Intraspecific violation of genetic colinearity and its implications in maize (original) (raw)
Abstract
Although allelic sequences can vary extensively, it is generally assumed that each gene in one individual will have an allelic counterpart in another individual of the same species. We report here that this assumption does not hold true in maize. We have sequenced over 100 kb from the bz genomic region of two different maize lines and have found dramatic differences between them. First, the retrotransposon clusters, which comprise most of the repetitive DNA in maize, differ markedly in make-up and location relative to the genes in the bz region. Second, and more importantly, the genes themselves differ between the two lines, demonstrating that genetic microcolinearity can be violated within the same species. Our finding has bearing on the underlying genetic basis of hybrid vigor in maize, and possibly other organisms, and on the measurement of genetic distances.
Comparative genetic mapping has revealed a remarkable degree of synteny or conservation of gene order among closely related plant species (1). The extensive conservation of gene content and order among grass chromosomes has even led to the proposal of a single progenitor genome structure for all grasses (2). Subsequent sequence analysis of orthologous regions in rice, sorghum, and maize has established that microcolinearity varies from one region of the genome to another. For example, the_a1-sh2_ orthologous regions of sorghum and rice contain the same genes, in the same order and orientation (3). These two grasses are thought to have diverged from a common ancestor around 50 million years ago (4). In contrast, the adh1 region of sorghum differs from that of maize, a much closer relative, by the presence of five additional genes that are interspersed with the nine colinear genes shared by both species (5). Hence, small rearrangements may be present even in largely colinear genomic regions. In general, comparative sequence analysis has revealed a much higher degree of diversity at the microstructural level than was predicted by genetic mapping studies of closely related plant species (6). Here, we report that microstructural diversity extends to even allelic regions of members of the same species.
We recently isolated a 230-kb bacterial artificial chromosome (BAC) contig of the bz region from McC, a maize line carrying the_Bz-McC_ allele used in several of our recombination studies (7, 8). We established that this bz allele is located in an unusually gene-dense region of the genome: 10 genes are found in a 32-kb stretch of DNA uninterrupted by retrotransposons (9). This compact packaging of adjacent genes was unexpected for maize, a species with a 2500-Mb genome and a large content of repetitive DNA (10, 11). Up to 80% of the maize genome is composed of retrotransposons (12), which are arranged principally in a nested type of organization (13) and have been found interspersed with genes in every large maize genomic region sequenced to date (5, 9, 14). Immediately proximal to the McC bz gene group lies a recombinationally inert and highly methylated 94-kb nested retrotransposon cluster (15), similar in structure to those first described in the vicinity of the_adh1_ locus (13).
We have now completed the sequence of the 230-kb BAC contig of McC and find that another nested retrotransposon cluster lies distal to the_Bz-McC_ gene group. We have also sequenced a 110-kb BAC contig from the bz region of the inbred B73, the line chosen for the large-scale sequencing of selected regions of the maize genome (available at http://www.nsf.gov/bio/dbi/dbi_pgr.htm). We find that the bz regions of these two maize lines differ not only in the make-up and pattern of interspersion of their retrotransposon clusters, but also, surprisingly, in the density and content of their genes.
Materials and Methods
BAC Isolation.
The isolation of the two adjacent Not_I Bz-McC BAC clones, which comprise a 230-kb contig of the bz region, has been described (16). Those BAC clones were isolated from a maize line carrying the Bz-McC allele that had been introgressed into the genetic background of the inbred W22 by repeated backcrossing. The_Bz-McC allele, named after B. McClintock, is the progenitor of the bz-m2(Ac) unstable mutation described by her (17) and most likely derives from a New England flint variety. The_Bz-B73_ BAC clones were isolated from a commercially available BAC library of the Corn Belt inbred B73 (Genome Systems, St. Louis), by using a probe from the stc1 gene, which is immediately distal to bz in the Bz-McC BAC clone (9).
DNA Sequencing and Assembly.
The BAC clones were sequenced by the shotgun sequence strategy (http://www.genome.ou.edu/proto.html) with some modifications, as described (15). Sequencing reactions were performed by using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and analyzed on ABI377 sequencing gels. The approximately ten-fold redundant sequences were assembled with the PHRED/PHRAP software (18). Contigs were extended and joined by custom specific primer walking to close the gaps. The reliability of the sequence was confirmed by the location of expected restriction sites.
