revealed that the S-layer glycoprotein is modified by a second novel group, diphytanylglyceryl phosphate, which is attached through an as yet uncharacterized linkage (218). Amino acid sequencing places the modification near an O-glycosylated Thr-rich stretch found in the C-terminal region of the protein, upstream of the single transmembrane domain (218). In Haloferax volcanii, lipid modification of the S-layer glycoprotein was also shown, although the chemical composition of the attached lipid is unknown, as is the site of attachment (99, 233).
Since haloarchaeal S-layer glycoproteins include a membrane-spanning domain (246, 421, 457), it is unclear why an additional membrane anchor in the form of a lipid would be required. Nonetheless, the attachment of the lipid moiety that takes place on the external surface of Haloferax volcanii and Halobacterium salinarum cells is responsible for the posttranslational, posttranslocational maturation of the S-layer glycoprotein in these strains, as detected through pulse-chase radiolabeling studies (99, 233). Furthermore, since other haloarchaeal S-layer glycoproteins also contain a sequence similar to that modified in Halobacterium salinarum (246, 421, 457), it would appear that such isoprenoid-based lipid modification of S-layer glycoproteins is a general trait of halophilic Archaea (218).
Acylated proteins.
Since some Archaea contain significant amounts of fatty acids (51, 127, 191) and completed archaeal genome sequences reveal the presence of genes involved in fatty acid biosynthesis and β-oxidation (342), it should not come as a surprise that the acylation of archaeal proteins has been reported. In Halobacterium cutirubrum and Methanobacterium thermoautotrophicum, subcellular fractionation and analytic chemical techniques were employed to show the acylation of several proteins (350). Chromatographic analyses identified palmitic and stearic acids as the main modifying agents, although lower levels of modification by myristic acid and other fatty acids were also observed. These acyl groups are thought to be linked to the protein via amide or ester bonds.
GPI-anchored proteins.
Glycosylphosphatidylinositol (GPI) anchors represent a carboxy-terminal posttranslational lipid-based modification used to tether eucaryal proteins to various membranes (176). The GPI anchor is added to target proteins using a preformed GPI-anchoring moiety which consists of a molecule of phosphatidylinositol linked at its myoinositol headgroup to ethanolamine phosphate through an aminoglycan bridge. This lipid is transferred to the newly exposed carboxy terminus of a nascent polypeptide. The modified protein is first synthesized as a membrane-anchored precursor that undergoes proteolytic processing upstream of its carboxy-terminal transmembrane domain. The cleaved protein is thus attached to the ethanolamine end of the preassembled GPI moiety.
Although widespread in the eucaryal domain, GPI-anchored proteins have not been observed in Bacteria (103). They have, however, been detected in Archaea. In Sulfolobus acidocaldarius, three proteins were identified that incorporate radiolabeled precursors of the GPI anchor moiety (224). One of these, a 185-kDa species, was also solubilized by the actions of a bacterial phosphatidylinositol-specific phospholipase C, a characteristic of GPI-anchored proteins (175). Although the other two Sulfolobus proteins were not released by the phospholipase, this is not inconsistent with GPI anchoring as phosphatidylinositol-specific phospholipase C-resistant GPI-anchored proteins have been reported (122, 365). Similarly, a typical archaeal ether-based phospholipid bearing the identical GPI anchor moiety head group as found in Eucarya was identified in Methanosarcina barkeri (310). Incubation of this lipid species with phosphatidylinositol-specific phospholipase C led to the release of the polar head group.
In addition to these biochemical studies, a bioinformatic analysis of available archaeal genome sequences predicts the presence of GPI-anchored proteins in other archaeal species (103). Moreover, many of the 19 enzymes known to participate in the biosynthesis of GPI anchors have been detected in archaeal genome sequences (104).
PROTEIN PHOSPHORYLATION
Like other forms of posttranslational modification considered in this review, the covalent attachment of phosphate groups to protein targets at any of a number of surface Asp, His, Ser, Thr, or Tyr residues can profoundly affect protein behavior. However, in contrast to N-glycosylation and, in most cases, lipid modification, covalent modification of proteins by phosphorylation is a reversible event. This property, combined with the major perturbation in protein structure that results from phosphorylation (189), has made this versatile form of posttranslational modification widely used when rapid and profound changes in protein behavior are called for (214, 215). As such, protein phosphorylation and dephosphorylation are most commonly exploited by the cell in adaptive pathways designed to present appropriate responses to various cues associated with a multitude of external and internal stimuli (173).
Although discovered in the 1950s (240), it took approximately 25 years for the first reports of phosphorylated proteins in Bacteria to appear (126, 459). Shortly thereafter, in 1980, the presence of phosphorylated proteins in Halobacterium salinarum was reported (413), confirming that Archaea too are capable of performing this posttranslational modification. With the subsequent availability of genome sequences, it became clear that Archaea also contain numerous kinases and phosphatases, enzymes responsible for protein phosphorylation and dephosphorylation, respectively (214, 215, 253).
Targets and Functions of Protein Phosphorylation in Archaea
The first examples of archaeal protein phosphorylation were reported when Halobacterium salinarum grown in the presence of 32P-labeled orthophosphate was shown to phosphorylate Ser and Thr residues of several protein species (413). The radiolabeling of 100- and 80-kDa proteins and, as shown later, an additional 62-kDa species (411) was, however, greatly diminished upon exposure to light. Moreover, the light-dependent dephosphorylation of these proteins could be linked to the proton motive force generated by the light-driven proton pump bacteriorhodopsin. In related studies (395), it was shown that growth in 32P-labeled orthophosphate-containing growth medium led to the appearance of serine- and threonine-phosphorylated proteins of 71, 52, 42, and 31.5 kDa in Sulfolobus acidocaldarius, in a growth-phase-dependent manner. Further examination revealed the existence of an additional 40-kDa Sulfolobus acidocaldarius phosphoprotein that was threonine-phosphorylated in the presence of [32P]polyphosphate (396). The first phosphoprotein with a known function to be identified in Archaea, however, was the methyltransferase activation protein from Methanosarcina barkeri, a key enzyme involved in the metabolic transformation of carbon dioxide to methane (81).
Although other phosphorylated proteins have been identified in Archaea (475), the observed phosphorylation cannot usually be attributed to a regulated protein kinase (see below), but rather reflects phosphorylated intermediates that appear during an enzyme's catalytic cycle. Such enzymes apparently include the alpha subunit of succinyl-coenzyme A synthase in Sulfolobus solfataricus (403) and Sulfolobus acidocaldarius glycogen synthase (52, 397). Nevertheless, examples of regulated protein phosphorylation in Archaea have been reported (Table 5) and are discussed below.
TABLE 5.
Archaeal proteins reported to be phosphorylated
| Protein | Species | Phosphorylated residue | Evidence for phosphorylation | Reference |
|---|---|---|---|---|
| CheA | Halobacterium salinarum | His | 32P incorporation | 374 |
| CheY | Asp | 32P incorporation | 374 | |
| Cdc6 | Methanobacterium thermoautotrophicum | Ser | 32P incorporation | 144 |
| Pyrobaculum aerophilum | Ser | 32P incorporation | 144 | |
| Sulfolobus solfataricus | Ser | 32P incorporation | 89 | |
| aIF2α | Pyrococcus horikoshii | Ser | 32P incorporation | 426 |
| Phenylalanyl-tRNA synthetase β-chain | Thermococcus kodakaraensis KOD1 | Tyr | Antiphosphotyrosine antibodies | 188 |
| Phosphomannomutase | Thermococcus kodakaraensis KOD1 | Tyr | Antiphosphotyrosine antibodies | 188 |
| RtcB | Thermococcus kodakaraensis KOD1 | Tyr | Antiphosphotyrosine antibodies | 188 |
| Zinc-dependent aminopeptidase | Sulfolobus solfataricus | 32P incorporation | 73 |
Phosphorylation of components involved in signal transduction.
Protein phosphorylation as part of an archaeal two-component signal transduction pathway was first shown for Halobacterium salinarum (373, 374). In Bacteria and a very limited number of Eucarya, two-component signal transduction response pathways are responsible for the appropriate response of the cell to a wide range of environmental conditions (234, 332, 423). The conformational changes that result upon ligand binding to the extracellular portion of a transmembrane receptor are transduced into the cell, where they lead to the modulation of sensor (histidine kinase, see below) and response regulator proteins. Such modulations ultimately activate the transcription of genes encoding compensatory proteins or affect the motion of the microorganism via motility structures. Transduction of the ligand binding event to sensor and response regulator proteins is achieved via a cascade of phosphorylation reactions. Hence, the detection of phosphorylated Halobacterium salinarum CheA and CheY, well-characterized sensor and response regulator proteins, respectively (114, 423), pointed to the presence of a two-component system in Archaea, charged with responding to various chemotactic and photactic stimuli (373, 374).
Protein phosphorylation in response to environmental change has also been observed in other archaeal species. Growth of Sulfolobus acidocaldarius in the presence of radiolabeled phosphate under limited-phosphate conditions revealed the existence of numerous phosphoproteins (319). In particular, the phosphorylation of a 36-kDa protein was augmented under phosphate starvation, hinting at a regulatory role in a cellular response pathway for this protein. In Haloferax volcanii, growth at elevated salt concentrations may lead to the appearance of several serine-phosphorylated proteins not detected during growth under optimal salt conditions (32). A threonine-phoshorylated 67-kDa membrane protein displaying serine kinase activity has been found in Sulfolobus solfataricus, although the pathway in which this protein participates remains to be defined (261, 264).
Phosphorylation of components involved in DNA replication, cell cycle regulation, and translation.
In addition to playing a role in signal transduction, protein phosphorylation has also been implicated in eucaryal DNA replication, cell cycle regulation, and protein translation (313, 314, 349). Similar roles for protein phosphoryation have also been observed in Archaea. In Methanobacterium thermoautotrophicum, Pyrobaculum aerophilum, and Sulfolobus solfataricus (89, 144), DNA-dependent serine autophosphorylation has been reported for the Cdc6 protein, an intiator protein that fulfills an essential role in DNA replication and is known to be phosphorylated in Eucarya (182, 212). The autophosphorylation of Cdc6 proteins reveals similarities between the archaeal and eucaryal replication processes, even though domain-specific differences in Cdc6 autophosphorylation have been noted (144). Protein phosphorylation also takes place during both eucaryal and archaeal protein translation. In vitro studies addressing the heterotrimeric archaeal initiation factor 2 complex (aIF2) from Pyrococcus horikoshii showed that the aIF2 α subunit could be phosphorylated (426), as is the case for the parallel eucaryal eIF2 α subunit (93, 251).
Phosphorylation of other proteins.
In other instances, archaeal phosphoproteins have been indentified in which the role of this posttranslational modification remains obscure. In Sulfolobus solfataricus, for example, a novel zinc-dependent aminopeptidase, originally isolated from cell lysates in complex with a chaperonin, was shown to be phosphorylated (73).
Finally, whereas the bulk of phosphorylated archaeal proteins experience modification of Asp, His, Ser, or Thr residues, it is now known that archaeal proteins can also undergo phosphorylation at Tyr residues. Using antiphosphotyrosine antibodies, tyrosine-phosphorylated proteins were first identified in cell extracts of Haloferax volcanii, Methanosarcina thermophila, and Sulfolobus solfataricus (401). In Thermococcus kodakaraensis KOD1, tyrosine-phosphorylated proteins recognized by antiphosphotyrosine antibodies were subsequently identified by N-terminal sequencing as RtcB, which is involved in RNA processing (128), the phenylalanyl-tRNA synthetase β-chain, and phosphomannomutase (188). Thus, long thought to be restricted to Eucarya (255) and later shown to occur also in Bacteria (77), proof for the existence of archaeal tyrosine phosphorylation shows this form of posttranslational modification to be ubiquitous across evolution (475).
Archaeal Protein Kinases and Phosphatases
In general, phosphorylated proteins do not contain readily recognizable sequence regions that allow their assignment as candidates for this posttranslational modification. In contrast, protein kinases and phosphatases, the enzymes responsible for the addition and removal, respectively, of orthophosphate groups from target proteins, contain conserved sequence motifs (213). Based on such motifs, protein kinases and phosphatases can be divided into several functional families (213). Thus, the availability of several archaeal genome sequences has allowed a catalogue of the potential protein kinases and phosphatases to be assembled (214, 215). A better understanding of the archaeal proteins should also provide insight into the relationship between eucaryal and bacterial kinases and phosphatases, which were once thought to be distinct (234, 253). For a more detailed examination of archaeal kinases and phosphatases, the reader is directed to a recent review of the subject (215).
Eucaryal protein kinases.
Members of the eucaryal protein kinase superfamily, an evolutionarily conserved group of proteins sharing a common core, serve as the major providers of protein serine/threonine/tyrosine kinase activity in Eucarya (154). Long considered to be restricted to Eucarya, homologues of eucaryal protein kinases were subsequently reported in Bacteria and more recently detected in Archaea (214). Initially, searches of the then-available archaeal gene sequences identified ORFs in Methanococcus thermolithotrophicus, Methanococcus vannielii, and Methanococcus voltae encoding proteins whose carboxy-terminal regions contain 9 of 11 subdomains associated with eucaryal protein kinases (400). In a later study (215), analysis of nine completed archaeal genomes revealed the presence of ORFs encoding polypeptides containing sequence motifs essential for eucaryal protein kinase activity in seven.
Gene-based studies of individual strains have also revealed the existence of eucaryal protein kinases in other Archaea, such as in Haloferax volcanii cells exposed to elevated salt levels, in which a salt-regulated gene putatively encoding a protein serine/threonine kinase was detected (32). Subsequent studies employing complete archaeal genome sequences, moreover, have expanded our knowledge of eucaryal protein kinases. In a comprehensive search based on a large number of completed genome sequences, including those of four Archaea, archaeal representatives of four novel putative protein kinase families were reported (253), such as the Rio1 family, comprising only archaeal and eucaryal members, or the ABC1 family, including only a single archaeal representative (from Methanobacterium thermoautotrophicum). Furthermore, the recent solution of the crystal structure of Archaeoglobus fulgidus Rio2 suggests that this protein defines a new family of protein kinases (245).
In addition to sequence-based analyses, archaeal homologues of eucaryal protein kinases have been examined at the protein level. Analysis of threonine-modified phosphoproteins in Sulfolobus solfataricus membranes following incubation with [γ-32P]ATP led to the identification of the protein encoded by ORF sso0469 (264). Sequence analysis revealed the presence of eukaryotic protein kinase motifs, while biochemical characterization of a recombinant version of the encoded protein revealed its ability to phosphorylate Ser residues of exogenous polypeptides in vitro. Similarly, SsoPK2, the product of Sulfolobus solfataricus ORF sso2387, also contains sequence motifs found in eucaryal protein kinases (263). Moreover, a recombinant form of the protein was able to phosphorylate itself as well as various exogenous targets, relying on that part of the protein homologous to eucaryal protein kinases, as revealed by mutagenesis approaches (263).
Histidine kinases.
Histidine kinases are elements of the two-component signal transduction pathway described above. In response to conformational changes experienced by upstream receptor-transducer teams, histidine kinase sensors use ATP to autophosphorylate His residues before transferring the phosphoryl group to Asp residues of downstream response regulators. The first example of an archaeal histidine kinase as part of a two-component system identified was Halobacterium salinarum CheA (373). A recombinant version of the haloarchaeal CheA histidine kinase was autophosphorylated upon addition of radiolabeled ATP and was subsequently able to transfer its phosphoryl group to an Asp residue of the Halobacterium salinarum CheY response regulator (374).
In later homology-based searches of nine completed archaeal genome sequences, histidine kinases were identified in four: Archaeoglobus fulgidus, Halobacterium sp. strain NRC-1, Methanobacterium thermoautotrophicum, and Pyrococcus horikoshii (215, 220, 234). Of these, Methanobacterium thermoautotrophicum and Archaeoglobus fulgidus contain the most histidine kinases (16 and 14, respectively) and response regulators (10 and 11, respectively). At the other extreme, Pyrococcus horikoshii contains only a single histidine kinase and two response regulators (corresponding to CheA and to CheY and CheB, respectively), while Aeropyrum pernix, Methanococcus jannaschii, and Thermoplasma acidophilum are not predicted to encode such proteins. The absence of Che proteins in Methanococcus jannaschii is noteworthy, given that this species is both flagellated and motile (436).
Protein serine/threonine phosphatases.
Protein serine/threonine phosphatases can be structurally and functionally grouped into the protein serine/threonine phosphatase (PPP) and the Mg2+ and Mn2+ protein phosphatase (PPM) families (21). PPP family members are mainly responsible for serine/threonine dephosphorylation in Eucarya and have also been reported in Bacteria (71, 213, 214). In contrast, members of the PPM family are the primary mediators of dephosphorylation in Bacteria, although this family encompasses several eucaryal protein phosphatase classes as well (37, 214). In Archaea, members of both protein serine/threonine phosphatase families have been identified in completed genome sequences and some have been studied at the protein level (213-215).
To date, three PPP family protein serine/threonine phosphatases have been characterized from Archaea. The genes encoding PP1-arch1, PP1-arch2, and Py-PP1 were cloned from Sulfolobus solfataricus (216, 252), Methanosarcina thermophila TM-1 (321, 403), and Pyrodictium abyssi TAG11 (268), respectively. In addition, other archaeal PPP family phosphorylases have been predicted following analysis of genome sequences, relying on the presence of conserved sequence motifs (24, 215). Such sequence comparisons revealed the archaeal enzymes to be more closely related to their eucaryal than their bacterial homologues (24). However, despite their sequence similarities to eucaryal PPP family members, archaeal PPP family protein serine/threonine phosphatases display a combination of eucaryal and bacterial features (215). Like their eucaryal counterparts, the archaeal enzymes specifically act upon protein-bound phosphoserine and phosphothreonine residues and, in the cases of PP1-arch2 and Py-PP1, are inhibited by toxic secondary metabolites such as okadaic acid (268, 321, 403). In contrast, the three archaeal PPP family members require the addition of metal ions such as Mn2+ for activity, as is the case for bacterial PPP family protein serine/threonine phosphatases (391). Finally, protein serine phosphatase activity has also been detected in extracts of Halobacterium salinarum (36) and Haloferax volcanii (320), but the enzymes responsible have not been identified.
A single ORF encoding a potential PPM family protein serine/threonine phosphatase was identified in the genome sequence of Thermoplasma volcanium. The putative protein includes all of the conserved sequence elements of PPM family members (209).
Protein tyrosine phosphatases.
While ORFs thought to encode protein tyrosine phosphatases have been detected in Archaeoglobus fulgidus, Methanococcus jannaschii, Methanobacterium thermoautotrophicum, Pyrococcus abyssi, Pyrococcus furiosus, Pyrococcus horikoshii, Sulfolobus solfataricus, and Thermococcus kodakaraensis KOD1 (215, 418), only the Thermococcus kodakaraensis KOD1 enzyme has been examined biochemically (188). A recombinant version acted on both free phosphotyrosine and phosphoserine, suggesting that it had dual specificity. Moreover, a mutant form of the enzyme was used to capture putative native substrates from a cell extract (188). In addition, studies performed with Halobacterium salinarum extracts detected protein serine/threonine phosphatase activity also able to hydrolyze phosphotyrosine, suggesting the responsible enzyme similarly had dual specificity (36).
Protein kinases and phosphatases of Thermoplasma acidophilum.
It should be noted that analysis of the genome of Thermoplasma acidophilum, using tools available today, has failed to detect the presence of any protein kinase or phosphatase (214). While it remains to be seen whether the current inability to recognize such proteins will be remedied in future with the development of more powerful bioinformatic prediction tools, it is also possible that Thermoplasma acidophilum contains novel archaea-specific kinases or phosphatases, or does not perform protein phosphorylation. Interestingly, genome analysis of two bacterial strains, a Buchnera sp. and Ricksettia prowazekii, also failed to detect ORFs encoding putative protein kinases or phosphatases (214), although the implications of these studies are at present unknown.
PROTEIN METHYLATION
Although methylation of nucleic acids is well known, in part due to a role in disease states such as cancer (86, 256, 424), a wide variety of proteins have also been reported to experience posttranslational methylation. This modification affects the amino group in the side chains of Ala, Arg, Glu, His, Lys, and Pro residues, the hydroxyl group in the side chains of Glu and Asp, and the thiol group of Cys residues (327). Enzyme-catalyzed addition of methyl groups from _S_-adenosylmethionine can either occur reversibly, as in _O_-methylation of carboxyl groups, or irreversibly, as in the _N_-methylation of amino-terminal or side chain nitrogen atoms (70).