Sequence Analysis.
The final sequence was divided into 5-kb fragments which served as queries to search the GenBank databases with the various BLAST programs (19). Programs from the Lasergene package (DNAstar, Madison, WI) were used for sequence comparisons and alignments.
Southern Blot Analysis.
Conditions for the preparation, digestion, and electrophoresis of high molecular weight DNA have been described (16). DNA was prepared from isolated nuclei of 4-week-old shoots and leaves. Agarose plugs containing approximately 10 μg of high molecular weight DNA from different inbred lines were digested to completion with_Not_I. The digested genomic DNAs were resolved in 1% agarose gels by pulsed-field gel electrophoresis (CHEF-DR II system, Bio-Rad). The gels were blotted to a Hybond+ nylon membrane (Amersham Pharmacia) and the membranes were hybridized with random-primer-labeled P32 probes for the various genes in the bz regions of McC and B73. Conditions for hybridization, high stringency washing, and exposure to x-ray film were standard.
Results
Structure of the Bz-McC and Bz-B73 Genomic Regions.
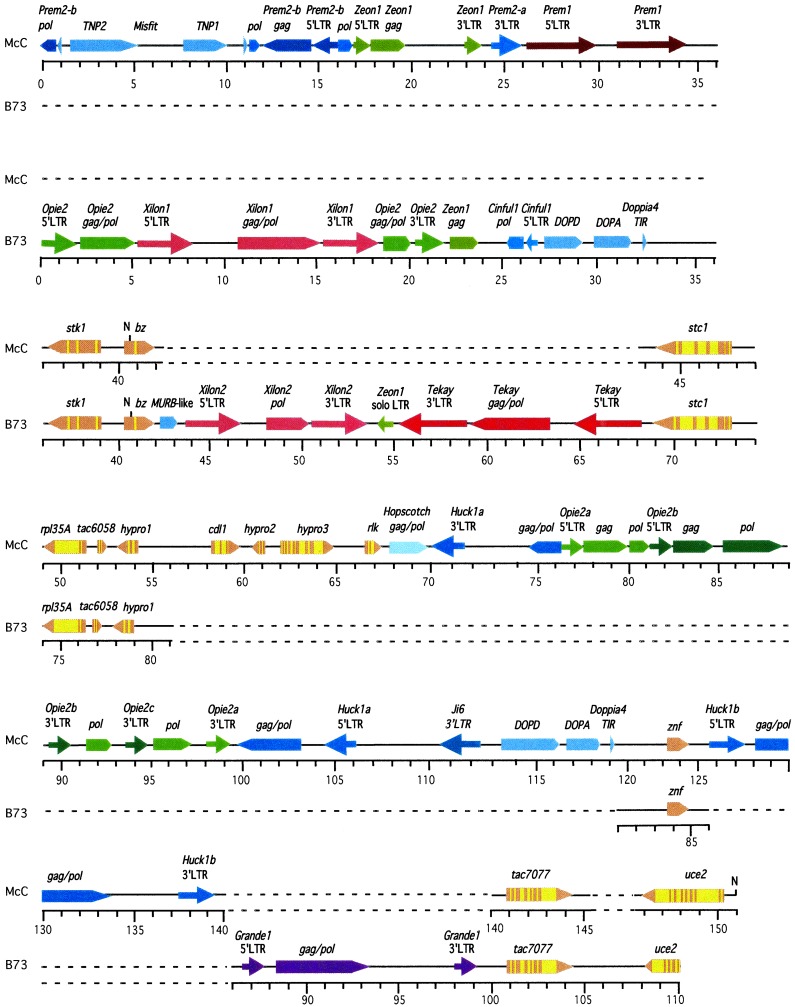
We have sequenced and analyzed the entire 230-kb BAC contig of the Bz-McC region (GenBank accession no. AF391808). The organization of the most distal 150 kb of the sequence is diagrammed in Fig. 1, with the centromere end at the top and the telomere end at the bottom. The 10-gene bz island, located between the 36- and 68-kb markers in Fig. 1, is separated from znf, the next predicted gene, by a 53-kb transposon block. The putative product of the intronless_znf_ gene is homologous to ring zinc finger proteins.znf is a solo gene separated by a single _Huck1b_retroelement from what appears to be another gene island. Two members of that gene island are present in our BAC clone: _tac7077_and uce2. tac7077 is a constitutively expressed gene that has no homology to any sequences in the database and was defined initially as the site of insertion of the transposed _Ac7077_element from bz (20). Its intron-exon structure was determined from a full-length cDNA isolated from immature tassels (21).uce2 is highly similar to several plant genes encoding a protein with homology to ubiquitin conjugating enzymes. Its intron-exon structure was inferred under the assumption that intron location is conserved among closely related plant genes.