As is the case with other posttranslational modification events considered in this review, analysis of protein methylation in Archaea has revealed novel forms of protein methylation as well as providing new insights into the biological role served by this posttranslational modification.
Protein Methylation in Response to External Stimuli
As described above, various external stimuli that modulate the motility of archaeal cells rely on phosphorylation of elements of the two-component signal transduction response pathway. Phosphorylation is not, however, the sole posttranslational modification experienced by proteins involved in taxis responses to environmental cues. As in Bacteria (90, 229, 236, 423), numerous proteins involved in the archaeal response to growth conditions also undergo methylation (see below). Methylation of taxis receptor or transducer proteins is thought to be responsible for adaptation, a form of cellular memory necessary for cells to be able to sense and move towards ever higher attractant concentrations or to recognize when motion is ocurring in the wrong direction, i.e., away from elevated attractant concentrations (423).
Three methylation-dependent taxis responses, phototaxis, chemotaxis, and aerotaxis, have been detected in Halobacterium salinarum, in which the archaeal response to environmental cues, as mediated through transducer proteins, has been well studied. In Halobacterium salinarum, the phototactic response is initiated by the excitation of the two retinal-containing photoreceptors, sensory rhodopsin I and sensory rhodopsin II (121, 239, 341, 412, 482, 488). These subsequently relay the excitatory signal to their respective transducer proteins, HtrI and HtrII. During phototaxis, these proteins undergo methylation, a posttranslational modification previously shown to modulate the life span of phototactic signals in Halobacterium salinarum, i.e., to play an adaptative role (164). Methylation of HtrII is also involved in the transducer role assumed by the protein during serine chemotaxis (171). The cytoplasmic transducer HtrXI undergoes methylation/demethylation in response to changes in extracellular histidine, aspartate and glutamate concentrations (43).
Arginine taxis in Halobacterium salinarum involves the methylatable soluble transducer Car, which monitors intracellular levels of the amino acid (417), while the methylation status of the membrane-bound transducer BasT affects chemotactic behavior towards leucine, isoleucine, valine, methionine, and cysteine (227). HtpIV, or CosT, the transducer for the haloarchaeal chemotaxis response towards trimethylammonium compounds, also experiences methylation (228). The aerotactic (oxygen gradient-sensing) response of Halobacterium salinarum was also shown to rely on methylation, in this case of the membrane-bound transducer HtrVIII (259). In contrast, aerotaxis in Bacteria such as Escherichia coli and Salmonella enterica serovar Typhimurium does not require transducer methylation (259). Most recently, MpcT, the transducer of membrane potential changes in Halobacterium salinarum (formerly known as HtrXIV) was shown to experience differential degrees of methylation (225).
Methylation of Methyl-Coenzyme M Reductase
In methanoarchaea, the final reaction in the release of methane is catalyzed by the enzyme methyl-coenzyme M reductase (434). Analysis of the crystal structure of the enzyme from Methanobacterium thermoautotrophicum revealed the presence of five modified amino acid residues in the α subunit of the hexameric enzyme, all situated near the active-site region (108). In addition to a thioglycine residue, the enzyme contains 1-_N_-methylhistidine, 5-(S)-methylarginine, 2-(S)-methylglutamine, and an _S_-methylcysteine residue (Fig. 5). Whereas 1-_N_-methylhistidine and _S_-methylcysteine have been detected in other proteins (70, 326) and a thiol-modified glycine residue has been identified in ThiS, one of the enzymes involved in thiamine biosynthesis in Escherichia coli (432), Methanobacterium thermoautotrophicum methyl-coenzyme M reductase is the first example of a 2-(S)-methylglutamine and 5-(S)-methylarginine. Previously, only _N_-methylglutamine and _N_-methylarginine had been reported (162, 492).
FIG. 5.
Methylated amino acids in Methanobacterium thermoautotrophicum methyl-coenzyme M reductase. A. 2-(S)-Methylglutamine. B. _S_-Methylcysteine. C. 5-(S)-Methylarginine. D. 1-_N_-Methylhistidine. In each case, the modifying methyl group is boxed.
The posttranslational modifications leading to the appearance of the four methylated amino acids in methyl-coenzyme M reductase involve the transfer of the methyl group of methionine, most likely in the _S_-adenosylmethionine form (387). The modifications are thought to occur before methyl-coenzyme M reductase assumes its quaternary structure, since the modified residues are buried deep inside the native enzyme, where they would be inaccessible to _S_-adenosylmethionine or methyltransferases, which catalyze protein methylation (108, 143). Furthermore, considering the differences in amino acid composition in the vicinities of the four methylated residues (387), it is probable that four different _S_-adenosylmethionine-dependent methyltransferases are involved in the modification reactions (308). Accordingly, multiple methyltransferases appear to be present in the genome sequence of Methanobacterium thermoautotrophicum (399).
In terms of function, methylation of His-257, which is involved in substrate binding, likely affects the substrate affinity of the enzyme (108). The thioglycine residue has been proposed to serve as a one-electron relay in the catalytic mechanism (434). The functional significance of the methylation of the other three modified residues, i.e., 5-(S)-methylarginine-271, 2-(S)-methylglutamine-400, and _S_-methylcysteine-452, remains unknown. However, analysis of methyl-coenzyme M reductase sequences in a wide range of methanarchaeal species reveals the absolute conservation of the five amino acid residues modified in the Methanobacterium thermoautotrophicum enzyme (311, 410). Moreover, the crystal structure of methyl-coenzyme M reductase from Methanosarcina barkeri also revealed the presence of thioglycine, _S_-methylcysteine, 1-_N_-methylhistidine, and 5-methylarginine residues, i.e., four of the five posttranslational modifications found in the Methanobacterium thermoautotrophicum enzyme, suggesting that such modifications are important for catalysis (143).
Methylated Proteins in Thermophilic Archaea
Methylated Lys residues have been detected in several thermophilic archaeal proteins, such as Sulfolobus acidocaldarius ferredoxin (289) and Sulfolobus solfataricus glutamate dehydrogenase (272), aspartate aminotransferase (485), and β-glycosidase (117). In the case of Sulfolobus solfataricus β-glycosidase, _N_-ɛ-methylation of specific Lys residues was associated with increased thermal stability as well as with a lower susceptibility to denaturation and aggregation, in comparison to the nonmethylated recombinant version of the enzyme produced in Escherichia coli (117). The methylated Lys residues found in the Sulfolobus solfataricus enzyme are not conserved in other mesophilic glycosidases belonging to glycosyl hydrolase family I, again pointing to a thermostabilizing role for this posttranslational modification.
Methylation of Archaeal DNA-Binding Proteins
Although grouped with Bacteria as prokaryotes (472), Archaea resemble Eucarya in many aspects, including that members of both domains contain histones, proteins involved in DNA packaging (355, 466). First demonstrated in Methanothermus fervidus (378, 415), over 30 archaeal histone sequences have since been identified (355). Archaeal histones are, however, apparently restricted to Euryarchaea, an archaeal subdomain, in which several different histone-encoding genes have been detected (355, 466). No archaeal histones have been observed in Crenarchaea, the other major archaeal subdomain. Instead, crenarchaeal species contain small, basic DNA-binding proteins thought to fulfill the same functions as histones, based on their physical properties (63, 64, 149, 355, 367, 466). In Sulfolobus, these can be grouped into 7-, 8-, and 10-kDa classes, with the 7-kDa proteins, referred to as the Sul7 family (466), predominating. Members of the Sul7 family in both Sulfolobus acidocaldarius and Sulfolobus solfataricus are modified by monomethylation of selected Lys residues to different extents in a strain-dependent manner (26, 63, 64, 97, 278, 317).
Given the modulation of eucaryal histone function that results from methylation (59, 238), it is likely that methylation of archaeal Sul7 proteins also affects their behavior. Indeed, the observation that methylation of Sul7 proteins increased during heat shock suggests that such posttranslational modification is of functional, although as yet undefined, significance (26). Sul7 methylation does not, however, affect DNA binding affinity, consistent with the positioning of methylated Lys residues on the surface of the Sul7d-DNA crystal rather than at the protein-DNA interface (25). Finally, it is somewhat ironic that while archaeal Sul7 proteins are methylated, no evidence for methylation of archaeal histones has appeared, in contrast to their eucaryal counterparts (355). This is due to the fact that archaeal histones lack the amino- and carboxy-terminal extensions that undergo this posttranslational modification in eucaryal histones (59). Indeed, analysis of archaeal genome sequences reveals homologues of only one of the components involved in the eucaryal histone modification event, i.e., the histone acetyltransferase Elp3 (355).
Methylation of Archaeal Ribosomal Proteins
Several bacterial ribosomal proteins, mainly found in the large 50S subunit, undergo methylation (55, 56). Of these, L11 is the major methylated ribosomal component. Analysis of Halobacterium cutirubrum and Sulfolobus solfataricus L11 proteins from cells grown in the presence of radiolabeled methionine and/or methylmethionine revealed that they are also methylated, albeit in a pattern distinct from that of the bacterial protein (9, 353, 354). Accordingly, genome searches have failed to identify an archaeal homologue of the bacterial L11 methyltransferase PrmA (45). The role of L11 methylation in both Bacteria and Archaea remains unknown.
DISULFIDE BONDS IN PROTEINS
In both Eucarya and Bacteria, secreted and extracellularly oriented membrane proteins are often stabilized by disulfide bonds, i.e., covalent links between the sulfhydryl groups of Cys residues in the same or different polypeptide chains. These can serve two roles. First, they can stabilize proteins by entropic destabilization of the unfolded conformation (78, 322, 463, 465). Second, they serve to limit damage to a protein resulting from oxidative or proteolytic agents, thereby enhancing protein lifetime. Accordingly, disulfide bonds are routinely employed by secretory and plasma membrane proteins in numerous organisms (315, 451).
The various compartments of the cell greatly differ in terms of redox potential and hence in their ability to catalyze disulfide bond formation. Accordingly, disulfide bond formation takes place in the endoplasmic reticulum of eucaryal cells (444) and in the periplasmic/extracellular compartment of bacterial cells (193, 351). In both locations, oxidative conditions favor disulfide bond formation and enzymes implicated in this posttranslational modification are found. Conversely, it had been generally accepted that proteins found in the reducing environment of the cytosol do not contain disulfide bonds, although it has recently become clear that a number of cytosolic proteins can contain specific and reversible disulfide bonds (see below). In such cases, the cyclic oxidation/reduction of a disulfide bond can control the activation/deactivation or otherwise modulate the activity of a protein (62, 80, 181, 325, 363). Indeed, controlled reduction of disulfide bonds has also been adopted by certain disulfide-containing secreted proteins and cell surface receptors (166). Nevertheless, the number of cytoplasmic proteins in Eucarya and Bacteria experimentally shown to contain disulfide bonds is limited. Archaea, however, do not follow this trend (269).
Disulfide Bonds in Cytoplasmic Archaeal Proteins
Unexpectedly, biochemical and structural characterization of many cytoplasmic archaeal proteins has revealed the presence of disulfide bonds. A disulfide bond was detected in Pyrococcus furiosus ferrodoxin, in which it plays a role in the redox cycle of the protein (141). The crystal structures of DNA polymerases from Thermococcus gorgonarius and Thermococcus sp. strain 9°N-7 revealed the presence of two disulfide bridges in each case (169, 368). The recombinant form of Aeropyrum pernix alcohol dehydrogenase was shown to contain a disulfide bond (152) as was Sulfolobus solfataricus glyceraldehyde-3-phosphate dehydrogenase (180). The three-dimensional structure of the TATA box-binding protein from the hyperthermophile Pyrococcus woesei revealed the presence of a disulfide bond not found in mesophilic versions of the protein (88). Indeed, in many of these examples, the presence of disulfide bonds is believed to contribute to the enhanced thermostability of the modified protein.
Disulfide bonds are also used by cytosolic archaeal proteins for the generation of higher-order structures. As revealed by X-ray crystallography and site-directed mutagenesis, a single intersubunit disulfide bridge is responsible for the dimeric nature of Sulfolobus solfataricus glycosyltrehalose trehalohydrolase (118) and pyrrolidone carboxyl peptidase from Thermococcus litoralis (394) and Pyrococcus furiosus (316). Similarly, ferric reductase from Archaeoglobus fulgidus was shown to be a homodimer, with a single disulfide bond serving to link the two subunits of the protein (60). In Pyrococcus horikoshii, oligomerization of isopropylmalate isomerase relies on intersubunit disulfide bridges (483). The homotetrameric structure of Pyrococcus abyssi tRNA (m1A) methyltransferase is also due to disulfide bonding (370). The nuclear magnetic resonance structure of Pyrobaculum aerophilum DsrC, the archaeal homologue of the γ subunit of dissimilatory sulfite reductase, responsible for the reduction of sulfite in sulfate-reducing bacteria, was also shown to contain two disulfide bonds (76). Disulfide bond formation is also responsible for the hexameric states of l-isoaspartyl-_O_-methyltransferase from Sulfolobus tokodaii (431) and of 5′-deoxy-5′-methylthioadenosine phosphorylase from Sulfolobus solfataricus (11, 47).
Despite the seemingly widespread presence of disulfide bonds in cytoplasmic archaeal proteins, it was only with the detection of three disulfide bonds in the crystal structure of Pyrobaculum aerophilum adenylosuccinate lyase (441) that the concept of the general use of disulfide bonds in cytoplasmic proteins in this and possibly other hyperthermophilic Archaea was proposed (269). Accordingly, computational analysis of completed archaeal genomic sequences, involving sequence-structure mapping approaches with subsequent analysis of the proximity of pairs of Cys residues, indicated that disulfide bonds are indeed prevalent in thermophilic and hyperthermophilic crenarchaeal cytoplasmic proteins, yet are not found in mesophilic versions of the same proteins (269). In this study, it was predicted that 44 and 40% of intracellular protein Cys residues in Pyrobaculum aerophilum and Aeropyrum pernix (both _T_opt ∼100°C), respectively, and approximately 30% of the Cys residues in Pyrococcus abyssi and Pyrococcocus horikoshii (both _T_opt ∼100°C) cytoplasmic polypeptides are found in disulfide bonds. In Archaeoglobus fulgidus (_T_opt ∼90°C), Methanobacterium thermoautotrophicum (_T_opt ∼80°C), and Methanococcus jannaschii (_T_opt ∼60°C), only 11 to 15% of the intracellular protein Cys content is predicted to participate in disulfide bonds. It thus appears that there exists a correlation between optimal growth temperature and the number of intracellular disulfide bond-containing proteins. Hence, disulfide bridge formation may well be one of many mechanisms known to enhance protein stability in Archaea. Interestingly, the same study (269) points to the presence of cytoplasmic disulfide bond-incorporating proteins in thermophilic Bacteria such as Aquifex aeolicus and Thermotoga maritima.
Disulfide Bonds in Extracellular Archaeal Proteins
The presence of disulfide bonds in archaeal secreted or membrane proteins has been reported in only a limited number of cases. Tetrabrachion, the major structural component of the Staphylothermus marinus S-layer, was reported to contain disulfide bonds based on the destabilizing effect of dithiothreitol treatment in the face of thermal and proteolytic challenges (345). Disulfide bonds have also been postulated to be present in halolysin R4, a serine protease secreted by Haloferax mediterranei, since mutagenesis of either of two Cys residues in a carboxy-terminal extension of the protein or complete removal of this domain drastically reduced both the amount and activity of the heterologously expressed protein (198). One explanation offered for these observations was that a putative disulfide bond, linking the two Cys residues in question, would assume a stablilizing role in the native protein. Possible disulfide bond formation involving Cys residues in the S-layer glycoprotein of Methanococcus jannaschii has been also been offered as an explanation for the thermostability of this protein, relative to other methanococcal S-layer glycoproteins which do not contain Cys residues (1). Such predictions, however, await experimental verification.
Enzymes Involved in Disulfide Bond Formation in Archaea
The reduced nature of the cytoplasm of eucaryal and bacterial cells (174, 363) and the seeming abundance of disulfide-bonded intracellular proteins in thermophilic and hyperthermophilic Archaea (269) raise questions concerning the redox state of the archaeal cytoplasm and the nature of the proteins that are involved in disulfide bond formation in these organisms.
In eucaryal and bacterial cells, the formation and redox states of disulfide bonds are mediated by protein disulfide oxidoreductases (168). Members of this ubiquitous protein family, which includes thioredoxins, glutaredoxins, disulfide bond formation (Dsb) proteins, and protein disulfide isomerases (PDI), have active sites containing the Cys-X-X-Cys sequence motif and the thioredoxin fold structural motif (273). DsbA is found in the bacterial periplasmic space and is involved in protein disulfide bond formation (193, 351), while PDI catalyzes protein disulfide bond formation, reduction, and rearrangement in the eucaryal endoplasmic reticulum (444, 471). Acting as strong reductants in various cellular processes (120, 348), both the thioredoxin system, involving two thioredoxins and thioredoxin reductase, and the glutaredoxin system, including three glutaredoxins and glutathione reductase, maintain intracellular disulfide bonds in the reduced state through NADPH-dependent pathways (168, 363).
To date, few archaeal protein disulfide oxidoreductases have been described (see below) and, considering the limited information available, it is too early to assign any of them a physiological role. What is known, however, points to the unique character of the archaeal proteins. For instance, Methanobacterium thermoautotrophicum contains a small protein (Mt0807) with a thioredoxin/glutaredoxin-like fold that exhibits sequence similarity to glutaredoxins, including the characteristic Cys-Pro-Tyr-Cys active-site motif (279). While its function was initially tentative, subsequent structural analysis and sensitive enzyme assays (10) revealed it to be a true thioredoxin. Nuclear magnetic resonance-based structural studies of another Methanobacterium thermoautotrophicum protein (Mt0895) revealed that it too contains a thioredoxin/glutaredoxin-like fold. This protein was originally annotated as a conserved hypothetical protein (31). The apparent absence of glutathione in Archaea (279, 301) together with the use of more precise structural analysis and activity assays led to the conclusion that Mt0895 is a thioredoxin. Structural and biochemical studies have shown that the same is true for Methanococcus jannaschii Mj0307 (54, 250). It has been suggested that proteins possessing a thioredoxin/glutaredoxin-like fold and a glutaredoxin-like active-site amino acid sequence but thioredoxin activity, such as Mt0895, Mt0807, and Mj0307, could belong to an ancient family predating the appearance of the present-day glutaredoxin and thioredoxin families that still exist in Archaea (10, 31).
As described above, the presence of disulfide bonds in noncytosplasmic archaeal proteins remains to be conclusively proven. If this posttranslational modification is indeed employed by such proteins, one can ask whether the introduction of disulfide bonds involves archaeal homologues of PDI or the Dsb proteins, which are used by Eucarya and Bacteria, respectively, for this purpose (193, 444). The available information points to the presence of PDI-like proteins in Archaea. The structure of a protein disulfide oxidoreductase from Pyrococcus furiosus, originally predicted by sequence analysis to be a glutaredoxin-like protein (151), revealed the presence of two domains, each organized into the characteristic thioredoxin/glutaredoxin fold and both containing the Cys-X-X-Cys active-site motif (356). This is reminiscent of eucaryal PDI, which also contains two thioredoxin/glutaredoxin folds (85). By contrast, thioredoxin, glutaredoxin, and DsbA contain a single thioredoxin/glutaredoxin fold each (273).
Subsequent biochemical characterization of the Pyrococcus furiosus protein revealed that it, like eucaryal PDI, also displays oxidative, reductive, and disulfide isomerase activities (339). In addition, a homologous protein had been purified earlier from Sulfolobus solfataricus (150) and was predicted to exist in other species, based upon examination of the genome sequences of hyperthermophilic Archaea (339). However, the homologous protein from Pyrococcus horikoshii together with a second protein identified as a thioredoxin reductase were shown to function as a thioredoxin system, mediating electron transfer from a thioredoxin reductase-like flavoprotein to a protein disulfide bond, suggesting a role for this protein other than as a disulfide bond-introducing PDI (205).
PROTEOLYTICALLY PROCESSED PROTEINS
Posttranslational protein modification also includes proteolytic cleavage of precursor forms of proteins. In Archaea, examples of proteolytic processing at the amino and carboxy termini, in addition to positions within a polypeptide chain, have been reported.
Archaeal Signal Sequences
In any cell, a subset of proteins must cross one or more membranes to realize their ultimate localization and fulfill their designated roles. Across evolution, such proteins are generally synthesized with a cleavable amino-terminal extension referred to as the signal sequence that is enzymatically removed once such proteins have traversed the membrane. Analysis of signal sequence composition in Archaea as well as their posttranslational removal reveals a mosaic of archaeal, eucaryal, and bacterial traits.