Figure 1.
Organization of genes and transposons in the bz genomic region of two different maize lines, McC and B73. The proximal end is at 0 kb; the distal end is at 151 kb in McC and at 110 kb in B73. Genes are shown as pentagons pointing in the direction of transcription; exons are in bronze, and introns are in yellow. Each transposon is in a different color. To facilitate the identification of interrupted retrotransposons, LTRs and encoded proteins of the same element are in the same color. Where a sequence in one line cannot be aligned with a sequence at the corresponding position in the other line, a dotted line is used to show the lack of alignment. The paired sequences of McC and B73 can be aligned only at the genes that they have in common:stk1, bz, stc1,rpl35A, tac6058, hypro1,znf, tac7077, and uce2. The McC chromosome contains three retrotransposon blocks, proximal to_stk1_, znf, and tac7077, respectively. The B73 chromosome contains two completely different retrotransposon blocks at the first and third of those locations plus a third cluster between bz and stc1, genes that are separated by less than 1 kb in McC. In addition, the genes_cdl1_, hypro2, hypro3, and_rlk_, which are found distal to hypro1 in McC, are missing entirely from B73. The location of cleavable_Not_I sites in McC and B73 genomic DNA is marked with an N. The McC sequence (GenBank accession no. AF391808) is a composite from two adjacent BAC clones centered around the _Not_I site in the Bz-McC allele (16). The B73 sequence (GenBank accession no. AF448416) is derived from two overlapping BACs isolated from a commercial library by using a _bz_probe.
We also have sequenced a 110-kb BAC contig from the _bz_region of the inbred B73. This line is the one chosen for the sequencing of selected regions of the maize genome and is presently the only one for which a complete BAC library is publicly available. We were prompted to investigate the organization of the _bz_region in this inbred after sizing and partially sequencing the_bz_-stc1 intergenic region in bz BACs from B73. Whereas the _bz_-stc1 intergenic region of McC is about 1.5 kb and consists mostly of small miniature repeat transposable element-like insertions (9), that of B73 was too large to be resolved in conventional gels and contained sequences typical of retrotransposons. This unexpected difference led us to sequence the entire B73 bz_-stc1 intergenic region and, subsequently, a 110-kb contig from two overlapping BACs of the_bz region (GenBank accession no. AF448416).
The sequence of the bz genomic region from B73 is presented in Fig. 1, below the corresponding sequence from McC. The sequences are considerably different and can only be aligned at the genes that they have in common. The lines differ in (i) the make-up and sizes of the retrotransposon blocks flanking common genes; (ii) the pattern of interspersion between gene islands and retrotransposon blocks; and most significantly, (iii) the number and content of the genes in the region. Following is a discussion of these differences, starting from the proximal end (position 0 in Fig. 1).
The sizes of the retrotransposon blocks proximal to bz_are similar in both lines (≈90 kb), but the transposons present in each block are completely different. Fig. 1 shows only the 35 kb closest to bz, the part of the cluster for which we have contiguous sequence data from both lines. Other than a short fragment of the gag gene from Zeon1, there are no common sequences. Both clusters show nesting. The McC retrotransposon nest has already been described (15). The B73 nest includes a new, apparently intact retrotransposon that we have named Xilon (after Xilonen, the Aztec goddess of young corn). The 1.5-kb_bz_-stc1 intergenic segment in McC (position 42–43.5) is replaced in B73 by a 26-kb retrotransposon block (position 42–69), which splits the bz gene island in two. The retrotransposons in this block are not nested; uninterrupted copies of_Xilon and Tekay are separated by a_Zeon1_ solo long terminal repeat (LTR). Flanking the block at the proximal end is a homologue of MuRB, one of the two genes present in the autonomous transposable element MuDR(22, 23).