Protein translocation in Archaea.
Translocation of extracytoplasmic proteins begins with their delivery to translocation sites in the membrane (42, 119, 298). Examples of both post- and cotranslational translocation have been found in Archaea. Chimeric signal sequence-bearing reporter proteins are secreted posttranslationally from transformed Haloferax volcanii cells (179). In addition, Haloferax volcanii has been reported to posttranslationally insert a chimeric protein containing the multispanning membrane protein bacterio-opsin (318). In contrast, cotranslational translocation, shown to be the general mode of membrane protein insertion in Haloferax volcanii (360), likely involves the archaeal signal recognition particle pathway (293, 494), as first reported for Halobacterium salinarum bacterio-opsin (83, 84, 148).
In Archaea, as across evolution, the Sec translocon is the major site for protein export (94). The SecY, SecE, and Sec61β proteins that form the core of the translocation apparatus are closer to their eucaryal than their bacterial homologues (49, 155, 223, 347, 357, 361). The recent solution of the three-dimensional structure of the Methanococcus jannaschii SecYEβ translocon has provided major insight into the translocation event across evolution, including the mode of translocon gating and mechanism of membrane protein insertion (446). The Sec translocon may also be involved in the translocation of archaeal flagellins, despite their distinct signal sequence composition (see below) (184, 436). In contrast to their bacterial counterparts, which cross the plasma membrane through the hollow core at the center of the growing flagellum (7, 265), archaeal flagellins are likely translocated across the membrane and only then added to the base of the growing motility structure, as gauged by the presence of unique cleavable N-terminal signal sequences in the archaeal proteins (20, 184, 436).
Archaea also use the twin-arginine targeting (Tat) pathway, a second protein export pathway (446). The Tat pathway can be distinguished from the Sec pathway by the unique composition of substrate signal sequences (see below) and by the ability of the Tat pathway to translocate folded or cofactor-incorporating proteins (29, 366). Although the Tat pathway is proposed to predominate in halophilic Archaea (35, 371), little is presently known of the workings of the Tat system in these or other organisms.
Genomic surveys of archaeal signal sequences.
Descriptions of archaeal signal sequences have largely relied on analysis of genome sequences, using computer-based tools originally designed to detect eucaryal or bacterial signal sequences (2, 16, 35, 94, 306, 371, 377). At best, these algorithms should be able to identify only those archaeal signal sequences bearing sufficient similarity to their eucaryal and bacterial counterparts. Archaeal signal sequences possessing domain-specific traits would, therefore, likely be overlooked in such searches. Thus, true characterization of archaeal signal sequences will have to wait for the number of experimentally verified targets to be extended well beyond the few experimentally verified sequences presently available. Nonetheless, such efforts have identified signal sequences recognized by the Sec and Tat pathways, archaeal flagellin-like signal sequences on both flagellin and nonflagellin proteins, as well as lipoprotein signal sequences (Fig. 6).
FIG. 6.
Schematic depiction of archaeal signal sequences. In each case, consensus sequence elements characteristic of that class of signal sequence are shown, where + corresponds to positively charged residues, x corresponds to any residue, and φ corresponds to a hydrophobic residue. Hydrophobic stretches of amino acid residues are portrayed in gray. The site of cleavage is denoted by the black wedge.
While the signal sequences of Sec pathway substrates can differ widely, they share common structural traits, such as a positively charged amino-terminal region leading to a hydrophobic core region that continues into an uncharged polar region terminating in the signal peptidase cleavage site (454). From examination of 10 genome sequences, it was concluded that predicted archaeal Sec signal sequences are more similar to their bacterial than their eucaryal counterparts (16, 307). The findings of this multigenome study (16) are in agreement with earlier studies addressing predicted signal sequences in Methanococcus jannaschii (306) and Solfolobus solfataricus (2), although differences exist. Nonetheless, as discussed below, apparent similarities in the mechanism of archaeal and eucaryal signal peptidase action (18, 100, 437) suggest similarities between the cleavage site regions of signal sequences in these two domains.
While sharing the same tripartite organization, Tat pathway signal sequences differ from those recognized by the Sec pathway in that the former include an extended amino-terminal region containing a highly conserved motif based on two Arg residues and a less hydrophobic core region (29, 366). Analysis of genome sequences predicts limited-use presence of Tat signal sequences in Archaea (2, 16, 94), with the apparent exception of halophilic Archaea (35, 371). Here, proteins bearing Tat signal sequences are predicted to greatly outnumber those synthesized with Sec signal sequences. The enhanced utilization of the Tat pathway by halophilic Archaea is thought to be a response to the highly saline cytoplasm in these strains, reportedly as high as 5 M (67, 132). It has been postulated that to overcome dangers to protein folding associated with maintaining a “loosely folded” conformation in a high-salt environment, as would be required for posttranslational translocation by the Sec pathway, reliance on the Tat pathway, capable of translocating folded protein substrates, is preferable.
In addition to Sec and Tat pathway signal sequences, archaeal proteins may be synthesized as precursors bearing other cleavable signal sequences. As first noted in Methanococcus voltae (113), archaeal flagellins are made as precursors bearing atypical short, positively charged signal sequences, reminiscent of signal sequences found in bacterial type IV prepilins (20, 184, 436). Unexpectedly, genome analysis predicted the presence of the same signal sequence in a set of 10 extracellular Sulfolobus solfataricus proteins, including six putative solute-binding proteins (2). Archaeal flagellin signal sequences have also been predicted to exist in other types of protein, including those assigned solute-binding roles, in Methanococcus jannaschii, Pyrococcus horikoshii, Sulfolobus shibatae, and Thermococcus litoralis (5). In contrast to Sec and Tat pathway signal sequences, cleavage of archaeal flagellin signal sequences by the appropriate signal peptidase (see below) occurs upstream, rather than downstream, of the hydrophobic core region.
As discussed above, sequence analysis studies have also predicted the existence of proteins synthesized with lipoprotein signal sequences in Archaea (4, 170, 228, 274), although experimental support for these predictions has yet to be presented.
Removal of archaeal signal sequences.
The signal sequences of proteins translocated by either the Sec or Tat pathway are removed by the actions of type I signal peptidases (82, 324). In Archaea, type I signal peptidases incorporate traits of both their eucaryal and bacterial counterparts. As in Eucarya, the archaeal signal peptidase does not rely on the catalytic Ser-Lys dyad employed by the bacterial enzyme and has replaced the conserved bacterial Lys with a His residue (100, 324, 437). At present, the catalytic mechanisms of both archaeal and eucaryal signal peptidases remain largely undefined (18, 447). On the other hand, in contrast to the eucaryal enzyme, which functions as part of a larger signal peptidase complex (477), both bacterial and archaeal signal peptidases apparently function independently (100, 324). Furthermore, certain archaeal signal peptidases incorporate a sequence domain of unknown function, referred to as domain II (323). This domain is found in the bacterial but not the eucaryal enzyme (100).
The limited experimental analysis of archaeal signal peptidase activity available comes from studies in which the gene encoding the enzyme from Methanococcus voltae was heterologously expressed in Escherichia coli (302). Isolated bacterial membranes then served as the source of the archaeal enzyme in an in vitro signal peptidase assay, using a truncated, poly-His-tagged version of the Methanococcus voltae S-layer protein as the substrate. Site-directed mutagenesis of the Methanococcus voltae enzyme identified three conserved residues, Ser-52, His-122, and Asp-148, essential for activity (18). The finding that a second conserved Asp residue (Asp-142) was not crucial for catalytic function suggests differences in the mechanisms of the archaeal and eucaryal signal peptidases, since Asp residues found at both corresponding positions are essential for activity of the Saccharomyces cerevisiae enzyme (324).
Type II signal peptidases are involved in the removal of signal sequences from lipoproteins (156). However, as noted above, no archaeal type II signal peptidase has been described, despite the apparent existence of archaeal lipoproteins (4, 170, 228, 274).
The unique signal sequences of archaeal flagellins are removed by the actions of a signal peptidase reminiscent of bacterial type IV prepilin peptidases (17, 75), exemplified by Pseudomonas aeruginosa PilD (419). Those translocated nonflagellar Sulfolobus solfataricus proteins bearing the archaeal flagellin signal sequence also rely on an archaeal version of the bacterial type IV prepilin peptidase, termed PibD (peptidase involved in biogenesis of prepilin-like proteins), for their processing (2, 6). Site-directed mutagenesis studies have begun to provide insight into the catalytic mechanism of the archaeal enzyme (6, 17).
Amino-Terminal Methionine Removal
In many instances, the initiator Met residue of a nascent polypeptide chain (or _N_-formyl-Met residue in Bacteria) is cleaved by the actions of methionine aminopeptidases (39, 137). While the reason for such processing remains unclear, several explanations, including facilitation of additional amino-terminal processing (13) and modulation of protein lifetime (39, 450), have been suggested. Indeed, methionine aminopeptidases are essential for the survival of Bacteria and yeasts (57, 257).
Methionine aminopeptidases are cobalt-dependent enzymes that can be divided into two groups, based on sequence comparison (14, 210). Type I methionine aminopeptidases are found in Eucarya and Bacteria, although the eucaryal enzyme includes an amino-terminal extension not found in its bacterial counterpart. Eucarya also contain a second isoform of the enzyme, referred to as type II methionine aminopeptidases. The two enzyme classes can be distinguished by the presence of an additional ∼60-amino-acid-residue carboxy-terminal stretch of unknown function in type II enzymes (14). Genome sequence analysis has revealed that Archaea contain only type II methionine aminopeptidases, although these lack an amino-terminal extension found in the eucaryal enzyme (427, 443). Examination of the crystal structure of the Pyrococcus furiosus enzyme confirmed the similarities of the catalytic domains of the two methionine aminopeptidase isoforms, despite their limited degree of sequence homology (74, 427).
Inteins in Archaeal Proteins
Inteins are genetic elements lying within a protein-encoding ORF that are transcribed and translated together with their host to yield an immature precursor protein (135, 260, 337). Self-splicing of inteins occurs at the posttranslational level, when the inteins excise themselves from the host protein, allowing the intein-bordering residues of the flanking segments of the host polypeptide to join through a peptide bond to yield the mature protein, which is now able to fold and function properly. Although first discovered in a yeast vacuolar ATPase (200) and found in proteins across evolution, inteins are most commonly observed in archaeal proteins; by spring 2005, approximately 200 inteins had been reported, with almost half being found in Archaea (references 343 and 346 and databases cited therein).
Inteins are most often found in enzymes involved in DNA replication and repair. Indeed, the first archaeal intein was found in a Thermococcus litoralis DNA polymerase (344). Inteins have subsequently been detected in numerous other archaeal DNA polymerases (48, 305, 408, 429, 476) but also in other proteins (68, 110, 359, 388) from various hyperthermophilic Archaea. This list includes Methanobacterium thermoautotrophicum ribonucleoside diphosphate reductase, which contains the smallest known intein to date (110). Examination of intein databases reveals that archaeal inteins are not restricted to hyperthermophilic proteins but are also predicted to exist in proteins from acidophiles such as Ferroplasma acidarmanus, Ferroplasma acidiphilum, and Picrophilus torridus, from the haloarchaea Halobacterium sp. strain NRC-1, Haloferax volcanii, and Haloarcula marismortui and from the Antarctica-derived methanogen Methanococcoides burtonii (343, 346).
Understanding the mechanics of the self-splicing reaction associated with intein excision began with experiments employing hyperthermophilic archaeal DNA polymerases (312, 337, 476). In fitting with the elevated growth temperatures of the host organism, intein splicing from these proteins occurs inefficiently at temperatures below 25°C. By inserting the coding sequence of the intein from Pyrococcus sp. strain GB-D DNA polymerase between genes encoding two foreign proteins, an intein-containing chimeric precursor was expressed in Escherichia coli at low temperatures (476). Subsequent splicing of the purified precursor could be initiated by raising the temperature. Such studies revealed that all the information needed for the splicing process is found within the sequences of the intein and flanking regions and that the excision reaction is catalyzed by the intein itself, without need for additional factors.
Since these pioneering studies, further examination of archaeal inteins has revealed the diversity of intein biochemistry and offered additional insight into this posttranslational modification. For instance, during the first of four steps involved in the intein self-splicing reaction, the conserved intein amino-terminal Ser or Cys residue undergoes an acyl rearrangement, resulting in the formation of a (thio)ester bond at the amino-terminal splice junction (312, 337). A Methanococcus jannaschii ATPase provided the first example of an intein bearing an amino-terminal Ala residue (140), leading to the description of an alternative splicing pathway (407). In the third step of the self-splicing reaction, cyclization of the intein carboxy-terminal Asn residue leads to peptide bond cleavage and subsequent excision of the intein (312, 337). This cyclization step is facilitated by the intein's penultimate His residue, which renders the carboxy-terminal Asn residue's carbonyl carbon more electrophilic (312, 337). Examination of intein cleavage from Methanococcus jannaschii phosphoenolpyruvate synthase and RNA polymerase subunit A′ has provided insight into how inteins lacking this penultimate His residue are processed (58). Furthermore, the presence of inteins in DNA polymerases from Halobacterium sp. strain NRC-1, Pyrococcus abyssi, and Pyrococcus horikoshii bearing carboxy-terminal Gln rather than Asn residues suggests that inteins may self-excise via mechanisms not involving side chain cyclization (58). Indeed, dissection of the self-splicing pathway of the Pyrococcus abyssi DNA polymerase II DP2 subunit intein failed to detect the formation of an intein intermediate containing a cyclized C-terminal Glu residue (288).
Carboxy-Terminal Maturation of Archaeal [NiFe] Hydrogenases
Examination of the Methanococcus voltae vhuU gene product, a component of a [NiFe] hydrogenase, revealed that the translated polypeptide was shorter than predicted by the gene sequence due to a carboxy-terminal cleavage of the protein (404, 405). Although first demonstrated in Methanococcus voltae, cleavage of a carboxy-terminal region downstream of an Asp-Pro-Cys-X-X-His sequence motif by a dedicated endopeptidase following nickel incorporation has since been shown to be a general feature of prokaryotic [NiFe] hydrogenases (53). Differences in the [NiFe] hydrogenase proteolytic maturation step do exist, however, between the bacterial and archaeal systems. In Escherichia coli and other Bacteria, the hydrogenase cleavage motif is followed by a stretch of 15 or more amino acid residues (53). By contrast, in the proteolytic processing of the Thermococcus kodakaraensis hydrogenase α subunit, only four amino acid residues were released from the carboxy terminus following the conserved cleavage motif (199).
Similarly short sequences are also thought to be released from the large subunit of hydrogenases in other archaeal strains, including Methanobacterium thermoautotrophicum (380), Pyrococcus furiosus (352), and Thermococcus litoralis (433). Moreover, in Methanobacterium thermoautotrophicum and Pyrococcus furiosus, the mature enzymes are proposed to terminate in an Arg rather than a His residue. Differences between predicted molecular weight and the smaller molecular mass measured by SDS-PAGE migration suggest that proteolytic maturation of the Pyrococcus furiosus enzymes does indeed occur (380, 433).
In EchE, the Methanosarcina barkeri homologue of the Escherichia coli hydrogenase 3 large subunit, the cleavage motif also terminates with an Arg residue, although in this case, proteolytic processing does not occur and the Arg thus corresponds to the terminal residue of the protein (241). In contrast, the homologous proteins in Methanococcus jannaschii and Methanobacterium thermautotrophicum also have Arg rather than His residues at this position and yet contain carboxy-terminal extensions that likely undergo proteolytic processing (241). Finally, although the maturation process experienced by archaeal hydrogenases has been less well characterized than the parallel process in Bacteria, archaeal homologues of bacterial enzymes involved in this posttranslational maturation process have been described (199, 428).
OTHER POSTTRANSLATIONAL MODIFICATIONS IN ARCHAEA
Protein Acetylation
The acetylation of selected Lys residues in a protein was first observed almost 40 years ago with eucaryal histones (129), in which this posttranslational modification acts to modulate transcription (474). Since then, acetylation has been reported to modulate the function of many eucaryal and a limited number of bacterial proteins (237, 416, 479). In Archaea, protein acetylation of so-called Alba proteins has been demonstrated by mass spectrometry. These are a family of small (10 kDa) DNA binding proteins first detected in Sulfolobus species (28, 96). They have since been identified in numerous other euryarchaeotal and crenarchaeotal species as well as in Eucarya (28, 458, 460, 466). Upon acetylation of Sulfolobus solfataricus Alba at the Lys-16 position, the affinity of the protein for DNA was lowered (28). In vitro experiments support a role for the Sulfolobus homologue of the eukaryotic histone deacetylase Sir2 in deacetylating Alba, although other deacetylases may act similarly (466).
The enzyme responsible for Alba acetylation has not been identified, although several possible candidates are evident in archaeal genome sequences (355, 466). In a second case, the amino terminus of halocyanin, the small blue copper protein from the haloalkaliphile Natronobacterium pharaonis, has also been proposed to undergo acetylation, in addition to lipid modification (see above), based on the results of mass spectroscopic studies (274).
Protein Ubiquitination
The proteasome is a multisubunit complex responsible for protein degradation in the cytoplasm of eucaryal (452) and archaeal (91, 276, 277) cells. In Eucarya, proteins are targeted for proteasomal degradation by the posttranslational covalent attachment of ubiquitin, a 76-amino-acid-residue polypeptide (69). At present, the putative posttranslational modification that leads to proteasome-mediated protein degradation in Archaea has not been defined. While some reports suggest the existence of ubiquitin in Archaea (275, 300, 473), no direct demonstration of archaeal ubiquitin has been provided, nor have analyses of archaeal genomes identified ubiquitin-encoding genes or genes encoding ubiquitin-transferring proteins. Stuctural studies, however, have revealed the existence of archaeal proteins bearing ubiquitin-like folds (33, 375). Nonetheless, it remains to be shown that these proteins participate in proteasome-mediated protein degradation in Archaea.
Hypusine-Containing Archaeal Protein
Hypusine [_N_-ɛ-(4-amino-2-hydroxybutyl)-l-lysine] is a nonstandard amino acid residue found in all Eucarya in a single protein, eukaryotic translation initiation factor 5A (eIF-5A) (329). Hypusine is irreversibly formed soon after the biogenesis of eIF-5A in a two-step posttranslational reaction (331). In the first step, catalyzed by deoxyhypusine synthase, the 4-aminobutyl moiety of the polyamine spermidine is transferred to the ɛ-amino group of a single specific Lys residue in the eIF-5A precursor protein to form an intermediate, deoxyhypusine. The 4-aminobutyl group of the intermediate undergoes hydroxylation by deoxyhypusine hydroxylase to yield hypusine. The presence of this hypusine residue is essential for eIF-5A function (50, 330, 331).
Hypusine also exists in Archaea, where it too is found exclusively in aIF-5A, the archaeal homologue of eIF-5A. This has been shown in Halobacterium cutirubrum, Methanococcus jannaschii, Pyrobaculum aerophilum, Pyrococcus horikoshii, Sulfolobus acidocaldarius, and Thermoplasma acidophilum (22, 221, 338, 386, 480). The ability to synthesize hypusine has been studied in Acidianus ambivalens, Pyrodictium occultum, and Thermoproteus tenax (23). The involvement of aIF-5A in archaeal cell growth and the archaeal cell cycle was shown by the arresting action of _N_1-guanyl-1,7-diaminoheptane, a hypusination inhibitor, in Halobacterium salinarum, Haloferax mediterranei, Sulfolobus acidocaldarius, and Sulfolobus solfataricus (183).
PROTEOME-WIDE ANALYSIS OF POSTTRANSLATIONAL MODIFICATIONS IN ARCHAEA
Most studies of posttranslational modification of archaeal proteins have thus far relied on individual genes or proteins, the choice of which has been largely guided by substrate availability. In the future, one can expect that our ever-improving ability to describe the entire genomic, transcriptomic, and proteomic profile of an organism will move the study of posttranslational protein modification from the scale of selected proteins to a cellwide perspective. At present, however, such attempts are limited by various factors, including the possible heterogeneity of posttranslational modifications experienced by a given gene product, the relative abundance of a given posttranslationally modified protein variant, and our ability to discern potential posttranslational modifications not encountered previously. Nevertheless, as better tools become available for the simultaneous analysis of the entire protein complement of a cell (187, 270), proteomewide description of posttranslational modifications will soon become routine.