The most remarkable difference occurs distal to_hypro1_, the sixth gene in the bz region that is colinear between McC and B73. The two sequences diverge completely until the next gene, znf, where colinearity resumes briefly. Four genes (cdl1, hypro2, hypro3, and_rlk_) are missing entirely in B73. These genes were identified by comparisons to sequences in the GenBank databases. They are homologous to other plant genes encoding the following predicted or confirmed proteins: cdl, cell division-like protein;hypro2 and hypro3, Arabidopsis_hypothetical proteins, and rlk, receptor-like kinase. All of these genes are members of small gene families in maize. By using a deletion of the entire bz region as a negative control, we have shown that at least three of them (hypro2,hypro3, and rlk) are expressed specifically in one or more maize tissues (9). Thus, different maize lines may carry a different complement of genes in certain locations of the genome that_a priori would have been considered allelic based on the composition of adjacent genes.
The retrotransposon blocks in the distal halves of the McC and B73 sequences are also completely different. The large, 53-kb nested transposon block that separates rlk and znf in McC (position 68–120 kb) is missing in B73. A single retrotransposon separates znf from the next gene island in both McC and B73, but the retrotransposons are different. Whereas a Huck1_retrotransposon is inserted between znf and_tac7077 in McC (labeled Huck1b in Fig. 1), a_Grande1_ retrotransposon is inserted at the equivalent location in B73. Colinearity between McC and B73 resumes at the most distal gene island in our sequence, which is defined by_tac7077_ and uce2.
Characterization of the bz Genomic Regions of Different Maize Lines.
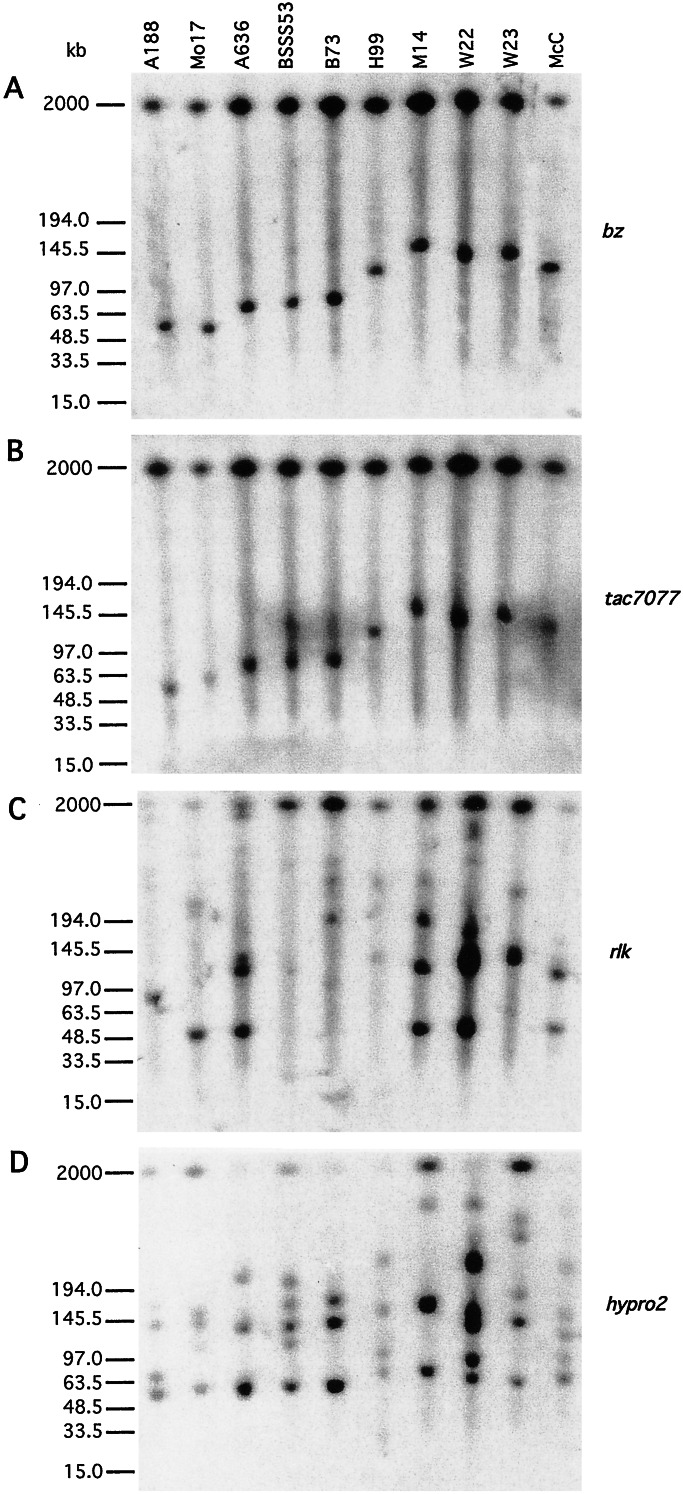
To confirm that the sequences cloned in BACs were, in fact, present in the genomes of McC and B73 and to survey the organization of the region in other inbred lines, we hybridized Southern blots containing large-sized DNA fragments from several inbreds with different probes of the region. The bz and tac7077 probes, at opposite ends of an McC Not_I BAC clone, detect the same_Not_I fragment in every inbred line but one (Fig.2 A and B). This finding suggests that most lines carry a bz and a_tac7077 gene in the same genomic fragment, within 50 to 140 kb of each other, based on the size of the smallest and largest common fragments detected by the two probes. The sizes of the fragments in McC and B73 are those expected based on their BAC sequences, confirming that the sequenced BACs were not cloning artifacts. Only in Mo17 did the two probes hybridize to different sized fragments, but this result could be due simply to the presence of a cleavable _Not_I site between bz and tac7077 in that inbred.