To date, several Archaea have been the subject of proteomic analysis. Such studies have provided novel insights into the adaptations adopted by extremophilic Archaea or have described technical advances in working with extremophilic proteomes (34, 66, 109, 123, 133, 134, 138, 167, 187, 192, 203, 295, 304, 491). Of these investigations, a number have focused on posttranslational modification of archaeal protein targets. In the first of a series of studies addressing the proteome of Methanococcus jannaschii, the appearance of a given gene product in multiple positions in a two-dimensional gel electrophoretic system was taken to reflect the posttranslational modification of that polypeptide (134). Accordingly, the multiple positions of Mj0324, which is annotated as an elongation factor (EF-1α), were assumed to correspond to isoforms modified by various degrees of phosphorylation, as had been observed with the eucaryal version of the protein (160). Mj0822, which is annotated as the S-layer glycoprotein, was also found in multiple positions in two-dimensional gels (134). In fact, protein spots corresponding to Mj0822 are among the most strongly stained by Coomassie blue, although the protein is resistant to silver staining, probably due to the presence of glycan moieties. Such differential staining of glycosylated proteins is well documented (186).
In a subsequent proteomic analysis of Methanococcus jannaschii as a function of growth conditions or growth stage, examination of peptide fragments derived from either Mj0891 or Mj0891, which are annotated as flagellin B1 and flagellin B2, respectively, revealed condition-specific changes in isoelectric point and abundance (133). Such modulations were proposed but not shown to result from differential degrees of posttranslational modification of the proteins.
More recent studies relying on advances in mass spectrometry for proteome analysis, which included elimination of intermediate proteolytic steps, resulted in a 100% sequence coverage of a set of 72 Methanococcus jannaschii proteins (123). Among these proteins, examples of protein acetylation and methylation, amino-terminal proteolytic processing, and disulfide bonds were detected. The applicability of this approach for the rapid determination of expected posttranslational modifications was shown when it was used to test the validity of histone acetylation in Methanosarcina acetivorans (125). Despite the proposed presence of a histone-modifying enzyme in this species, no histone modification was detected.
CONCLUSIONS
Archaea have proven to be a valuable resource in the search for new information on posttranslational protein modification. In several cases, Archaea have provided the first prokaryotic examples of modifications once thought to be restricted to Eucarya. The glycosylation of the Halobacterium salinarum S-layer glycoprotein represents one such example. In other cases, Archaea present previously unknown variations on a given posttranslational protein modification theme, such as the methylation profile of methyl-coenzyme M reductase or the unique lipid moieties attached to haloarchaeal proteins. Elucidation of the enzymatic steps involved in the archaeal version of a particular posttranslational modification event, such as signal sequence cleavage or intein splicing, has dramatically enhanced our understanding of the mechanistics of many posttranslational modifications.
With the advent of the proteomic era, when one can determine the protein profile of individual cells, complete physiological systems, and even entire organisms in response to a myriad of conditions, the protein complexity arising from posttranslational modifications should become even more obvious. If past contributions are any indication, the study of Archaea will continue to expand understanding of the scope, the roles, and the biogenesis of posttranslational modifications that a protein can experience. Such information could provide insight into the strategies adopted by Archaea in the face of the extreme environments in which they can exist. One immediate benefit of relating posttranslational protein modifications to protein structure, stability, and function, together with enhanced tools for the manipulation of archaeal protein expression and secretion, will be the utilization of enzymes from extremophilic Archaea tailored for a broad spectrum of biotechnological and industrial applications.
Acknowledgments
This work was supported by the Israel Science Foundation-Charles H. Revson Foundation (grant 433/03 to J.E.) and the National Science Foundation (BES-0317911 and MCB 0129841 to M.A.).
We thank Frank E. Jenney, Jr., for critical reading of the manuscript and the two anonymous reviewers for valuable comments and suggestions.
REFERENCES
- 1.Akca, E., H. Claus, N. Schultz, G. Karbach, B. Schlott, T. Debaerdemaeker, J. P. Declercq, and H. Konig. 2002. Genes and derived amino acid sequences of S-layer proteins from mesophilic, thermophilic, and extremely thermophilic methanococci. Extremophiles 6**:**351-358. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., and A. J. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177**:**209-216. [DOI] [PubMed] [Google Scholar]
- 3.Albers, S. V., M. G. L. Elferink, R. L. Charlebois, W. Sensen, A. J. M. Driessen, and W. N. Konings. 1999. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J. Bacteriol. 181**:**4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albers, S. V., S. M. Koning, W. N. Konings, and A. J. Driessen. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36**:**5-15. [DOI] [PubMed] [Google Scholar]
- 5.Albers, S. V., W. N. Konings, and A. J. Driessen. 1999. A unique short signal sequence in membrane-anchored proteins of Archaea. Mol. Microbiol. 31**:**1595-1596. [DOI] [PubMed] [Google Scholar]
- 6.Albers, S. V., Z. Szabo, and A. J. Driessen. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185**:**3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5**:**160-165. [DOI] [PubMed] [Google Scholar]
- 8.Amano, T., T. Wakagi, and T. Oshima. 1993. An ecto-enzyme from Sulfolobus acidocaldarius strain 7 which catalyzes hydrolysis of inorganic pyrophosphate, ATP, and ADP: purification and characterization. J. Biochem. (Tokyo) 114**:**329-333. [DOI] [PubMed] [Google Scholar]
- 9.Amaro, A. M., and C. A. Jerez. 1984. Methylation of ribosomal proteins in bacteria: evidence of conserved modification of the eubacterial 50S subunit. J. Bacteriol. 158**:**84-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amegbey, G. Y., H. Monzavi, B. Habibi-Nazhad, S. Bhattacharyya, and D. S. Wishart. 2003. Structural and functional characterization of a thioredoxin-like protein (Mt0807) from Methanobacterium thermoautotrophicum. Biochemistry 42**:**8001-8010. [DOI] [PubMed] [Google Scholar]
- 11.Appleby, T. C., I. I. Mathews, M. Porcelli, G. Cacciapuoti, and S. E. Ealick. 2001. Three-dimensional structure of a hyperthermophilic 5′-deoxy-5′-methylthioadenosine phosphorylase from Sulfolobus solfataricus. J. Biol. Chem. 276**:**39232-39242. [DOI] [PubMed] [Google Scholar]
- 12.Apweiler, R., H. Hermjakob, and N. Sharon. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473**:**4-8. [DOI] [PubMed] [Google Scholar]
- 13.Arfin, S. M., and R. A. Bradshaw. 1988. Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry 27**:**7979-7984. [DOI] [PubMed] [Google Scholar]
- 14.Arfin, S. M., R. L. Kendall, L. Hall, L H. Weaver, A. E. Stewart, B. W. Matthews, and R. A. Bradshaw. 1995. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc. Natl. Acad. Sci. USA 92**:**7714-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chiu, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14**:**2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides—a comparative survey at the genome level. Protein Sci. 12**:**1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50**:**1339-1347. [DOI] [PubMed] [Google Scholar]
- 18.Bardy, S. L., S. Y. Ng, D. S. Carnegie, and K. F. Jarrell. 2005. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae. J. Bacteriol. 187**:**1188-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardy, S., S. Y. Ng, and K. F. Jarrell. 2003. Prokaryotic motility structures. Microbiology 149**:**295-304. [DOI] [PubMed] [Google Scholar]
- 20.Bardy, S. L., S. Y. Ng, and K. F. Jarrell. 2004. Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7**:**41-51. [DOI] [PubMed] [Google Scholar]
- 21.Barford, D., A. K. Das, and M. P. Egloff. 1998. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27**:**133-164. [DOI] [PubMed] [Google Scholar]
- 22.Bartig, D., K. Lemkemeier, J. Frank, F. Lottspeich, and F. Klink. 1992. The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur. J. Biochem. 204**:**751-758. [DOI] [PubMed] [Google Scholar]
- 23.Bartig, D., H. Schümann, and F. Klink. 1990. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. Syst. Appl. Microbiol. 13**:**112-116. [Google Scholar]
- 24.Barton, G. J., P. T. Cohen, and D. Barford. 1994. Conservation analysis and structure prediction of the protein serine/threonine phosphatases. Sequence similarity with diadenosine tetraphosphatase from Escherichia coli suggests homology to the protein phosphatases. Eur. J. Biochem. 220**:**225-237. [DOI] [PubMed] [Google Scholar]
- 25.Baumann, H., S. Knapp, A. Karshikoff, R. Ladenstein, and T. Härd. 1995. DNA-binding Surface of the Sso7d Protein from Sulfolobus solfataricus. J. Mol. Biol. 247**:**840-846. [DOI] [PubMed] [Google Scholar]
- 26.Baumann, H., S. Knapp, T. Lundback, R. Ladenstein, and T. Hard. 1994. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Biol. 1**:**808-819. [DOI] [PubMed] [Google Scholar]
- 27.Bayley, D. P., M. L. Kalmokoff, and K. F. Jarrell. 1993. Effect of bacitracin on flagellar assembly and presumed glycosylation of the flagellins of Methanococcus deltae. Arch. Microbiol. 160**:**179-185. [Google Scholar]
- 28.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296**:**148-151. [DOI] [PubMed] [Google Scholar]
- 29.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47**:**187-254. [DOI] [PubMed] [Google Scholar]
- 30.Bernfield, M., M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68**:**729-777. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya, S., B. Habibi-Nazhad, G. Amegbey, C M. Slupsky, A. Yee, C. Arrowsmith, and D. S. Wishart. 2002. Identification of a novel archaebacterial thioredoxin: determination of function through structure. Biochemistry 41**:**4760-4770. [DOI] [PubMed] [Google Scholar]
- 32.Bidle, K. A. 2003. Differential expression of genes influenced by changing salinity using RNA arbitrarily primed PCR in the archaeal halophile Haloferax volcanii. Extremophiles 7**:**1-7. [DOI] [PubMed] [Google Scholar]
- 33.Bienkowska, J. R., H. Hartman, and T. F. Smith. 2003. A search method for homologs of small proteins. Ubiquitin-like proteins in prokaryotic cells? Protein Eng. 16**:**897-904. [DOI] [PubMed] [Google Scholar]
- 34.Blonder, J., T. P. Conrads, L. R. Yu, A. Terunuma, G. M. Janini, H. J. Issaq, J. C. Vogel, and T. D. Veenstra. 2004. A detergent- and cyanogen bromide-free method for integral membrane proteomics: application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics 4**:**31-45. [DOI] [PubMed] [Google Scholar]
- 35.Bolhuis, A. 2002. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology 148**:**3335-3346. [DOI] [PubMed] [Google Scholar]
- 36.Bonet, M. L., F. I. Llorca, and E. Cadenas. 1992. Alkaline p-nitrophenylphosphate phosphatase activity from Halobacterium halobium. Selective activation by manganese and effect of other divalent cations. Int. J. Biochem. 24**:**839-845. [DOI] [PubMed] [Google Scholar]
- 37.Bork, P., N. P. Brown, H. Hegyi, and J. Shultz. 1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 5**:**1421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher, Y., M. Kamekura, and W. F. Doolittle. 2004. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52**:**515-527. [DOI] [PubMed] [Google Scholar]
- 39.Bradshaw, R. A., W. W. Brickey, and K. W. Walker. 1998. N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 23**:**263-267. [DOI] [PubMed] [Google Scholar]
- 40.Braun, V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415**:**335-377. [DOI] [PubMed] [Google Scholar]
- 41.Bröckl, G., M. Behr, S. Fabry, R. Hensel, H. Kaudewitz, E. Biendl, and H. König. 1991. Analysis and nucleotide sequence of the genes encoding the surface-layer glycoproteins of the hyperthermophilic methanogens Methanothermus fervidus and Methanothermus sociabilis. Eur. J. Biochem. 199**:**147-152. [DOI] [PubMed] [Google Scholar]
- 42.Brodsky, J. L. 1996. Posttranslational protein translocation: not all hsc70s are created equal. Trends Biochem. Sci. 21**:**122-126. [PubMed] [Google Scholar]
- 43.Brooun, A., W. Zhang, and M. Alam. 1997. Primary structure and functional analysis of the soluble transducer protein HtrXI from the archaeon Halobacterium salinarium. J. Bacteriol. 179**:**2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown, S. H., and R. M. Kelly. 1993. Characterization of amylolytic enzymes, having both (α)-1,4 and (α)-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl. Environ. Microbiol. 59**:**2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bujnicki, J. M. 2000. Sequence, structural, and evolutionary analysis of prokaryotic ribosomal protein L11 methyltransferases. Acta Microbiol. Pol. 49**:**19-28. [PubMed] [Google Scholar]
- 46.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426**:**239-257. [DOI] [PubMed] [Google Scholar]
- 47.Cacciapuoti, G., M. Porcelli, C. Bertoldo, M. De Rosa, and V. Zappia. 1994. Purification and characterization of extremely thermophilic and thermostable 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus. Purine nucleoside phosphorylase activity and evidence for intersubunit disulfide bonds. J. Biol. Chem. 269**:**24762-24769. [PubMed] [Google Scholar]
- 48.Cambon-Bonavita, M. A., P. Schmitt, M. Zieger, J. M. Flaman, F. Lesongeur, G. Raguenes, D. Bindel, N. Frisch, Z. Lakkis, D. Dupret, G. Barbier, and J. Querellou. 2000. Cloning, expression, and characterization of DNA polymerase I from the hyperthermophilic archaea Thermococcus fumicolans. Extremophile 4**:**215-225. [DOI] [PubMed] [Google Scholar]
- 49.Cao, T. B., and M. H. Saier Jr. 2003. The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys. Acta 1609**:**115-125. [DOI] [PubMed] [Google Scholar]
- 50.Caraglia, M., M. Marra, G. Giuberti, A. M. D'Alessandro, A. Budillon, S. del Prete, A. Lentini, S. Beninati, and A. Abbruzzese. 2001. The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids 2**:**91-104. [DOI] [PubMed] [Google Scholar]
- 51.Carballeira, N. M., M. Reyes, A. Sostre, H. Huang, M. F. Verhagen, and M. W. Adams. 1997. Unusual fatty acid compositions of the hyperthermophilic archaeon Pyrococcus furiosus and the bacterium Thermotoga maritima. J. Bacteriol. 179**:**2766-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardona, S., F. Remonsellez, N. Guiliani, and C. A. Jerez. 2001. The glycogen-bound polyphosphate kinase from Sulfolobus acidocaldarius is actually a glycogen synthase. Appl. Environ. Microbiol. 67**:**4773-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9**:**228-237. [DOI] [PubMed] [Google Scholar]
- 54.Cave, J. W., H. S. Cho, A. M. Batchelder, H. Yokota, R. Kim, and D. E. Wemmer. 2001. Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii. Protein Sci. 10**:**384-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang, C. N., and F. N. Chang. 1974. Methylation of ribosomal proteins in vitro. Nature 251**:**731-733. [DOI] [PubMed] [Google Scholar]
- 56.Chang, C. N., and N. Chang. 1975. Methylation of the ribosomal proteins in Escherichia coli. Nature and stoichiometry of the methylated amino acids in 50S ribosomal proteins. Biochemistry 14**:**468-477. [DOI] [PubMed] [Google Scholar]
- 57.Chang, S. Y., E. C. McGary, and S. Chang. 1989. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol. 171**:**4071-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen, L., J. Benner, and F. B. Perler. 2000. Protein splicing in the absence of an intein penultimate histidine. J. Biol. Chem. 275**:**20431-20435. [DOI] [PubMed] [Google Scholar]
- 59.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103**:**263-271. [DOI] [PubMed] [Google Scholar]
- 60.Chiu, H. J., E. Johnson, I. Schroder, and D. C. Rees. 2001. Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+. Structure (Cambridge) 9**:**311-319. [DOI] [PubMed] [Google Scholar]
- 61.Cho, C. W., S. H. Lee, J. Choi, S. J. Park, D. J. Ha, H. J. Kim, and C. W. Kim. 2003. Improvement of the two-dimensional gel electrophoresis analysis for the proteome study of Halobacterium salinarum. Proteomics 3**:**2325-2329. [DOI] [PubMed] [Google Scholar]
- 62.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105**:**103-113. [DOI] [PubMed] [Google Scholar]
- 63.Choli, T., P. Henning, B. Wittmann-Liebold, and R. Reinhardt. 1988. Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the Archaebacterium, Sulfolobus solfataricus. Biochim. Biophys. Acta 950**:**193-120. [DOI] [PubMed] [Google Scholar]
- 64.Choli, T., B. Wittmann-Liebold, and R. Reinhardt. 1988. Microsequence analysis of DNA-binding proteins 7a, 7b, and 7e from the archaebacterium Sulfolobus acidocaldarius. J. Biol. Chem. 263**:**7087-7093. [PubMed] [Google Scholar]
- 65.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327**:**482-486. [DOI] [PubMed] [Google Scholar]
- 66.Christendat, D., A. Yee, A. Dharamsi, Y. Kluger, A. Savchenko, J. R. Cort, V. Booth, C. D. Mackereth, V. Saridakis, I. Ekiel, G. Kozlov, K. L. Maxwell, N. Wu, L. P. McIntosh, K. Gehring, M. A. Kennedy, A. R. Davidson, E. F. Pai, M. Gerstein, A. M. Edwards, and C. H. Arrowsmith. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7**:**903-909. [DOI] [PubMed] [Google Scholar]
- 67.Christian, J. H., J. A. Waltho. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta 65**:**506-508. [DOI] [PubMed] [Google Scholar]
- 68.Chute, I. C., Z. Hu, and X. Q. Liu. 1998. A topA intein in Pyrococcus furiosus and its relatedness to the r-gyr intein of Methanococcus jannaschii. Gene 210**:**85-92. [DOI] [PubMed] [Google Scholar]
- 69.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22**:**442-451. [DOI] [PubMed] [Google Scholar]
- 70.Clarke, S. 1993. Protein methylation. Curr. Opin. Cell Biol. 5**:**977-983. [DOI] [PubMed] [Google Scholar]
- 71.Cohen, P. T. W. 1997. Novel protein serine/ threonine phosphatases: variety is the spice of life. Trends Biochem. Sci. 22**:**245-251. [DOI] [PubMed] [Google Scholar]
- 72.Cohen-Krausz, S., and S. Trachtenberg. 2002. The structure of the archaebacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J. Mol. Biol. 321**:**383-395. [DOI] [PubMed] [Google Scholar]
- 73.Condo, I., D. Ruggero, R. Reinhardt, and P. Londei. 1998. A novel aminopeptidase associated with the 60 kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 29**:**775-785. [DOI] [PubMed] [Google Scholar]
- 74.Copik, A. J., B. P. Nocek, S. I. Swierczek, S. Ruebush, S. B. Jang, L. Meng, V. M. D'Souza, J. W. Peters, B. Bennett, and R. C. Holz. 2005. EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus. Biochemistry 44**:**121-129. [DOI] [PubMed] [Google Scholar]
- 75.Correia, J. D., and K. F. Jarrell. 2000. Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae and other methanogens. J. Bacteriol. 182**:**855-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cort, J. R., S. V. Mariappan, C. Y. Kim, M. S. Park, T. S. Peat, G. S. Waldo, T. C. Terwilliger, and M. A. Kennedy. 2001. Solution structure of Pyrobaculum aerophilum DsrC, an archaeal homologue of the gamma subunit of dissimilatory sulfite reductase. Eur. J. Biochem. 268**:**5842-5850. [DOI] [PubMed] [Google Scholar]
- 77.Cortay, J. C., B. Duclos, and A. J. Cozzone. 1986. Phosphorylation of a bacterial protein at tyrosine. J. Mol. Biol. 187**:**305-308. [DOI] [PubMed] [Google Scholar]
- 78.Creighton, T. E. 1988. Disulphide bonds and protein stability. Bioessays 8**:**57-63. [DOI] [PubMed] [Google Scholar]
- 79.Cruden, D., R. Sparling, and A. J. Markovetz. 1989. Isolation and ultrastructure of the flagella of Methanococcus thermolithotrophicus and Methanospirillum hungatei. Appl. Environ. Microbiol. 55**:**1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cumming, R. C., N. L. Andon, P. A. Haynes, M. Park, W. H. Fischer, and D. Schubert. 2004. Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem. 279**:**21749-21758. [DOI] [PubMed] [Google Scholar]
- 81.Daas, P. J., R. W. Wassenaar, P. Willemsen, R. J. Theunissen, J. T. Keltjens, C. van der Drift, and G. D. Vogels. 1996. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J. Biol. Chem. 271**:**22339-22345. [DOI] [PubMed] [Google Scholar]
- 82.Dalbey, R. E., M. O. Lively, S. Bron, and J. M. van Dijl. 1997. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6**:**1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dale, H., C. M. Angevine, and M. P. Krebs. 2000. Ordered membrane insertion of an archaeal opsin in vivo. Proc. Natl. Acad. Sci. USA 97**:**7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dale, H., and M. P. Krebs. 1999. Membrane insertion kinetics of a protein domain in vivo. The bacterioopsin n terminus inserts co-translationally. J. Biol. Chem. 274**:**22693-22698. [DOI] [PubMed] [Google Scholar]
- 85.Darby, N. J., J. Kemmink, and T. E. Creighton. 1996. Identifying and characterizing a structural domain of protein disulfide isomerase. Biochemistry 35**:**10517-10528. [DOI] [PubMed] [Google Scholar]
- 86.Das, P. M., and R. Singal. 2004. DNA methylation and cancer. J. Clin. Oncol. 22**:**4632-4642. [DOI] [PubMed] [Google Scholar]
- 87.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73**:**241-268. [DOI] [PubMed] [Google Scholar]
- 88.DeDecker, B. S., R. O'Brien, P. J. Fleming, J. H. Geiger, S. P. Jackson, and P. B. Sigler. 1996. The crystal structure of a hyperthermophilic archaeal TATA-box binding protein. J. Mol. Biol. 264**:**1072-1084. [DOI] [PubMed] [Google Scholar]
- 89.De Felice, M., L. Esposito, B. Pucci, F. Carpentieri, M. De Falco, M. Rossi, and F. M. Pisani. 2003. Biochemical characterization of a CDC6-like protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 278**:**46424-46431. [DOI] [PubMed] [Google Scholar]
- 90.DeFranco, A. L., and D. E. Koshland, Jr. 1980. Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 77**:**2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Mot, R., I. Nagy, J. Walz, and W. Baumeister. 1999. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 7**:**88-92. [DOI] [PubMed] [Google Scholar]
- 92.De Rosa, M., A. Gambacorta, and A. Gliozzi. 1986. Structure, biosynthesis and physicochemical properties of archaebacterial lipids. Microbiol. Rev. 50**:**70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dever, T. E. 1999. Translation initiation: adept at adapting. Trends Biochem. Sci. 24**:**398-403. [DOI] [PubMed] [Google Scholar]
- 94.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185**:**1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doig, P., N. Kinsella, P. Guerry, and T. J. Trust. 1996. Characterization of a posttranslational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol. 19**:**379-387. [DOI] [PubMed] [Google Scholar]
- 96.Edmondson, S. P., M. A. Kahsai, R. Gupta, and J. W. Shriver. 2004. Characterization of Sac10a, a hyperthermophile DNA-binding protein from Sulfolobus acidocaldarius. Biochemistry 43**:**13026-13036. [DOI] [PubMed] [Google Scholar]
- 97.Edmondson, S. P., and J. W. Shriver. 2001. DNA binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 334**:**129-145. [DOI] [PubMed] [Google Scholar]
- 98.Eichler, J. 2000. Novel glycoproteins of the halophilic archaeon Haloferax volcanii. Arch. Microbiol. 173**:**445-448. [DOI] [PubMed] [Google Scholar]
- 99.Eichler, J. 2001. Posttranslational modification of the S-layer glycoprotein occurs following translocation across the plasma membrane of the haloarchaeon Haloferax volcanii. Eur. J. Biochem. 268**:**4366-4373. [DOI] [PubMed] [Google Scholar]
- 100.Eichler, J. 2002. Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eucaryal enzymes. J. Mol. Evol. 54**:**411-415. [DOI] [PubMed] [Google Scholar]
- 101.Eichler, J. 2003. Facing extremes: archaeal surface-layer (glyco)proteins. Microbiology 149**:**3347-3351. [DOI] [PubMed] [Google Scholar]
- 102.Eichler, J., I. Silman, and L. Anglister. 1992. G2-acetylcholinesterase is presynaptically localized in Torpedo electric organ. J. Neurocytol. 21**:**707-716. [DOI] [PubMed] [Google Scholar]
- 103.Eisenhaber, B., P. Bork, and F. Eisenhaber. 2001. Posttranslational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng. 14**:**17-25. [DOI] [PubMed] [Google Scholar]
- 104.Eisenhaber, B., S. Maurer-Stroh, M. Novatchkova, G. Schneider, and F. Eisenhaber. 2003. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and posttranslational transfer to proteins. Bioessays 25**:**367-385. [DOI] [PubMed] [Google Scholar]
- 105.Elbein, A. D. 1981. The tunicamycins: useful tools for studies on glycoproteins. Trends Biochem. Sci. 6**:**219-221. [Google Scholar]
- 106.Elferink, M. G. L., S. V. Albers, W. N. Konings, and A. J. M. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39**:**1494-1503. [DOI] [PubMed] [Google Scholar]
- 107.Erickson, P. R., and M. C. Herzberg. 1993. Evidence for the covalent linkage of carbohydrate polymers to a glycoprotein from Streptococcus sanguis. J. Biol. Chem. 268**:**23780-23783. [PubMed] [Google Scholar]
- 108.Ermler, U., W. Grabarse, S. Shima, M. Goubeaud, and R. K. Thauer. 1997. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278**:**1457-1462. [DOI] [PubMed] [Google Scholar]
- 109.Evans, E. C., T. Horn, M. A. Wagner, M. Eschenbrenner, C. V. Mujer, and V. G. DelVecchio. 2003. Isolation protocol for two-dimensional-polyacrylamide gel electrophoresis analysis of Haloferax volcanii proteome. BioTechniques 35**:**478-482. [DOI] [PubMed] [Google Scholar]
- 110.Evans, T. C., Jr., J. Benner, and M. Q. Xu. 1999. The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J. Biol. Chem. 274**:**3923-3926. [DOI] [PubMed] [Google Scholar]
- 111.Evdokimov, A. G., D. E. Anderson, K. M. Rautzahn, and D. S. Waugh,. 2000. Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein. J. Mol. Biol. 305**:**891-904. [DOI] [PubMed] [Google Scholar]
- 112.Faguy, D. M., D. P. Bayley, A. S. Kostyukova, N. A. Thomas, and K. F. Jarrell. 1996. Isolation and characterization of flagella and flagellin proteins from the thermoacidophilic archaea Thermoplasma volcanium and Sulfolobus shibatae. J. Bacteriol. 178**:**902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Faguy, D. M., K. F. Jarrell, J. Kuzio, and M. L. Kalmokoff. 1994. Molecular analysis of archaeal flagellins: Similarity to the type IV pilin-transport superfamily widespread in bacteria. Can. J. Microbiol. 40**:**67-71. [DOI] [PubMed] [Google Scholar]
- 114.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13**:**457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farazi, T. A., G. Waksman, and J. I. Gordon. 2001. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276**:**39501-39504. [DOI] [PubMed] [Google Scholar]
- 116.Faulkner, A., X. Chen, J. Rush, B. Horazdovsky, C. J. Waechter, G. M. Carman, and P. C. Sternweis. 1999. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274**:**14831-14837. [DOI] [PubMed] [Google Scholar]
- 117.Febbraio, F., A. Andolfo, F. Tanfani, R. Briante, F. Gentile, S. Formisano, C. Vaccaro, A. Scire, E. Bertoli, P. Pucci, and R. Nucci. 2004. Thermal stability and aggregation of Sulfolobus solfataricus beta-glycosidase are dependent upon the N-epsilon-methylation of specific lysyl residues: critical role of in vivo posttranslational modifications. J. Biol. Chem. 279**:**10185-10194. [DOI] [PubMed] [Google Scholar]
- 118.Feese, M. D., Y. Kato, T. Tamada, M. Kato, T. Komeda, Y. Miura, M. Hirose, K. Hondo, K. Kobayashi, and R. Kuroki. 2000. Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus. J. Mol. Biol. 301**:**451-464. [DOI] [PubMed] [Google Scholar]
- 119.Fekkes, P., and A. J. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63**:**161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fernandes, A. P., and A. Holmgren. 2004. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal 6**:**63-74. [DOI] [PubMed] [Google Scholar]
- 121.Ferrando-May, E, M. Krah, W. Marwan, and D. Oesterhelt. 1993. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 12**:**2999-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Field, M. C., A. K. Menon, and G. A. M. Cross. 1991. A glycosylphosphatidylinositol protein anchor from procyclic stage Trypanosoma brucei: lipid structure and biosynthesis. EMBO J. 10**:**2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Forbes, A. J., S. M. Patrie, G. K. Taylor, Y. B. Kim, L. Jiang, and N. L. Kelleher. 2004. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. USA 101**:**2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fukada, Y., T. Takao, H. Ohguro, T. Yoshizawa, T. Akino, and Y. Shimonishi. 1990. Farnesylated gamma-subunit of photoreceptor G protein indispensable for GTP-binding. Nature 346**:**658-660. [DOI] [PubMed] [Google Scholar]
- 125.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12**:**532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garnak, M., and H. C. Reeves. 1979. Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science 203**:**1111-1112. [DOI] [PubMed] [Google Scholar]
- 127.Gattinger, A., M. Schloter, and J. C. Munch. 2002. Phospholipid etherlipid and phospholipid fatty acid fingerprints in selected euryarchaeotal monocultures for taxonomic profiling, FEMS Microbiol. Lett. 213**:**133-139. [DOI] [PubMed] [Google Scholar]
- 128.Genschik, P., E. Billy, M. Swianiewicz, and W. Filipowicz. 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in eucarya, bacteria and archaea. EMBO J. 16**:**2955-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gershey, E. L., G. Vidali, and V. G. Allfrey. 1968. Chemical studies of histone acetylation. The occurrence of epsilon-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 243**:**5018-5022. [PubMed] [Google Scholar]
- 130.Giam, C. Z., T. Chai, S. Hayashi, and H. C. Wu. 1984. Prolipoprotein modification and processing in Escherichia coli. A unique secondary structure in prolipoprotein signal sequence for the recognition by glyceryl transferase. Eur. J. Biochem. 141**:**331-337. [DOI] [PubMed] [Google Scholar]
- 131.Gilson, E., G. Alloing, T. Schmidt, J. P. Claverys, R. Dudler, and M. Hofnung. 1988. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 7**:**3971-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ginzburg, M., L. Sachs, and B. Z. Ginzburg. 1970. Ion metabolism in a Halobacterium. I. Influence of age of culture on intracellular concentrations. J. Gen. Physiol. 55**:**187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giometti, C. S., C. I. Reich, S. L. Tollaksen, G. Babnigg, H. Lim, J. R. Yates 3rd, and G. J. Olsen. 2001. Structural modifications of Methanococcus jannaschii flagellin proteins revealed by proteome analysis. Proteomics 1**:**1033-1042. [DOI] [PubMed] [Google Scholar]
- 134.Giometti, C. S., C. Reich, S. Tollaksen, G. Babnigg, H. Lim, W. Zhu, J. Yates, and G. Olsen. 2002. Global analysis of a “simple” proteome: Methanococcus jannaschii. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 782**:**227-243. [DOI] [PubMed] [Google Scholar]
- 135.Gogarten, J. P., A. G. Senejani, O. Zhaxybayeva, L. Olendzenski, and E. Hilario. 2002. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56**:**263-287. [DOI] [PubMed] [Google Scholar]
- 136.Goldman, S., K. Hecht, H. Eisenberg, and M. Mevarech. 1990. Extracellular Ca2+-dependent inducible alkaline phosphatase from the extremely halophilic archaebacterium Haloarcula marismorturi. J. Bacteriol. 172**:**7065-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gonzales, T., and J. Robert-Baudouy. 1996. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 18**:**319-344. [DOI] [PubMed] [Google Scholar]
- 138.Goodchild, A., M. Raftery, N. F. Saunders, M. Guilhaus, and R. Cavicchioli. 2004. Biology of the cold adapted archaeon, Methanococcoides burtonii determined by proteomics using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 3**:**1164-1176. [DOI] [PubMed] [Google Scholar]
- 139.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50**:**659-671. [DOI] [PubMed] [Google Scholar]
- 140.Gorbalenya, A. E. 1998. Non-canonical inteins. Nucleic Acids Res. 26**:**1741-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gorst, C. M., Z. H. Zhou, K. Ma, Q. Teng, J. B. Howard, M. W. Adams, and G. N. La Mar. 1995. Participation of the disulfide bridge in the redox cycle of the ferredoxin from the hyperthermophile Pyrococcus furiosus: 1H nuclear magnetic resonance time resolution of the four redox states at ambient temperature. Biochemistry 34**:**8788-8795. [DOI] [PubMed] [Google Scholar]
- 142.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86**:**5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grabarse, W., F. Mahlert, S. Shima, R. K. Thauer, and U. Ermler. 2000. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J. Mol. Biol. 303**:**329-344. [DOI] [PubMed] [Google Scholar]
- 144.Grabowski, B., and Z. Kelman. 2001. Autophosphorylation of archaeal Cdc6 homologues is regulated by DNA. J. Bacteriol. 183**:**5459-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Greller, G., R. Riek, and W. Boos. 2001. Purification and characterization of the heterologously expressed trehalose/maltose ABC transporter complex of the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 268**:**4011-4018. [DOI] [PubMed] [Google Scholar]
- 146.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171**:**6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Grogan, D. W. 1996. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can. J. Microbiol. 42**:**1163-1171. [Google Scholar]
- 148.Gropp, R., F. Gropp, and M. C. Betlach. 1992. Association of the halobacterial 7S RNA to the polysome correlates with expression of the membrane protein bacterioopsin. Proc. Natl. Acad. Sci. USA 89**:**1204-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grote, M., J. Dijk, and R. Reinhard. 1986. Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 873**:**405-413. [Google Scholar]
- 150.Guagliardi, A., D. de Pascale, R. Cannio, V. Nobile, S. Bartolucci, and M. Rossi. 1995. The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270**:**5748-5755. [DOI] [PubMed] [Google Scholar]
- 151.Guagliardi, A., V. Nobile, S. Bartolucci, and M. Rossi. 1994. A thioredoxin from the extreme thermophilic Archaeon Sulfolobus solfataricus Int. J. Biochem. 26**:**375-380. [Google Scholar]
- 152.Guy, J. E., M. N. Isupov, and J. A. Littlechild. 2003. The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix. J. Mol. Biol. 331**:**1041-1051. [DOI] [PubMed] [Google Scholar]
- 153.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63**:**133-139. [DOI] [PubMed] [Google Scholar]
- 154.Hanks, S. K., and T. Hunter. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9**:**576-596. [PubMed] [Google Scholar]
- 155.Hartmann, E., T. Sommer, S. Prehn, D. Gorlich, S. Jentsch, and T. A. Rapoport. 1994. Evolutionary conservation of components of the protein translocation complex. Nature 367**:**654-657. [DOI] [PubMed] [Google Scholar]
- 156.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22**:**451-471. [DOI] [PubMed] [Google Scholar]
- 157.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73**:**1019-1049. [DOI] [PubMed] [Google Scholar]
- 158.Helenius, J., and M. Aebi. 2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13**:**171-178. [DOI] [PubMed] [Google Scholar]
- 159.Helenius, J., D. T. W. Ng, C. L. Marolda, P. Walter, M. A. Valvano, and M. Aebi. 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415**:**447-450. [DOI] [PubMed] [Google Scholar]
- 160.Hershey, J. W. 1990. Overview: phosphorylation and translation control. Enzyme 44**:**17-27. [DOI] [PubMed] [Google Scholar]
- 161.Hettmann, T., C. L. Schmidt, S. Anemuller, U. Zahringer, H. Moll, A. Petersen, and G. Schafer. 1998. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 273**:**12032-12040. [DOI] [PubMed] [Google Scholar]
- 162.Heurgue-Hamard, V., S. Champ, A. Engstrom, M. Ehrenberg, and R. H. Buckingham. 2002. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21**:**769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8**:**67-113. [DOI] [PubMed] [Google Scholar]
- 164.Hildebrand, E., and A. Schimz. 1990. The lifetime of photosensory signals in Halobacterium halobium and its dependence on protein methylation. Biochim. Biophys. Acta 1052**:**96-105. [DOI] [PubMed] [Google Scholar]
- 165.Hirschberg, C. B., and M. D. Snider. 1987. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 56**:**63-87. [DOI] [PubMed] [Google Scholar]
- 166.Hogg, P. J. 2003. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28**:**210-214. [DOI] [PubMed] [Google Scholar]
- 167.Holden, J. F., F. L. Poole, S. L. Tollaksen, C. S. Giometti, H. Lim, J. R. Yates, and M. W. W. Adams. 2001. Identification of membrane proteins in the hyperthermophilic archaeon Pyrococcus furiosus using proteomics and prediction programs. Comp. Funct. Gen. 2**:**275-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264**:**13963-13966. [PubMed] [Google Scholar]
- 169.Hopfner, K. P., A. Eichinger, R. A. Engh, F. Laue, W. Ankenbauer, R. Huber, and B. Angerer. 1999. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl. Acad. Sci. USA 96**:**3600-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Horlacher, R., K. B. Xavier, H. Santos, J. DiRuggiero, M. Kossmann, and W. Boos. 1988. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180**:**680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hou, S., A. Brooun, H. S. Yu, T. Freitas, and M. Alam. 1998. Sensory rhodopsin II transducer HtrII is also responsible for serine chemotaxis in the archaeon Halobacterium salinarum. J. Bacteriol. 180**:**1600-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huber, R., T. Wilharm, D. Huber, A. Trincone, S. Burggraf, H. Konig, R. Rachel, I. Rockinger, H. Fricke, and K. O. Stetter. 1992. Aquifex pyrophilus gen. nov., sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxiding bacteria. Syst. Appl. Microbiol. 15**:**340-351. [Google Scholar]
- 173.Hunter, T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signalling. Cell 80**:**225-236. [DOI] [PubMed] [Google Scholar]
- 174.Hwang, C., A. J. Sinskey, and H. F. Lodish. 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257**:**1496-1502. [DOI] [PubMed] [Google Scholar]
- 175.Ikezawa, H. 1991. Bacterial PIPLCs—unique properties and usefulness in studies on GPI anchors. Cell Biol. Int. Rep. 15**:**1115-1131. [DOI] [PubMed] [Google Scholar]
- 176.Ikezawa, H. 2002. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25**:**409-417. [DOI] [PubMed] [Google Scholar]
- 177.Imhoff, J. F., D. J. Kushner, S. C. Kushwaha, and M. Kates. 1982. Polar lipids in phototrophic bacteria of the Rhodospirillaceae and Chromatiaceae families. J. Bacteriol. 150**:**1192-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Iniguez-Lluhi, J. A., M. I. Simon, J. D. Robishaw, and A. G. Gilman. 1992. G protein beta gamma subunits synthesized in Sf9 cells. Functional characterization and the significance of prenylation of gamma. J. Biol. Chem. 267**:**23409-23417. [PubMed] [Google Scholar]
- 179.Irihimovitch, V., and J. Eichler. 2003. Posttranslational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278**:**12881-12887. [DOI] [PubMed] [Google Scholar]
- 180.Isupov, M. N., T. M. Fleming, A. R. Dalby, G. S. Crowhurst, P. C. Bourne, and J. A. Littlechild. 1999. Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 291**:**651-660. [DOI] [PubMed] [Google Scholar]
- 181.Jakob, U., W. Muse, M. Eser, and J. C. Bardwell. 1999. Chaperone activity with a redox switch. Cell 96**:**341-352. [DOI] [PubMed] [Google Scholar]
- 182.Jallepalli, P. V., G. W. Brown, M. Muzi-Falconi, D. Tien, and T. J. Kelly. 1997. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 11**:**2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jansson, B. P., L. Malandrin, and H. E. Johansson. 2000. Cell cycle arrest in archaea by the hypusination inhibitor N(1)-guanyl-1,7-diaminoheptane. J. Bacteriol. 182**:**1158-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jarrell, K. F., D. P. Bayley, and A. S. Kostyukova. 1996. The archaeal flagellum: a unique motility structure. J. Bacteriol. 178**:**5057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jarrell, K. F., and S. F. Koval. 1989. Ultrastructure and biochemistry of Methanococcus voltae. Crit. Rev. Microbiol. 17**:**53-87. [DOI] [PubMed] [Google Scholar]
- 186.Jay, G. D., D. J. Culp, and M. R. Jahnke. 1990. Silver staining of extensively glycosylated proteins on sodium dodecyl sulfate-polyacrylamide gels: enhancement by carbohydrate-binding dyes. Anal. Biochem. 185**:**324-330. [DOI] [PubMed] [Google Scholar]
- 187.Jensen, O. N. 2004. Modification-specific proteomics: characterization of posttranslational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8**:**33-41. [DOI] [PubMed] [Google Scholar]
- 188.Jeon, S. J., S. Fujiwara, M. Takagi, T. Tanaka, and T. Imanaka. 2002. Tk-PTP, protein tyrosine/serine phosphatase from hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Commun. 295**:**508-514. [DOI] [PubMed] [Google Scholar]
- 189.Johnson, L. N., and D. Barford. 1993. The effects of phosphorylation on the structure and function of proteins. Annu. Rev. Biophys. Biomol. Struct. 22**:**199-232. [DOI] [PubMed] [Google Scholar]
- 190.Jones, C. J., and S. Aizawa. 1991. The bacterial flagellum and flagellar motor: structure, assembly and function. Adv. Microb. Physiol. 32**:**109-172. [DOI] [PubMed] [Google Scholar]
- 191.Jones, W. J., and G. U. Holzer. 1991. The polar and neutral lipid composition of Methanosphaera stadtmanae. Syst. Appl. Microbiol. 14**:**130-134. [Google Scholar]
- 192.Joo, W. A., and C. W. Kim. 2005. Proteomics of halophilic archaea. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 815**:**237-250. [DOI] [PubMed] [Google Scholar]
- 193.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72**:**111-135. [DOI] [PubMed] [Google Scholar]
- 194.Kahn, R. A., P. Randazzo, T. Serafini, O. Weiss, C. Rulka, J. Clark, M. Amherdt, P. Roller, L. Orci, and J. E. Rothman. 1992. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J. Biol. Chem. 267**:**13039-13046. [PubMed] [Google Scholar]
- 195.Kalmokoff, M. L., and K. F. Jarrell. 1991. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J. Bacteriol. 173**:**7113-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kalmokoff, M. L., S. F. Koval, and K. F. Jarrell. 1992. Relatedness of the flagellins from methanogens. Arch. Microbiol. 157**:**481-487. [DOI] [PubMed] [Google Scholar]
- 197.Kamekura, M. M. Dyall-Smith, V. Upasani, A. Ventosa, and M. Kates. 1997. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int. J. Syst. Bacteriol. 47**:**853-857. [DOI] [PubMed] [Google Scholar]
- 198.Kamekura, M., Y. Seno, and M. Dyall-Smith. 1996. Halolysin R4, a serine proteinase from the halophilic archaeon Haloferax mediterranei; gene cloning, expression and structural studies. Biochim. Biophys. Acta 1294**:**159-167. [DOI] [PubMed] [Google Scholar]
- 199.Kanai, T., S. Ito, and T. Imanaka. 2003. Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185**:**1705-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kane, P. M., C. T. Yamashiro, D. F. Wolczyk, N. Neff, M. Goebl, and T. H. Stevens. 1990. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science 250**:**651-657. [DOI] [PubMed] [Google Scholar]
- 201.Kang, M. S., and A. D. Elbein. 1979. Incorporation of glucose into lipid-linked saccharides in aorta and its inhibition by amphomycin. Arch. Biochem. Biophys. 198**:**304-313. [DOI] [PubMed] [Google Scholar]
- 202.Kang, M. S., J. P. Spencer, and A. D. Elbein. 1978. Amphomycin inhibition of mannose and GlcNAc incorporation into lipid-linked saccharides. J. Biol. Chem. 253**:**8860-8866. [PubMed] [Google Scholar]
- 203.Karadzic, I. M., and J. A. Maupin-Furlow. 2005. Improvement of two-dimensional gel electrophoresis proteome maps of the haloarchaeon Haloferax volcanii. Proteomics 5**:**354-359. [DOI] [PubMed] [Google Scholar]
- 204.Kärcher, U., H. Schröder, E. Haslinger, G. Allmaier, R. Schreiner, F. Wieland, A. Haselbeck, and H. König. 1993. Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus. J. Biol. Chem. 268**:**26821-26826. [PubMed] [Google Scholar]
- 205.Kashima, Y., and K. Ishikawa. 2003. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii. Arch. Biochem. Biophys. 418**:**179-185. [DOI] [PubMed] [Google Scholar]
- 206.Kates, M. 1978. The phytanyl ether-linked polar lipids and isoprenoid neutral lipids of extremely halophilic bacteria. Prog. Chem. Fats Other Lipids 15**:**301-342. [DOI] [PubMed] [Google Scholar]
- 207.Kates, M. 1992. Archaebacterial lipids: structure, biosynthesis and function. Biochem. Soc. Symp. 58**:**51-72. [PubMed] [Google Scholar]
- 208.Kates, M. 1993. Membrane lipids of archaea, p. 261-295. In M. Kates, D. J. Kushner and A. T. Matheson, (ed.), The biochemistry of archaea (archaebacteria). Elsevier Science, Amsterdam, The Netherlands.