Figure 2.
Composition of the bz genomic region in different maize lines. _Not_I-digested genomic DNA from 10 different maize lines was separated by CHEF gel electrophoresis, blotted to Nylon membranes, hybridized to the four probes shown, and washed under high-stringency conditions. (A) bz. (B) tac7077. (C)rlk. (D) hypro2.
To determine what other genes from the McC or B73 bz regions were present in the Not_I fragments that hybridized to_bz in the other inbreds, we hybridized the same membrane to probes from genes found either in both inbreds (znf) or exclusively in McC (hypro2 and rlk). The_znf_ probe hybridizes to the same Not_I genomic fragment as bz in all inbreds (data not shown). The_rlk and hypro2 probes detect small gene families in most lines (Fig. 2 C and D). This observation agrees with the sequence variability observed earlier among_hypro2_ cDNA homologs from a maize tassel cDNA library (9). However, rlk and hypro2 hybridize to a_Not_I fragment of the same size as bz only in McC, W22, W23, and Mo17. Therefore, the other inbreds resemble B73 in lacking the hypro2 and rlk genes between_bz_ and tac7077, although they appear to have copies of these sequences elsewhere in the genome. A summary of the CHEF gel hybridization data are presented in Table1. Four hybridization patterns can be distinguished. Only in McC, W22, and W23 do all probes hybridize to the same size fragment. In the other inbreds, one or more genes are missing from the _Not_I genomic fragment that contains _bz_and tac7077.
Table 1.
Summary of CHEF gel hybridization patterns in 10 U.S. maize lines
| DNA source | Probes | Hybridization group | ||||
|---|---|---|---|---|---|---|
| bz | hyp2 | rlk | znf | tac7077 | ||
| McC | +* | + | + | + | + | 1 |
| W23 | + | + | + | + | + | 1 |
| W22 | + | + | + | + | + | 1 |
| M14 | + | − | − | + | + | 2 |
| H99 | + | − | − | + | + | 2 |
| B73 | + | − | − | + | + | 2 |
| BSSS53 | + | − | − | + | + | 2 |
| A636 | + | − | − | + | + | 2 |
| Mo17 | + | − | + | + | D‡ | 3 |
| A188 | + | + | − | + | + | 4 |
Discussion
We have found that the bz genomic regions of two North American maize lines differ extensively in the organization and content not only of the intergenic retrotransposon clusters in the region, but also of the genes themselves. A central issue raised by these findings, with potential practical implications, is the meaning of allelism in a region such as the one where bz resides. Of the 10 genes identified originally in the McC bz genomic region, only the proximal 6 have allelic counterparts in B73. Therefore, a B73/McC hybrid is hemizygous for 4 of the 10 genes in the region. Clearly, this type of variability is only possible for genes with relatively minor quantitative effects. In agreement with this, all 4 genes are members of small gene families, and their absence from the bz region in some lines may be partially compensated for by duplicate copies located elsewhere. Extensive chromosomal duplications are well known in maize; it has been estimated that as many as one-third of the genes are present in multiple copies (24).