- 209.Kawashima, T., N. Amano, H. Koike, S. Makino, S. Higuchi, Y. Kawashima-Ohya, K. Watanabe, M. Yamazaki, K. Kanehori, T. Kawamoto, T. Nunoshiba, Y. Yamamoto, H. Aramaki, K. Makino, and M. Suzuki. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 97**:**14257-14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Keeling, P. J., and W. F. Doolittle. 1996. Methionine aminopeptidase-1: the MAP of the mitochondrion? Trends Biochem. Sci. 21**:**285-286. [PubMed] [Google Scholar]
- 211.Kehry, M., C. Sibley, J. Schilling, and L. Hood. 1979. Amino acid sequence of a mouse immunoglobulin mu chain. Proc. Natl. Acad. Sci. USA 76**:**2832-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69**:**829-880. [DOI] [PubMed] [Google Scholar]
- 213.Kennelly, P. J. 2001. Protein phosphatases—a phylogenetic perspective. Chem. Rev. 101**:**2291-2312. [DOI] [PubMed] [Google Scholar]
- 214.Kennelly, P. J. 2002. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206**:**1-8. [DOI] [PubMed] [Google Scholar]
- 215.Kennelly, P. J. 2003. Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem. J. 370**:**373-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kennelly, P. J., K. A. Oxenrider, J. Leng, J. S. Cantwell, and N. Zhao. 1993. Identification of a serine/threonine-specific protein phosphatase from the archaebacterium Sulfolobus solfataricus. J. Biol. Chem. 268**:**6505-6510. [PubMed] [Google Scholar]
- 217.Kessel, M., I. Wildhaber, S. Cohen, and W. Baumeister. 1988. Three-dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead Sea. EMBO J. 7**:**1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kikuchi, A., H. Sagami, and K. Ogura. 1999. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J. Biol. Chem. 274**:**18011-18016. [DOI] [PubMed] [Google Scholar]
- 219.Kim, B. K., T. D. Pihl, J. N. Reeve, and L. Daniels. 1995. Purification of the copper response extracellular proteins secreted by the copper-resistant methanogen Methanobacterium bryantii BKYH and cloning, sequencing, and transcription of the gene encoding these proteins. J. Bacteriol. 177**:**7178-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kim, D., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147**:**1197-1212. [DOI] [PubMed] [Google Scholar]
- 221.Kim, K. K., L. W. Hung, H. Yokota, R. Kim, and S. H. Kim. 1998. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution. Proc. Natl. Acad. Sci. USA 95**:**10419-10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kimura, Y., and T. C. Stadtman. 1995. Glycine reductase selenoprotein A is not a glycoprotein: the positive periodic acid-Schiff reagent test is the result of peptide bond cleavage and carbonyl group generation. Proc. Natl. Acad. Sci. USA 92**:**2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kinch, L. N., M. H. Saier, Jr., and N. V. Grishin. 2002. Sec61beta—a component of the archaeal protein secretory system. Trends Biochem. Sci. 27**:**170-171. [DOI] [PubMed] [Google Scholar]
- 224.Kobayashi, T., R. Nishizaki, and H. Ikezawa. 1997. The presence of GPI-linked protein(s) in an archaeobacterium, Sulfolobus acidocaldarius, closely related to eukaryotes. Biochim. Biophys. Acta 1334**:**1-4. [DOI] [PubMed] [Google Scholar]
- 225.Koch, M. K., and D. Oesterhelt. 2005. MpcT is the transducer for membrane potential changes in Halobacterium salinarum. Mol. Microbiol. 55**:**1681-1694. [DOI] [PubMed] [Google Scholar]
- 226.Koga, Y., M. Nishihara, H. Morii, and M. Akagawa-Matsushita. 1993. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol. Rev. 57**:**164-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kokoeva, M. V., and D. Oesterhelt. 2000. BasT, a membrane-bound transducer protein for amino acid detection in Halobacterium salinarum. Mol. Microbiol. 35**:**647-656. [DOI] [PubMed] [Google Scholar]
- 228.Kokoeva, M. V., K. F. Storch, C. Klein, and D. Oesterhelt. 2002. A novel mode of sensory transduction in archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 21**:**2312-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kondoh, H., C. B. Ball, and J. Adler. 1979. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 76**:**260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Koning, S. M., M. G. Elferink, W. N. Konings, and A. J. Driessen. 2001. Cellobiose uptake in the hyperthermophilic archaeon Pyrococcus furiosus is mediated by an inducible, high-affinity ABC transporter. J. Bacteriol. 183**:**4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Koning, S. M., W. N. Konings, and A. J. M. Driessen. 2002. Biochemical evidence for the presence of two α-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1**:**19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Konrad, Z., and J. Eichler. 2002. Protein glycosylation in Haloferax volcanii: partial characterization of a 98-kDa glycoprotein. FEMS Microbiol. Lett. 209**:**197-202. [DOI] [PubMed] [Google Scholar]
- 233.Konrad, Z., and J. Eichler. 2002. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochem. J. 366**:**959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2000. Evolution of two-component signal transduction. Mol. Biol. Evol. 17**:**1956-1970. [DOI] [PubMed] [Google Scholar]
- 235.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54**:**631-664. [DOI] [PubMed] [Google Scholar]
- 236.Kort, E. N., M. F. Goy, S. H. Larsen, and J. Adler. 1975. Methylation of a membrane protein involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 72**:**3939-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19**:**1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12**:**198-209. [DOI] [PubMed] [Google Scholar]
- 239.Krah, M., W. Marwan, A. Vermeglio, and D. Oesterhelt. 1994. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 13**:**2150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Krebs, E. G., and E. H. Fischer. 1989. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. 1956. Biochim. Biophys. Acta 1000**:**302-309. [PubMed] [Google Scholar]
- 241.Kunkel, A., J. A. Vorholt, R. K. Thauer, and R. Hedderich. 1998. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur. J. Biochem. 252**:**467-476. [DOI] [PubMed] [Google Scholar]
- 242.Kuntz, C., J. Sonnenbichler, I. Sonnenbichler, M. Sumper, and R. Zeitler. 1997. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology 7**:**897-904. [DOI] [PubMed] [Google Scholar]
- 243.Kushwaha, S. C., M. B. Gochnauer, D. J. Kushner, and M. Kates. 1974. Pigments and isoprenoid compounds in extremely and moderately halophilic bacteria. Can. J. Microbiol. 20**:**241-245. [DOI] [PubMed] [Google Scholar]
- 244.Lanyi, J. K. 2004. Bacteriorhodopsin. Annu. Rev. Physiol. 66**:**665-688. [DOI] [PubMed] [Google Scholar]
- 245.LaRonde-LeBlanc, N., and A. Wlodawer. 2004. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure (Cambridge) 12**:**1585-1594. [DOI] [PubMed] [Google Scholar]
- 246.Lechner, J., and M. Sumper. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262**:**9724-9729. [PubMed] [Google Scholar]
- 247.Lechner, J., and F. Wieland. 1989. Structure and biosynthesis of prokaryotic glycoproteins. Annu. Rev. Biochem. 58**:**173-194. [DOI] [PubMed] [Google Scholar]
- 248.Lechner, J., F. Wieland, and M. Sumper. 1985. Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J. Biol. Chem. 260**:**860-866. [PubMed] [Google Scholar]
- 249.Lechner, J., F. Wieland, and M. Sumper. 1985. Transient methylation of dolichyl oligosaccharides is an obligatory step in halobacterial sulfated glycoprotein biosynthesis. J. Biol. Chem. 260**:**8984-8989. [PubMed] [Google Scholar]
- 250.Lee, D. Y., B. Y. Ahn, and K. S. Kim. 2000. A thioredoxin from the hyperthermophilic archaeon Methanococcus jannaschii has a glutaredoxin-like fold but thioredoxin-like activities. Biochemistry 39**:**6652-6659. [DOI] [PubMed] [Google Scholar]
- 251.Lee, J. H., S. K. Choi, A. Roll-Mecak, S. K. Burley, and T. E. Dever. 1999. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. USA 96**:**4342-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Leng, J., A. J. Cameron, S. Buckel, and P. J. Kennelly. 1995. Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J. Bacteriol. 177**:**6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Leonard, C. J., L. Aravind, and E. V. Koonin. 1998. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 8**:**1038-1047. [DOI] [PubMed] [Google Scholar]
- 254.Levine, M. J., and R. G. Spiro. 1979. Isolation from glomerular basement membrane of a glycopeptide containing both asparagine-linked and hydroxylysine-linked carbohydrate units. J. Biol. Chem. 254**:**8121-8124. [PubMed] [Google Scholar]
- 255.Levitzki, A., and A. Gazit. 1995. Tyrosine kinase inhibition: an approach to drug development. Science 267**:**1782-1788. [DOI] [PubMed] [Google Scholar]
- 256.Li, L. C., S. T. Okino, and R. Dahiya. 2004. DNA methylation in prostate cancer. Biochim. Biophys. Acta 1704**:**87-102. [DOI] [PubMed] [Google Scholar]
- 257.Li, X., and Y. H. Chang. 1995. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA 92**:**12357-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lin, X., and J. Tang. 1990. Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J. Biol. Chem. 265**:**1490-1495. [PubMed] [Google Scholar]
- 259.Lindbeck, J. C., E. A. Goulbourne, Jr., M. S. Johnson, and B. L. Taylor. 1995. Aerotaxis in Halobacterium salinarium is methylation-dependent. Microbiology 141**:**2945-2953. [DOI] [PubMed] [Google Scholar]
- 260.Liu, X. Q. 2000. Protein-splicing intein: genetic mobility, origin, and evolution. Annu. Rev. Genet. 34**:**61-76. [DOI] [PubMed] [Google Scholar]
- 261.Lower, B. H., K. M. Bischoff, and P. J. Kennelly. 2000. The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase activity that preferentially phosphorylates threonine residues. J. Bacteriol. 182**:**3452-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lower, B. H., and P. J. Kennelly. 2002. The membrane-associated protein-serine/threonine kinase from Sulfolobus solfataricus is a glycoprotein. J. Bacteriol. 184**:**2614-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lower, B. H., and P. J. Kennelly. 2003. Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J. Bacteriol. 185**:**3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lower, B. H., M. B. Potters, and P. J. Kennelly. 2004. A phosphoprotein from the archaeon Sulfolobus solfataricus with protein-serine/threonine kinase activity. J. Bacteriol. 186**:**463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57**:**77-100. [DOI] [PubMed] [Google Scholar]
- 266.Madern, D, C. Ebel, and G. Zaccai. 2000. Halophilic adaptation of enzymes. Extremophiles 4**:**91-98. [DOI] [PubMed] [Google Scholar]
- 267.Magnuson, K., S. Jackowski, C. O. Rock, and J. E. Cronan. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57**:**522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mai, B., G. Frey, R. V. Swanson, E. J. Mathur, and K. O. Stetter. 1998. Molecular cloning and functional expression of a protein-serine/threonine phosphatase from the hyperthermophilic archaeon Pyrodictium abyssi TAG11. J. Bacteriol. 180**:**4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mallick, P., D. R. Boutz, D. Eisenberg, and T. O. Yeates. 2002. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl. Acad. Sci. USA 99**:**9679-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mann, M., and O. N. Jensen. 2003. Proteomic analysis of posttranslational modifications. Nat. Biotechnol. 21**:**255-261. [DOI] [PubMed] [Google Scholar]
- 271.Mann, R. K., and P. A. Beachy. 2004. Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73**:**891-923. [DOI] [PubMed] [Google Scholar]
- 272.Maras, B., V. Consalvi, R. Chiaraluce, L. Politi, M. De Rosa, F. Bossa, R. Scandurra, and D. Barra. 1992. The protein sequence of glutamate dehydrogenase from Sulfolobus solfataricus, a thermoacidophilic archaebacterium. Is the presence of N-epsilon-methyllysine related to thermostability? Eur. J. Biochem. 203**:**81-87. [DOI] [PubMed] [Google Scholar]
- 273.Martin, J. L. 1995. Thioredoxin—a fold for all reasons. Structure 3**:**245-250. [DOI] [PubMed] [Google Scholar]
- 274.Mattar, S., B. Scharf, S. B. Kent, K. Rodewald, D. Oesterhelt, and M. Engelhard. 1994. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem. 269**:**14939-14945. [PubMed] [Google Scholar]
- 275.Maupin-Furlow, J. A., and J. G. Ferry. 1995. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J. Biol. Chem. 270**:**28617-28622. [DOI] [PubMed] [Google Scholar]
- 276.Maupin-Furlow, J. A., S. J. Kaczowka, M. S. Ou, and H. L. Wilson. 2001. Archaeal proteasomes: proteolytic nanocompartments of the cell. Adv. Appl. Microbiol. 50**:**279-338. [DOI] [PubMed] [Google Scholar]
- 277.Maupin-Furlow, J. A., H. L. Wilson, S. J. Kaczowka, and M. S. Ou. 2000. Proteasomes in the archaea: from structure to function. Front. Biosci. 5**:**D837-D865. [DOI] [PubMed] [Google Scholar]
- 278.McAfee, J. G., S. P. Edmondson, P. K. Datta, J. W. Shriver, and R. Gupta. 1995. Gene cloning, expression, and characterization of the Sac7 proteins from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry 34**:**10063-10077. [DOI] [PubMed] [Google Scholar]
- 279.McFarlan, S. C., C. A. Terrell, and H. P. Hogenkamp. 1992. The purification, characterization, and primary structure of a small redox protein from Methanobacterium thermoautotrophicum, an archaebacterium. J. Biol. Chem. 267**:**10561-10569. [PubMed] [Google Scholar]
- 280.Mengele, R., and M. Sumper. 1992. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 267**:**8182-8185. [PubMed] [Google Scholar]
- 281.Mescher, M. F., U. Hansen, and J. L. Strominger. 1976. Formation of lipid-linked sugar compounds in Halobacterium salinarium. Presumed intermediates in glycoprotein synthesis. J. Biol. Chem. 251**:**7289-7294. [PubMed] [Google Scholar]
- 282.Mescher, M. F., and J. L. Strominger. 1976. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc. Natl. Acad. Sci. USA 73**:**2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mescher, M. F., and J. L. Strominger. 1976. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 251**:**2005-2014. [PubMed] [Google Scholar]
- 284.Mescher, M. F., and J. L. Strominger. 1978. Glycosylation of the surface glycoprotein of Halobacterium salinarium via a cyclic pathway of lipid-linked intermediates. FEBS Lett. 89**:**37-41. [DOI] [PubMed] [Google Scholar]
- 285.Messner, P. 2004. Prokaryotic glycoproteins: unexplored but important. J. Bacteriol. 186**:**2517-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Meyer, W., and G. Schafer. 1992. Characterization and purification of a membrane-bound archaebacterial pyrophosphatase from Sulfolobus acidocaldarius. Eur. J. Biochem. 207**:**741-746. [DOI] [PubMed] [Google Scholar]
- 287.Milligan, G., M. Parenti, and A. I. Magee. 1995. The dynamic role of palmitoylation in signal transduction. Trends Biochem. Sci. 20**:**181-186. [DOI] [PubMed] [Google Scholar]
- 288.Mills, K. V., J. S. Manning, A. M. Garcia, and L. A. Wuerdeman. 2004. Protein splicing of a Pyrococcus abyssi intein with a C-terminal glutamine. J. Biol. Chem. 279**:**20685-20691. [DOI] [PubMed] [Google Scholar]
- 289.Minami, Y., S. Wakabayashi, K. Wada, H. Matsubara, L. Kerscher, and D. Oesterhelt. 1985. Amino acid sequence of a ferredoxin from thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. Presence of an N6-monomethyllysine and phyletic consideration of archaebacteria. J. Biochem. (Tokyo) 97**:**745-753. [DOI] [PubMed] [Google Scholar]
- 290.Mizanur, R. M., C. J. Zea, and N. L. Pohl. 2004. Unusually broad substrate tolerance of a heat-stable archaeal sugar nucleotidyltransferase for the synthesis of sugar nucleotides. J. Am. Chem. Soc. 126**:**15993-15998. [DOI] [PubMed] [Google Scholar]
- 291.Moens, S., K. Michiels, and J. van der Leyden. 1995. Glycosylation of the flagellin of the polar flagellum of Azospirillum brasilense, a gram-negative nitrogen-fixing bacterium. Microbiology 141**:**2651-2657. [Google Scholar]
- 292.Moens, S., and J. Vanderleyden. 1997. Glycoproteins in prokaryotes. Arch. Microbiol. 168**:**169-175. [DOI] [PubMed] [Google Scholar]
- 293.Moll, R. G. 2004. The archaeal signal recognition particle: steps toward membrane binding. J. Bioenerg. Biomembr. 36**:**47-53. [DOI] [PubMed] [Google Scholar]
- 294.Moll, R. G., and G. Schafer. 2004. Novel functional aspects of the membrane-bound exo-pyrophosphatase of the hyperthermoacidophilic archaeon Sulfolobus are provided by analysis of its gene and the adjacent gene cluster. J. Bioenerg. Biomembr. 36**:**143-150. [DOI] [PubMed] [Google Scholar]
- 295.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97**:**11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Mulloy, B., and M. J. Forster. 2000. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 10**:**1147-1156. [DOI] [PubMed] [Google Scholar]
- 297.Muñiz, M., and H. Riezman. 2000. Intracellular transport of GPI-anchored proteins. EMBO J. 19**:**10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Nagai, K., C. Oubridge, A. Kuglstatter, E. Menichelli, C. Isel, and L. Jovine. 2003. Structure, function and evolution of the signal recognition particle. EMBO J. 22**:**3479-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nakamura, S., S. Mizutani, H. Wakai, H. Kawasaki, R. Aono, and K. Horikoshi. 1995. Purification and partial characterization of cell surface glycoprotein from extremely halophilic archaeon Haloarcula japonica strain TR-1. Biotechnol. Lett. 17**:**705-706. [Google Scholar]
- 300.Nercessian, D., R. E. de Castro, and R. D. Conde. 2002. Ubiquitin-like proteins in halobacteria. J. Basic Microbiol. 42**:**277-283. [DOI] [PubMed] [Google Scholar]
- 301.Newton, G. L., and B. Javor. 1985. γ-Glutamylcysteine and thiosulfate are the major low-molecular-weight thiols in halobacteria. J. Bacteriol. 161**:**438-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ng, S. Y., and K. F. Jarrell. 2003. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae. J. Bacteriol. 185**:**5936-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97**:**12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Nichols, D. S., M. R. Miller, N. W. Davies, A. Goodchild, M. Raftery, and R. Cavicchioli. 2004. Cold adaptation in the Antarctic archaeon Methanococcoides burtonii involves membrane lipid unsaturation. J. Bacteriol. 186**:**8508-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Niehaus, F., B. Frey, and G. Antranikian. 1997. Cloning and characterisation of a thermostable alpha-DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. TY. Gene 204**:**153-158. [DOI] [PubMed] [Google Scholar]
- 306.Nielsen, H., Brunak, S., and von Heijne, G. 1999. Machine learning approaches for the prediction of signal peptides and other learning sorting signals. Protein Eng. 12**:**3-9. [DOI] [PubMed] [Google Scholar]
- 307.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10**:**1-6. [DOI] [PubMed] [Google Scholar]
- 308.Niewmierzycka, A., and S. Clarke. 1999. _S_-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel arginine methyltransferase. J. Biol. Chem. 274**:**814-824. [DOI] [PubMed] [Google Scholar]
- 309.Nilsson, I., D. J. Kelleher, Y. Miao, Y. Shao, G. Kreibich, R. Gilmore, G. von Heijne, and A. E. Johnson. 2003. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 161**:**715-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Nishihara, M., M. Utagawa, H. Akutsu, and Y. Koga. 1992. Archaea contain a novel diether phosphoglycolipid with a polar head group identical to the conserved core of eucaryal glycosyl phosphatidylinositol. J. Biol. Chem. 267**:**12432-12435. [PubMed] [Google Scholar]
- 311.Nölling, J., A. Elfner, J. R. Palmer, V. J. Steigerwald, T. D. Pihl, J. A. Lake, and J. N. Reeve. 1996. Phylogeny of Methanopyrus kandleri based on methyl coenzyme M reductase operons. Int. J. Syst. Bacteriol. 46**:**1170-1173. [DOI] [PubMed] [Google Scholar]
- 312.Noren, C. J., J. Wang, and F. B. Perler. 2000. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. 39**:**450-466. [PubMed] [Google Scholar]
- 313.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20**:**214-220. [DOI] [PubMed] [Google Scholar]
- 314.Nurse, P. M. 2002. Nobel Lecture: Cyclin dependent kinases and cell cycle control. Biosci. Rep. 22**:**487-499. [DOI] [PubMed] [Google Scholar]
- 315.O'Connor, B. D., and T. O. Yeates. 2004. GDAP: a web tool for genome-wide protein disulfide bond prediction. Nucleic Acids Res. 32(Web Server issue)**:**W360-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ogasahara, K., N. N. Khechinashvili, M. Nakamura, T. Yoshimoto, and K. Yutani. 2001. Thermal stability of pyrrolidone carboxyl peptidases from the hyperthermophilic archaeon, Pyrococcus furiosus. Eur. J. Biochem. 268**:**3233-3242. [DOI] [PubMed] [Google Scholar]
- 317.Oppermann, U. C., S. Knapp, V. Bonetto, R. Ladenstein, and H. Jornvall. 1998. Isolation and structure of repressor-like proteins from the archaeon Sulfolobus solfataricus. Copurification of RNase A with Sso7c. FEBS Lett. 432**:**141-144. [DOI] [PubMed] [Google Scholar]
- 318.Ortenberg, R., and M. Mevarech. 2000. Evidence for posttranslational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J. Biol. Chem. 275**:**22839-22846. [DOI] [PubMed] [Google Scholar]
- 319.Osorio, G., and C. A. Jerez. 1996. Adaptive response of the archaeon Sulfolobus acidocaldarius BC65 to phosphate starvation. Microbiology 142**:**1531-1536. [DOI] [PubMed] [Google Scholar]
- 320.Oxenrider, K. A., and P. J. Kennelly. 1993. A protein-serine phosphatase from the halophilic archaeon Haloferax volcanii. Biochem. Biophys. Res. Commun. 194**:**1330-1335. [DOI] [PubMed] [Google Scholar]
- 321.Oxenrider, K. A., M. E. Rasche, M. V. Thorsteinsson, and P. J. Kennelly. 1993. Inhibition of an archaeal protein phosphatase activity by okadaic acid, microcystin-LR, or calyculin A. FEBS Lett. 331**:**291-295. [DOI] [PubMed] [Google Scholar]
- 322.Pace, C. N., G. R. Grimsley, J. A. Thomson, and B. J. Barnett. 1988. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J. Biol. Chem. 263**:**11820-11825. [PubMed] [Google Scholar]
- 323.Paetzel, M., R. E. Dalbey, and N. C. J. Strynadka. 1998. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature 396**:**186-190. [DOI] [PubMed] [Google Scholar]
- 324.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102**:**4549-4580. [DOI] [PubMed] [Google Scholar]
- 325.Paget, M. S., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 37**:**91-121. [DOI] [PubMed] [Google Scholar]
- 326.Paik, W. K., and S. Kim. 1971. Protein methylation. Science 174**:**114-119. [DOI] [PubMed] [Google Scholar]
- 327.Paik, W. K., and S. Kim. 1986. Enzymology of protein methylation Yonsei Med. J. 27**:**159-177. [DOI] [PubMed] [Google Scholar]
- 328.Paltauf, F. 1994. Ether lipids in biomembranes. Chem. Phys. Lipids 74**:**101-139. [DOI] [PubMed] [Google Scholar]
- 329.Park, M. H., H. L. Cooper, and J. E. Folk. 1981. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 78**:**2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Park, M. H., E. C. Wolff, and J. E. Folk. 1993. Is hypusine essential for eukaryotic cell proliferation? Trends Biochem. Sci. 12**:**475-479. [DOI] [PubMed] [Google Scholar]
- 331.Park, M. H., E. C. Wolff, and J. E. Folk. 1993. Hypusine: its posttranslational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 4**:**95-104. [PubMed] [Google Scholar]
- 332.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26**:**71-112. [DOI] [PubMed] [Google Scholar]
- 333.Parodi, A. J. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348**:**1-13. [PMC free article] [PubMed] [Google Scholar]
- 334.Patel, G. B., and G. D. Sprott. 1999. Archaeobacterial ether lipid liposomes (archaeosomes) as novel vaccine and drug delivery systems. Crit. Rev. Biotechnol. 19**:**317-357. [DOI] [PubMed] [Google Scholar]
- 335.Paul, G., F. Lottspeich, and F. Wieland. 1986. Asparaginyl-_N_-acetylgalactosamine. Linkage unit of halobacterial glycosaminoglycan. J. Biol. Chem. 261**:**1020-1024. [PubMed] [Google Scholar]
- 336.Paul, P., T. M. Lutz, C. Osborn, S. Kyosseva, A. D. Elbein, H. Towbin, A. Radominska, R. R. Drake. 1993. Synthesis and characterization of a new class of inhibitors of membrane-associated UDP-glycosyltransferases. J. Biol. Chem. 268**:**12933-12938. [PubMed] [Google Scholar]
- 337.Paulus, H. 2000. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69**:**447-496. [DOI] [PubMed] [Google Scholar]
- 338.Peat, T. S., J. Newman, G. S. Waldo, J. Berendzen, and T. C. Terwilliger. 1998. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution. Structure 6**:**1207-1214. [DOI] [PubMed] [Google Scholar]
- 339.Pedone, E., B. Ren, R. Ladenstein, M. Rossi, and S. Bartolucci. 2004. Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a member of a novel protein family related to protein disulfide-isomerase. Eur. J. Biochem. 271**:**3437-3448. [DOI] [PubMed] [Google Scholar]
- 340.Pellerin, P., B. Fournet, and P. Debeire. 1990. Evidence for the glycoprotein nature of the cell sheath of _Methanosaeta_-like cells in the culture of Methanothrix soehngenii strain FE. Can. J. Microbiol. 36**:**631-636. [Google Scholar]
- 341.Perazzona, B., and J. L. Spudich. 1999. Identification of methylation sites and effects of phototaxis stimuli on transducer methylation in Halobacterium salinarum. J. Bacteriol. 181**:**5676-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Pereto, J., P. Lopez-Garcia, and D. Moreira. 2004. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 29**:**469-477. [DOI] [PubMed] [Google Scholar]
- 343.Perler, F. B. 2002. InBase, the intein database. Nucleic Acids Res. 30**:**383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Perler, F. B., D. G. Comb, W. E. Jack, L. S. Moran, B. Qiang, R. B. Kucera, J. Benner, B. E. Slatko, D. O. Nwankwo, S. K. Hempstead, C. K. S. Carlow, and H. Jannasch. 1992. Intervening sequences in an Archaea DNA polymerase gene. Proc. Natl. Acad. Sci. USA 89**:**5577-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Peters, J., M. Nitsch, B. Kühlmorgen, R. Golbik, A. Lupas, J. Kellermann, H. Engelhardt, J. P. Pfander, S. Müller, K. Goldie, A. Engel, K. O. Stetter, and W. Baumeister. 1995. Tetrabrachion: a filamentous archaebacterial surface protein assembly of unusual structure and extreme stability. J. Mol. Biol. 245**:**385-401. [DOI] [PubMed] [Google Scholar]
- 346.Pietrokovski, S. 2001. Intein spread and extinction in evolution. Trends Genet. 17**:**465-472. [DOI] [PubMed] [Google Scholar]
- 347.Pohlschroder, M., K. Dilks, N. J. Hand, and W. R. Rose. 2004. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol. Rev. 28**:**3-24. [DOI] [PubMed] [Google Scholar]
- 348.Powis, G., D. Mustacich, and A. Coon. 2000. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic. Biol. Med. 29**:**312-322. [DOI] [PubMed] [Google Scholar]
- 349.Proud, C. G. 1992. Protein phosphorylation in translational control. Curr. Top. Cell Regul. 32**:**243-369. [DOI] [PubMed] [Google Scholar]
- 350.Pugh, E. L., and M. Kates. 1994. Acylation of proteins of the archaebacteria Halobacterium cutirubrum and Methanobacterium thermoautotrophicum. Biochim. Biophys. Acta 1196**:**38-44. [DOI] [PubMed] [Google Scholar]
- 351.Raina, S., and D. Missiakas. 1997. Making and breaking disulfide bonds. Annu. Rev. Microbiol. 51**:**179-202. [DOI] [PubMed] [Google Scholar]
- 352.Rakhely, G., Z. H. Zhou, M. W. Adams, and K. L. Kovacs. 1999. Biochemical and molecular characterization of the [NiFe] hydrogenase from the hyperthermophilic archaeon, Thermococcus litoralis. Eur. J. Biochem. 266**:**1158-1165. [DOI] [PubMed] [Google Scholar]
- 353.Ramirez, C., L. C. Shimmin, P. P. Dennis, and A. T. Matheson. 1991. Comparison of the structure of archaebacterial ribosomal proteins equivalent to proteins L11 and L1 from Escherichia coli ribosomes. Protein Seq. Data Anal. 4**:**75-79. [PubMed] [Google Scholar]
- 354.Ramirez, C., L. C. Shimmin, C. H. Newton, A. T. Matheson, and P. P. Dennis. 1989. Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can. J. Microbiol. 35**:**234-244. [DOI] [PubMed] [Google Scholar]
- 355.Reeve, J. N. 2003. Archaeal chromatin and transcription. Mol. Microbiol. 48**:**587-598. [DOI] [PubMed] [Google Scholar]
- 356.Ren, B., G. Tibbelin, D. de Pascale, M. Rossi, S. Bartolucci, and R. Ladenstein. 1998. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat. Struct. Biol. 5**:**602-611. [DOI] [PubMed] [Google Scholar]
- 357.Rensing, S. A., and U. G. Maier. 1994. The SecY protein family: comparative analysis and phylogenetic relationships. Mol. Phylogenet. Evol. 3**:**187-191. [DOI] [PubMed] [Google Scholar]
- 358.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451**:**1-16. [DOI] [PubMed] [Google Scholar]
- 359.Riera, J., F. T. Robb, R. Weiss, and M. Fontecave. 1997. Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl. Acad. Sci. USA 94**:**475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Ring, G., and J. Eichler. 2004. In the archaeon Haloferax volcanii, membrane protein biogenesis and protein synthesis rates are affected by decreased ribosomal-binding to the translocon. J. Biol. Chem. 279**:**53160-53166. [DOI] [PubMed] [Google Scholar]
- 361.Ring, G., and J. Eichler. 2004. Extreme secretion: protein translocation across the archaeal plasma membrane. J. Bioenerg. Biomembr. 36**:**35-45. [DOI] [PubMed] [Google Scholar]
- 362.Rip, J. W., C. A. Rupar, K. Ravi, and K. K. Carroll. 1985. Distribution, metabolism and function of dolichol and polyprenols. Prog. Lipid Res. 24**:**269-309. [DOI] [PubMed] [Google Scholar]
- 363.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55**:**21-48. [DOI] [PubMed] [Google Scholar]
- 364.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50**:**285-315. [DOI] [PubMed] [Google Scholar]
- 365.Roberts, W. L., J. J. Myher, A. Kuksis, M. G. Low, and T. L. J. Rosenberry. 1988. Lipid analysis of the glycoinositol phospholipid membrane anchor of human erythrocyte acetylcholinesterase. Palmitoylation of inositol results in resistance to phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 263**:**18766-18775. [PubMed] [Google Scholar]
- 366.Robinson, C., and A. Bolhuis. 2004. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim. Biophys. Acta 1694**:**135-147. [DOI] [PubMed] [Google Scholar]
- 367.Robinson, H., Y. G. Gao, B. S. McCrary, S. P. Edmondson, J. W. Shriver, and A. H. Wang. 1998. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392**:**202-205. [DOI] [PubMed] [Google Scholar]
- 368.Rodriguez, A. C., H. W. Park, C. Mao, and L. S. Beese. 2000. Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J. Mol. Biol. 299**:**447-462. [DOI] [PubMed] [Google Scholar]
- 369.Rolin, S., J. Hanocq-Quertier, F. Paturiaux-Hanocq, D. Nolan, D. Salmon, H. Webb, M. Carrington, P. Voorheis, and E. Pays. 1996. Simultaneous but independent activation of adenylate cyclase and glycosylphosphatidylinositol-phospholipase C under stress conditions in Trypanosoma brucei. J. Biol. Chem. 271**:**10844-10852. [DOI] [PubMed] [Google Scholar]
- 370.Roovers, M., J. Wouters, J. M. Bujnicki, C. Tricot, V. Stalon, H. Grosjean, and L. Droogmans. 2004. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32**:**465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Rose, R. W., T. Bruser, J. C. Kissinger, and M. Pohlschroder. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45**:**943-950. [DOI] [PubMed] [Google Scholar]
- 372.Rudolph, J., B. Nordmann, K. F. Storch, H. Gruenberg, K. Rodewald, and D. Oesterhelt. 1996. A family in halobacterial transducer proteins. FEMS Microbiol. Lett. 139**:**161-168. [DOI] [PubMed] [Google Scholar]
- 373.Rudolph, J., and D. Oesterhelt. 1995. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarum. EMBO J. 14**:**667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Rudolph, J., N. Tolliday, C. Schmitt, S. C. Schuster, and D. Oesterhelt. 1995. Phosphorylation in halobacterial signal transduction. EMBO J. 14**:**4249-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Rudolph, M. J., M. M. Wuebbens, K. V. Rajagopalan, and H. Schindelin. 2001. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat. Struct. Biol. 8**:**42-46. [DOI] [PubMed] [Google Scholar]
- 376.Sagami, H., A. Kikuchi, and K. Ogura. 1994. Novel isoprenoid modified proteins in halobacteria. Biochem. Biophys. Res. Commun. 203**:**972-978. [DOI] [PubMed] [Google Scholar]
- 377.Saleh, M. T., M. Fillon, P. J. Brennan, and J. T. Belisle. 2001. Identification of putative exported/secreted proteins in prokaryotic proteomes. Gene 269**:**195-204. [DOI] [PubMed] [Google Scholar]
- 378.Sandman, K., J. A. Krzychi, B. Dobrinski, R. Lurz, and J. N. Reeve. 1990. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. USA 87**:**5788-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein: transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269**:**19701-19706. [PubMed] [Google Scholar]
- 380.Sapra, R., M. F. Verhagen, and M. W. Adams. 2000. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182**:**3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Schaffer, C., M. Graninger, and P. Messner. 2001. Prokaryotic glycosylation. Proteomics 1**:**248-261. [DOI] [PubMed] [Google Scholar]
- 382.Schäffer, C., and P. Messner. 2001. Glycobiology of surface layer proteins. Biochimie 83**:**591-599. [DOI] [PubMed] [Google Scholar]
- 383.Schaffer, C., and P. Messner. 2004. Surface-layer glycoproteins: an example for the diversity of bacterial glycosylation with promising impacts on nanobiotechnology. Glycobiology 14**:**31R-42R. [DOI] [PubMed] [Google Scholar]
- 384.Schirm, M., S. K. Arora, A. Verma, E. Vinogradov, P. Thibault, R. Ramphal, and S. M. Logan. 2004. Structural and genetic characterization of glycosylation of type-a flagellin in Pseudomonas aeruginosa. J. Bacteriol. 186**:**2523-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Schreiner, R., E. Schnabel, and F. Wieland. 1994. Novel N-glycosylation in eukaryotes: laminin contains the linkage unit beta-glucosylasparagine. J. Cell Biol. 124**:**1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Schumann, H., and F. Klink. 1989. Archaebacterial protein contains hypusine, a unique amino-acid characteristic for eukaryotic translation initiation factor-4D. Syst. Appl. Microbiol. 11**:**103-107. [Google Scholar]
- 387.Selmer, T., J. Kahnt, M. Goubeaud, S. Shima, W. Grabarse, U. Ermler, and R. K. Thauer. 2000. The biosynthesis of methylated amino acids in the active site region of methyl-coenzyme M reductase. J. Biol. Chem. 275**:**3755-3760. [DOI] [PubMed] [Google Scholar]
- 388.Senejani, A. G., E. Hilario, and J. P. Gogarten. 2001. The intein of the Thermoplasma A-ATPase A subunit: structure, evolution and expression in E. coli. BMC Biochem. 2**:**13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Sergenova, I. S., Y. Y. Polosina, A. S. Kostyukova, A. L. Metlina, M. G. Pyatibratov, and O. V. Fedorov. 1995. Flagella of halophilic archaea: biochemical and genetic analysis. Biochemistry (Moscow) 60**:**953-957. [PubMed] [Google Scholar]
- 390.Shahmohammadi, H. R., E. Asgarani, H. Terato, T. Saito, Y. Ohyama, K. Gekko, O. Yamamoto, and H. Ide. 1998. Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents. J. Radiat. Res. (Tokyo) 39**:**251-262. [DOI] [PubMed] [Google Scholar]
- 391.Shi, L. 2004. Manganese-dependent protein O-phosphatases in prokaryotes and their biological functions. Front. Biosci. 9**:**1382-1397. [DOI] [PubMed] [Google Scholar]
- 392.Silberstein, S., and R. Gilmore. 1996. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 10**:**849-858. [PubMed] [Google Scholar]
- 393.Sinensky, M. 2000. Recent advances in the study of prenylated proteins. Biochim. Biophys. Acta 1484**:**93-106. [DOI] [PubMed] [Google Scholar]
- 394.Singleton, M. R., M. N. Isupov, and J. A. Littlechild. 1999. X-ray structure of pyrrolidone carboxyl peptidase from the hyperthermophilic archaeon Thermococcus litoralis. Structure 7**:**237-244. [DOI] [PubMed] [Google Scholar]
- 395.Skorko, R. 1984. Protein phosphorylation in the archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 145**:**617-622. [DOI] [PubMed] [Google Scholar]
- 396.Skorko, R. 1989. Polyphosphate as a source of phosphoryl group in protein modification in the archaebacterium Sulfolobus acidocaldarius. Biochimie 71**:**1089-1093. [DOI] [PubMed] [Google Scholar]
- 397.Skorko, R., J. Osipiuk, and K. O. Stetter. 1989. Glycogen-bound polyphosphate kinase from the archaebacterium Sulfolobus acidocaldarius. J. Bacteriol. 171**:**5162-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Smit, A., and A. Mushegian. 2000. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 10**:**1468-1484. [DOI] [PubMed] [Google Scholar]