Table 1 shows that the 10 lines surveyed fall into 4 groups with respect to the genic content of the bz region. The lines in each of the two groups with more than one member are known to be related (25). Thus, group 2 inbreds B73, BSSS53, and A636 are derived from Iowa Stiff Stalk Synthetic, a synthetic population representing germplasm of the Reid Yellow Dent open pollinated cultivar. The two other inbreds in this group, M14 and H99, also have that cultivar in their lineage, although H99 has a complex pedigree and is generally considered a Lancaster Sure Crop type in hybrid combinations (26). Similarly, group 1 inbreds W22 and W23 share the cultivar Golden Glow as a common ancestor. Mo17, the only member of group 3, is a representative of the Lancaster Sure Crop germplasm. Interestingly, hybrids between B73 and Mo17 show pronounced heterosis and illustrate the heterotic pattern (Lancaster Sure Crop by Reid Yellow Dent) that has received greatest use in U.S. Corn Belt breeding programs (27). The genetic basis of heterosis in corn has been explained by one of two primary models. According to the dominance model, heterosis is due to the action of loci showing partial or complete dominance; according to the overdominance model, it is due to the action of loci at which the heterozygote is superior to either homozygote. Our findings provide a simple molecular basis for the dominance model, which is also the one supported by the preponderance of the quantitative genetics evidence (27). In different maize lines, genes that are members of gene families and are, therefore, expected to have quantitative, rather than qualitative, effects, may be present or absent in certain regions of the genome. Lines lacking different genes would complement one another and show hybrid vigor, whereas lines lacking mostly the same genes would not complement and, in breeding terminology, would fall in the same heterotic group. Our model also accounts for the severe inbreeding depression observed in maize, because inbreeding would result in the progressive loss of functional genes at many genomic locations. Finally, if different genes occurred at the same location in the two homologs, it would not be possible to assemble all of the genes at that location in one chromosome by crossing over, which would explain why it has not been possible to “fix” hybrid vigor in maize.
Our findings also have bearing on measurements of genetic recombination. The bz mutants used in earlier studies of intragenic recombination are derivatives of the Bz-McC and_Bz-W22_ alleles, which lie in large gene islands and are flanked by the stk1 and stc1 genes (ref. 9, and H.F. and H.K.D., unpublished data). Those studies showed that the_bz_ locus is very highly recombinogenic. Instead,Bz-B73 lies in a two-gene “island” and is flanked at its 3′ end by a 25-kb retrotransposon block. If recombination within_bz_ is affected by the neighboring sequences, it will likely be lower in heterozygotes in which one or both heteroalleles are derived from Bz-B73. Intergenic distances also may differ. As in many organisms, recombination in maize seems to occur mainly in genes (15, 28). Therefore, genetic distances between genes will be affected by the number of genes in the intervening region that have allelic counterparts in both homologs. The variability in estimates of genetic distances for a particular region, often seen in maize, may be attributable in part to the type of microstructural heterogeneity reported here at the bz locus.
Differences in the retrotransposon blocks that flank genes were anticipated from the restriction polymorphisms that flank different_Adh1_ alleles in regions corresponding to retrotransposons in the sequenced Adh1-F allele (12). These differences may account partly for the observation that adh1 mutations derived from different progenitor alleles recombine poorly (29). On the other hand, variability in the pattern of interspersion of retrotransposon blocks within gene islands was not expected. This finding implies that the size and make-up of gene islands is a polymorphic trait within maize. The variable size and location of retrotransposon blocks in the bz region of different inbreds is reminiscent of the variability in knob positions observed among different maize lines (30). Clusters of retrotransposons have, in fact, been detected in polymorphic heterochromatic knobs in maize (31) and Arabidopsis (32), and it is likely that the smaller heterochromatic chromomeres in maize also contain such clusters.
Our study, involving a limited number of lines from the U.S. Corn Belt, shows that the bz genomic region varies extensively within maize. The lack of any similarity in the make-up of the retrotransposon blocks and the differences in their pattern of interspersion within genes suggest independent origins for the McC and B73 bz regions. The retrotransposon explosion that led to a doubling of the size of the maize genome is estimated to have occurred within the last 2–3 million years (33). Our data suggest that this amplification happened separately in different individuals of the population from which modern maize eventually evolved, and that at least part of this variability was preserved through the “domestication bottleneck” of maize (34). It has been estimated that U.S. inbreds contain roughly 77% of the level of genetic diversity within maize (35) and only ≈40% of that of its wild progenitor, represented today by Zea mays, subsp.parviglumis (36). Based on the large variability encountered in our limited sample, we can expect that the bz region will be highly variable among maize lines and even more so among its wild relatives.