- 399.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179**:**7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Smith, R. F., and K. Y. King. 1995. Identification of a eukaryotic-like protein kinase gene in archaebacteria. Protein Sci. 4**:**126-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Smith, S. C., P. J. Kennelly, and M. Potts. 1997. Protein-tyrosine phosphorylation in the archaea. J. Bacteriol. 179**:**2418-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Solow, B., K. M. Bischoff, M. J. Zylka, and P. J. Kennelly. 1998. Archael phosphoproteins. Identification of a hexosephosphate mutase and the alpha-subunit of succinyl-CoA synthetase in the extreme acidothermophile Sulfolobus solfataricus. Protein Sci. 7**:**105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Solow, B., J. C. Young, and P. J. Kennelly. 1997. Gene cloning and expression and characterization of a toxin-sensitive protein phosphatase from the methanogenic archaeon Methanosarcina thermophila TM-1. J. Bacteriol. 179**:**5072-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Sorgenfrei, O., D. Linder, M. Karas, and A. Klein. 1993. A novel very small subunit of a selenium containing [NiFe] hydrogenase of Methanococcus voltae is postranslationally processed by cleavage at a defined position. Eur. J. Biochem. 213**:**1355-1358. [DOI] [PubMed] [Google Scholar]
- 405.Sorgenfrei, O., S. Muller, M. Pfeiffer, I. Sniezko, and A. Klein. 1997. The [NiFe] hydrogenases of Methanococcus voltae: genes, enzymes and regulation. Arch. Microbiol. 167**:**189-195. [DOI] [PubMed] [Google Scholar]
- 406.Southam, G., M. L. Kalmoko, K. F. Jarrell, S. F. Koval, and T. J. Beveridge. 1990. Isolation, characterization and cellular insertion of the flagella from two strains of the archaebacterium Methanospirillum hungatei. J. Bacteriol. 172**:**3221-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Southworth, M. W., J. Benner, and F. B. Perler. 2000. An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. EMBO J. 19**:**5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Southworth, M. W., H. Kong, R. B. Kucera, J. Ware, H. W. Jannasch, and F. B. Perler. 1996. Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9°N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 93**:**5281-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Spiro, R. G. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12**:**43R-56R. [DOI] [PubMed] [Google Scholar]
- 410.Springer, E., M. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45**:**554-559. [DOI] [PubMed] [Google Scholar]
- 411.Spudich, E. N., and J. L. Spudich. 1981. Photosensitive phosphoproteins in Halobacteria: regulatory coupling of transmembrane proton flux and protein dephosphorylation. J. Cell Biol. 91**:**895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Spudich, E. N., T. Takahashi, and J. L. Spudich. 1989. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc. Natl. Acad. Sci. USA 86**:**7746-7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Spudich, J. L., and W. Stoeckenius. 1980. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J. Biol. Chem. 255**:**5501-5503. [PubMed] [Google Scholar]
- 414.Stanley, P., V. Koronakis, and C. Hughes. 1998. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. Rev. 62**:**309-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Starich, M. R., K. Sandman, J. N. Reeve, and M. F. Summers. 1996. NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone. J. Mol. Biol. 255**:**187-203. [DOI] [PubMed] [Google Scholar]
- 416.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64**:**435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Storch, K. F., J. Rudolph, D. Oesterhelt. 1999. Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J. 18**:**1146-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Stravopodis, D. J., and N. C. Kyrpides. 1999. Identification of protein-tyrosine phophatases in archaea. J. Mol. Evol. 48**:**625-627. [DOI] [PubMed] [Google Scholar]
- 419.Strom, M. S., D. N. Nunn, and S. Lory. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA 90**:**2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Sumper, M. 1987. Halobacterial glycoprotein biosynthesis. Biochim. Biophys. Acta 906**:**69-79. [DOI] [PubMed] [Google Scholar]
- 421.Sumper, M., E. Berg, R. Mengele, and I. Strobel. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172**:**7111-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177**:**1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68**:**301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Szyf, M., P. Pakneshan, and S. A. Rabbani. 2004. DNA demethylation and cancer: therapeutic implications. Cancer Lett. 211**:**133-143. [DOI] [PubMed] [Google Scholar]
- 425.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3**:**225-237. [DOI] [PubMed] [Google Scholar]
- 426.Tahara, M., A. Ohsawa, S. Saito, and M. Kimura. 2004. In vitro phosphorylation of initiation factor 2 alpha (aIF2 alpha) from hyperthermophilic archaeon Pyrococcus horikoshii OT3. J. Biochem. (Tokyo) 135**:**479-485. [DOI] [PubMed] [Google Scholar]
- 427.Tahirov, T. H., H. Oki, T. Tsukihara, K. Ogasahara, K. Yutani, K. Ogata, Y. Izu, S. Tsunasawa, and I. Kato. 1988. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J. Mol. Biol. 284**:**101-124. [DOI] [PubMed] [Google Scholar]
- 428.Takacs, M., G. Rakhely, and K. L. Kovacs. 2001. Molecular characterization and heterologous expression of hypCD, the first two [NiFe] hydrogenase accessory genes of Thermococcus litoralis. Arch. Microbiol. 176**:**231-235. [DOI] [PubMed] [Google Scholar]
- 429.Takagi, M., M. Nishioka, H. Kakihara, M. Kitabayashi, H. Inoue, B. Kawakami, M. Oka, and T. Imanaka. 1997. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 63**:**4504-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57**:**320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Tanaka, Y., K. Tsumoto, Y. Yasutake, M. Umetsu, M. Yao, H. Fukada, I. Tanaka, and I. Kumagai. 2004. How oligomerization contributes to the thermostability of an archaeon protein. Protein L-isoaspartyl-_O_-methyltransferase from Sulfolobus tokodaii. J. Biol. Chem. 279**:**32957-32967. [DOI] [PubMed] [Google Scholar]
- 432.Taylor, S. V., N. L. Kelleher, C. Kinsland, H. J. Chiu, C. A. Costello, A. D. Backstrom, F. W. McLafferty, and T. P. Begley. 1998. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273**:**16555-16560. [DOI] [PubMed] [Google Scholar]
- 433.Tersteegen, A., and R. Hedderich. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Transcription of the operons and sequence analysis of the deduced proteins. Eur. J. Biochem. 264**:**930-943. [DOI] [PubMed] [Google Scholar]
- 434.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology 144**:**2377-2406. [DOI] [PubMed] [Google Scholar]
- 435.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276**:**34862-34870. [DOI] [PubMed] [Google Scholar]
- 436.Thomas, N. A., S. L. Bardy, and K. F. Jarrell. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25**:**147-174. [DOI] [PubMed] [Google Scholar]
- 437.Tjalsma, H., A. Bolhuis, M. L. van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. J. Quax, G. Venema, S. Bron, and J. M. van Dijl. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12**:**2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Tornabene, T. G., M. Kates, E. Gelpi, and J. Oro. 1969. Occurrence of squalene, di- and tetrahydrosqualenes, and vitamin MK8 in an extremely halophilic bacterium, Halobacterium cutirubrun. J. Lipid Res. 10**:**294-303. [PubMed] [Google Scholar]
- 439.Tornabene, T. G., T. A. Langworthy, G. Holzer, and J. Oro. 1979. Squalenes, phytanes and other isoprenoids as major neutral lipids of methanogenic and thermoacidophilic “archaebacteria.” J. Mol. Evol. 13**:**73-83. [DOI] [PubMed] [Google Scholar]
- 440.Tornabene, T. G., R. S. Wolfe, W. E. Balch, G. Holzer, G. E. Fox, and J. Oro. 1978. Phytanyl-glycerol ethers and squalenes in the archaebacterium Methanobacterium thermoautotrophicum. J. Mol. Evol. 11**:**259-266. [DOI] [PubMed] [Google Scholar]
- 441.Toth, E. A., C. Worby, J. E. Dixon, E. R. Goedken, S. Marqusee, and T. O. Yeates. 2000. The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds. J. Mol. Biol. 301**:**433-450. [DOI] [PubMed] [Google Scholar]
- 442.Trombetta, E. S. 2003. The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology 13**:**77R-91R. [DOI] [PubMed] [Google Scholar]
- 443.Tsunasawa, S., Y. Izu, M. Miyagi, and I. Kato. 1997. Methionine aminopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus: molecular cloning and overexpression in Escherichia coli of the gene, and characteristics of the enzyme. J. Biochem. (Tokyo) 122**:**843-850. [DOI] [PubMed] [Google Scholar]
- 444.Tu, B. P., and J. S. Weissman. 2004. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164**:**341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3**:**363-379. [DOI] [PubMed] [Google Scholar]
- 446.Van den Berg, B., W. M. Clemons, Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427**:**36-44. [DOI] [PubMed] [Google Scholar]
- 447.van Valkenburgh, C., X. Chen, C. Mullins, H. Fang, and N. Green. 1999. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J. Biol. Chem. 274**:**11519-11525. [DOI] [PubMed] [Google Scholar]
- 448.Varki, A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3**:**97-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Varki, A. 1998. Factors controlling the glycosylation potential of the Golgi apparatus. Trends Cell Biol. 8**:**34-40. [DOI] [PubMed] [Google Scholar]
- 450.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93**:**12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Vinayagam, A., G. Pugalenthi, T. Rajesh, and R. Sowdhamini. 2004. DSDBASE: a consortium of native and modelled disulphide bonds in proteins. Nucleic Acids Res. 32**:**D200-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68**:**1015-1068. [DOI] [PubMed] [Google Scholar]
- 453.Voisin, S., R. Scott Houliston, J. Kelly, J. R. Brisson, D. Watson, S. L. Bardy, K. F. Jarrell, and S. M. Logan. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280**:**16586-16593. [DOI] [PubMed] [Google Scholar]
- 454.von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115**:**195-201. [DOI] [PubMed] [Google Scholar]
- 455.Voorhorst, W. G., R. I. Eggen, A. C. Geerling, C. Platteeuw, R. J. Siezen, and W. M. Vos. 1996. Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 271**:**20426-20431. [DOI] [PubMed] [Google Scholar]
- 456.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298**:**1790-1793. [DOI] [PubMed] [Google Scholar]
- 457.Wakai, H., S. Nakamura, H. Kawasaki, K. Takada, S. Mizutani, R. Aono, and K. Horikoshi. 1997. Cloning and sequencing of the gene encoding the cell surface glycoprotein of Haloarcula japonica strain TR-1. Extremophiles 1**:**29-35. [DOI] [PubMed] [Google Scholar]
- 458.Wang, G., R. Guo, M. Bartlam, H. Yang, H. Xue, Y. Liu, L. Huang, and Z. Rao. 2003. Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii. Protein Sci. 12**:**2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Wang, J. Y., and D. E. Koshland, Jr. 1978. Evidence for protein kinase activities in the prokaryote Salmonella typhimurium. J. Biol. Chem. 253**:**7605-7608. [PubMed] [Google Scholar]
- 460.Wardleworth, B. N., R. J. Russell, S. D. Bell, G. L. Taylor, and M. F. White. 2002. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 21**:**4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Webb, H., N. Carnall, L. Vanhamme, S. Rolin, J. Van Den Abbeele, S. Welburn, E. Pays, and M. Carrington. 1997. The GPI-phospholipase C of Trypanosoma brucei is nonessential but influences parasitemia in mice. J. Cell Biol. 139**:**103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Wedegaertner, P. B., P. T. Wilson, and H. R. Bourne. 1995. Lipid modifications of trimeric G-proteins. J. Biol. Chem. 270**:**503-506. [DOI] [PubMed] [Google Scholar]
- 463.Wedemeyer, W. J., E. Welker, M. Narayan, and H. A. Scheraga. 2000. Disulfide bonds and protein folding. Biochemistry 39**:**4207-4216. [DOI] [PubMed] [Google Scholar]
- 464.Weinberg, M. V., G. J. Schut, S. Brehm, S. Datta, and M. W. W. Adams. 2005. Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins J. Bacteriol. 187**:**336-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Wetzel, R. 1987. Harnessing disulfide-bonds using protein engineering. Trends Biochem. Sci. 12**:**478-482. [Google Scholar]
- 466.White, M. F., and S. D. Bell. 2002. Holding it together: chromatin in the Archaea. Trends Genet. 18**:**621-626. [DOI] [PubMed] [Google Scholar]
- 467.Whitfeld, C. 1995. Biosynthesis of lipopolysaccharide O antigens, Trends Microbiol. 3**:**178-185. [DOI] [PubMed] [Google Scholar]
- 468.Wieland, F. 1988. Structure and biosynthesis of prokaryotic glycoproteins. Biochimie 70**:**1493-1504. [DOI] [PubMed] [Google Scholar]
- 469.Wieland, F., W. Dompert, G. Bernhardt, and M. Sumper. 1980. Halobacterial glycoprotein saccharides contain covalently linked sulphate. FEBS Lett. 120**:**110-114. [DOI] [PubMed] [Google Scholar]
- 470.Wieland, F., G. Paul, and M. Sumper. 1985. Halobacterial flagellins are sulfated glycoproteins. J. Biol. Chem. 260**:**15180-15185. [PubMed] [Google Scholar]
- 471.Wilkinson, B., and H. F. Gilbert. 2004. Protein disulfide isomerase. Biochim. Biophys. Acta 1699**:**35-44. [DOI] [PubMed] [Google Scholar]
- 472.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74**:**5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Wolf, S., F. Lottspeich, and W. Baumeister. 1993. Ubiquitin found in the archaebacterium Thermoplasma acidophilum. FEBS Lett. 326**:**42-44. [DOI] [PubMed] [Google Scholar]
- 474.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25**:**619-623. [DOI] [PubMed] [Google Scholar]
- 475.Wurgler-Murphy, S. M., D. M. King, and P. J. Kennelly. 2004. The phosphorylation site database: a guide to the serine-, threonine-, and/or tyrosine-phosphorylated proteins in prokaryotic organisms. Proteomics 4**:**1562-1570. [DOI] [PubMed] [Google Scholar]
- 476.Xu, M. Q., M. W. Southworth, F. B. Mersha, L. J. Hornstra, and F. B. Perler. 1993. In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell 75**:**1371-1377. [DOI] [PubMed] [Google Scholar]
- 477.YaDeau, J. T., C. Klein, and G. Blobel. 1991. Yeast signal peptidase contains a glycoprotein and the Sec11 gene product. Prot. Natl. Acad. Sci. USA. 88**:**517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Yang, L. L., and A. Haug. 1979. Purification and partial characterization of a procaryotic glycoprotein from the plasma membrane of Thermoplasma acidophilum. Biochim. Biophys. Acta 556**:**265-277. [DOI] [PubMed] [Google Scholar]
- 479.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26**:**1076-1087. [DOI] [PubMed] [Google Scholar]
- 480.Yao, M., A. Ohsawa, S. Kikukawa, I. Tanaka, and M. Kimura. 2003. Crystal structure of hyperthermophilic archaeal initiation factor 5A: a homologue of eukaryotic initiation factor 5A (eIF-5A). J. Biochem. (Tokyo) 133**:**75-81. [DOI] [PubMed] [Google Scholar]
- 481.Yao, R., A. J. Macario, and E. de Macario. 1992. Immunochemical differences among Methanosarcina mazei S-6 morphologic forms. J. Bacteriol. 174**:**4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Yao, V. J., and J. L. Spudich. 1992. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc. Natl. Acad. Sci. USA 89**:**11915-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Yasutake, Y., M. Yao, N. Sakai, T. Kirita, and I. Tanaka. 2004. Crystal structure of the Pyrococcus horikoshii isopropylmalate isomerase small subunit provides insight into the dual substrate specificity of the enzyme. J. Mol. Biol. 344**:**325-333. [DOI] [PubMed] [Google Scholar]
- 484.Zahringer, U., H. Moll, T. Hettmann, Y. A. Knirel, and G. Schafer. 2000. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur. J. Biochem. 267**:**4144-4149. [DOI] [PubMed] [Google Scholar]
- 485.Zappacosta, F., G. Sannia, L. A. Savoy, G. Marino, and P. Pucci. 1994. Posttranslational modifications in aspartate aminotransferase from Sulfolobus solfataricus. Detection of N-epsilon-methyllysines by mass spectrometry. Eur. J. Biochem. 222**:**761-767. [DOI] [PubMed] [Google Scholar]
- 486.Zeitler, R., E. Hochmuth, R. Deutzmann, and M. Sumper. 1998. Exchange of Ser-4 for Val, Leu or Asn in the sequon Asn-Ala-Ser does not prevent N-glycosylation of the cell surface glycoprotein from Halobacterium halobium. Glycobiology 8**:**1157-1164. [DOI] [PubMed] [Google Scholar]
- 487.Zhang, W., A. Brooun, J. McCandless, P. Banda, and M. Alam. 1996. Signal transduction in the Archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc. Natl. Acad. Sci. USA 93**:**4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Zhang, W., A. Brooun, M. M. Mueller, and M. Alam. 1996. The primary structures of the Archaeon Halobacterium salinarium blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc. Natl. Acad. Sci. USA 93**:**8230-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Zhu, B. C., R. R. Drake, H. Schweingruber, and R. A. Laine. 1995. Inhibition of glycosylation by amphomycin and sugar nucleotide analogs PP36 and PP55 indicates that Haloferax volcanii beta-glucosylates both glycoproteins and glycolipids through lipid-linked sugar intermediates: evidence for three novel glycoproteins and a novel sulfated dihexosyl-archaeol glycolipid. Arch. Biochem. Biophys. 319**:**355-364. [DOI] [PubMed] [Google Scholar]
- 490.Zhu, B. C., and R. A. Laine. 1996. Dolichyl-phosphomannose synthase from the archae Thermoplasma acidophilum. Glycobiology 6**:**811-816. [DOI] [PubMed] [Google Scholar]
- 491.Zhu, W., C. I. Reich, G. J. Olsen, C. S. Giometti, and J. R. Yates 3rd. 2004. Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis. J. Proteome Res. 3**:**538-548. [DOI] [PubMed] [Google Scholar]
- 492.Zobel-Thropp, P., J. D. Gary, and S. Clark. 1998. Delta-_N_-methylarginine is a novel posttranslational modification of arginine residues in yeast proteins. J. Biol. Chem. 273**:**29283-29286. [DOI] [PubMed] [Google Scholar]
- 493.Zufferey, R., R. Knauer, P. Burda, I. Stagljar, S. te Heesen, L. Lehle, and M. Aebi. 1995. STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14**:**4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Zwieb, C., and J. Eichler. 2002. Getting on target: The archaeal signal recognition particle. Archaea 1**:**27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]