How did these deletions arise? The fact that the set of genes missing from one line is next to a retrotransposon block in the other would implicate the retrotransposons in the deletion process, although no clear mechanism is discernible from a comparison of the sequences. The simplest scenario, deletion by intrachromosomal recombination between the 5′ and 3′ LTRs of a retroelement, would leave behind a solo LTR flanked by the same 5-bp sequence, a common structure in the_Rar_ region of barley (37). However, the only LTR of this type that we found is the Zeon1 solo LTR in the middle of the retrotransposon block separating bz and _stc1_in B73. Alternatively, although retrotransposon blocks are most likely recombinationally inert in present-day maize (15), the deletions could have been generated by unequal crossing over between related retrotransposon sequences in the wild ancestors of maize. The occurrence of DNA transposon sequences, such as MuRB and the CACTA family members Misfit and Doppia4 (22, 23, 38,39), within and flanking some retrotransposon blocks, raises the possibility that they, too, could have been involved in the deletion process, because complex DNA transposons are known to induce chromosome breakage (40–42). The presence of the same Doppia4_-related element at the termini of two different retrotransposon blocks in different locations of the McC and B73 bz genomic regions is particularly suggestive. A Doppia element also has been implicated in the origination of the complex rearrangement seen in the_R-r:std allele (43).
Ancient large-scale duplication followed by selective gene loss have been proposed to be the major factors in the evolution of dicot family genomes (44). The same factors, compressed over a much shorter evolutionary time frame, seem to have been involved in the evolution of the maize genome (4, 24). The retrotransposon multiplication that occurred within the last 3 million years (33) may have accelerated the process of gene loss within maize, generating the intraspecific variation in gene content that we uncovered at bz.
There is no reason to believe that the type of variability found in the bz genomic region is unique. It may be a feature of other plant species that have extensive duplications in their genomes, particularly of those that show strong inbreeding depression, like alfalfa and the brassicas. The region covered by our study is known to be nonvital, because deletions that include it and extend beyond the distal marker sh1 are homozygous viable, although compromised in vigor (45). Similar variability can be expected of other regions of the maize genome that contain nonessential genes, such as_R_, which controls the tissue specificity of anthocyanin pigmentation, and Rp1, which is involved in rust resistance. In fact, a high level of structural polymorphism among different geographic accessions has already been uncovered at both of these loci (46–48). Sequencing of large chromosomal clones of the same genomic region in different lines of maize and its relatives will help to determine how extensive the plus/minus type of variation described here is in maize.
Acknowledgments
We thank William Tracy, Ben Burr, and Matt Cowperthwaite for their valuable suggestions and comments on the manuscript, Victor Llaca for use of the B73 BAC filters, and Michele Morgante and Scott Tingey for the dot-plot analysis of the two complete sequences. H.F. acknowledges a leave of absence from Wuhan University while performing this work. This work was supported by National Science Foundation Grant MCB 99-04646.
Abbreviations
BAC
bacterial artificial chromosome
LTR
long terminal repeat
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF391808 andAF448416).
See commentary on page 9093.
References
- 1.Paterson A H, Bowers J E, Burow M D, Draye X, Elsik C G, Jiang C X, Katsar C S, Lan T H, Lin Y R, Ming R, Wright R J. Plant Cell. 2000;12:1523–1540. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, SanMiguel P, Bennetzen J L. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaut B S, Doebley J. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tikhonov A P, SanMiguel P J, Nakajima Y, Gorenstein N M, Bennetzen J L, Avramova Z. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennetzen J L. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dooner H K. Genetics. 1986;113:1021–1036. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dooner H K, Martinez-Ferez I M. Plant Cell. 1997;9:1633–1646. doi: 10.1105/tpc.9.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu H, Park W, Yan X, Zheng Z, Shen B, Dooner H K. Proc Natl Acad Sci USA. 2001;98:8903–8908. doi: 10.1073/pnas.141221898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett M D, Smith J B. Philos Trans R Soc London B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 11.Hake S, Walbot V. Chromosoma. 1980;79:251–270. [Google Scholar]
- 12.SanMiguel P, Bennetzen J L. Ann Bot (London) 1998;82:37–44. [Google Scholar]
- 13.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 14.Song R, Llaca V, Linton E, Messing J. Genome Res. 2001;11:1817–1825. doi: 10.1101/gr.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu H, Zheng Z, Dooner H K. Proc Natl Acad Sci USA. 2002;99:1082–1087. doi: 10.1073/pnas.022635499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu H, Dooner H K. Genome Res. 2000;10:866–873. doi: 10.1101/gr.10.6.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClintock B. Carnegie Inst Washington, Year Book. 1955;54:245–255. [PubMed] [Google Scholar]
- 18.Green P. DOE Human Genome Program Contractor-Grantee Workshop V. Washington, DC: U.S. Dept. of Energy, Office of Science, Office of Biological and Environmental Research; 1996. p. 157. [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dooner H K, Belachew A. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowperthwaite M, Park W, Xu Z, Yan X, Maurais S, Dooner H K. Plant Cell. 2002;14:713–726. doi: 10.1105/tpc.010468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donlin M J, Lisch D, Freeling M. Plant Cell. 1995;7:1989–2000. doi: 10.1105/tpc.7.12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hershberger R J, Benito M I, Hardeman K J, Warren C, Chandler V L, Walbot V. Genetics. 1995;140:1087–1098. doi: 10.1093/genetics/140.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaut B S. Gen Research. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes J T, Behr C F, Coors J G, Tracy W F. Compilation of North American Maize Breeding Germplasm. Madison, WI: Crop Sci. Soc. Amer.; 1993. [Google Scholar]
- 26.Gerdes J T, Tracy W F. Crop Sci. 1993;33:334–337. [Google Scholar]
- 27.Hallauer A R, Russell W A, Lamkey K R. In: Corn and Corn Improvement. Sprague G F, Dudley J W, editors. Madison, WI: Amer. Soc. Agron.; 1988. pp. 463–564. [Google Scholar]
- 28.Puchta H, Hohn B. Trends Plant Sci. 1996;1:340–348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 29.Freeling M. Genetics. 1976;83:707–719. doi: 10.1093/genetics/83.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClintock B, Kato T A, Blumenschein A. Chromosome Constitution of Races of Maize. Chapingo, Mexico: Colegio de Postgraduados; 1981. [Google Scholar]
- 31.Ananiev E V, Phillips R L, Rines H W. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arabidopsis Sequencing Consortium. Cell. 2000;100:377–386. [PubMed] [Google Scholar]
- 33.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nature Gen. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 34.Eyre-Walker A, Gaut R L, Hilton H, Feldman D L, Gaut B S. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenaillon M I, Sawkins M C, Long A D, Gaut R L, Doebley J F, Gaut B S. Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaut B S, Le Thierry d'Ennequin M, Peek A S, Sawkins M C. Proc Natl Acad Sci USA. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirasu K, Schulman A H, Lahaye T, Schulze-Lefert P. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gierl A, Saedler H, Peterson P A. Annu Rev Genet. 1989;23:71–85. doi: 10.1146/annurev.ge.23.120189.000443. [DOI] [PubMed] [Google Scholar]
- 39.Bercury S D, Panavas T, Irenze K, Walker E L. Plant Mol Biol. 2001;47:341–351. doi: 10.1023/a:1011606529513. [DOI] [PubMed] [Google Scholar]
- 40.McClintock B. Cold Spring Harbor Symp Quant Biol. 1952;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Doring H-P, Nelsen-Salz B, Garber R, Tillmann E. In: Plant Genetics. Freeling M, editor. New York: Liss; 1985. pp. 355–367. [Google Scholar]
- 42.Dooner H K, Belachew A. Genetics. 1991;129:855–862. doi: 10.1093/genetics/129.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker E L, Robbins T P, Bureau T E, Kermicle J, Dellaporta S L. EMBO J. 1995;14:2350–2363. doi: 10.1002/j.1460-2075.1995.tb07230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ku H M, Vision T, Liu J, Tanksley S D. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mottinger J P. Genetics. 1970;64:259–271. doi: 10.1093/genetics/64.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins T P, Walker E L, Kermicle J L, Alleman M, Dellaporta S L. Genetics. 1991;129:271–283. doi: 10.1093/genetics/129.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eggleston W B, Alleman M, Kermicle J L. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]