In vivo activity of nuclease-resistant siRNAs (original) (raw)
Abstract
Chemical modifications have been incorporated into short interfering RNAs (siRNAs) without reducing their ability to inhibit gene expression in mammalian cells grown in vitro. In this study, we begin to assess the potential utility of 2′-modified siRNAs in mammals. We demonstrate that siRNA modified with 2′-flouro (2′-F) pyrimidines are functional in cell culture and have a greatly increased stability and a prolonged half-life in human plasma as compared to 2′-OH containing siRNAs. Moreover, we show that the 2′-F containing siRNAs are functional in mice and can inhibit the expression of a target gene in vivo. However, even though the modified siRNAs have greatly increased resistance to nuclease degradation in plasma, this increase in stability did not translate into enhanced or prolonged inhibitory activity of target gene reduction in mice following tail vein injection. Thus, this study shows that 2′-F modified siRNAs are functional in vivo, but that they are not necessarily more potent than unmodified siRNAs in animals.
Keywords: RNAi, siRNA, modified nucleotides, luciferase
INTRODUCTION
RNA interference (RNAi) is a highly conserved biological response to double-stranded RNA that mediates post-transcriptional sequence-specific gene silencing (for review, see Hannon 2002; McManus and Sharp 2002; Dykxhoorn et al. 2003). The RNAi phenomenom was originally discovered in Caenorhabditis elegans in which delivery of only a few molecules of double-stranded RNA (dsRNA) per cell can engender gene-specific silencing throughout the entire organism and its progeny (Fire et al. 1998). The mechanism of RNAi involves the generation of short dsRNAs, termed short interfering RNAs (siRNAs), approximately 21 nucleotides in length by the enzyme Dicer (Hamilton and Baul-combe 1999; Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001). This observation led to the discovery that direct delivery of siRNAs into mammalian cells can trigger gene-specific silencing (Elbashir et al. 2001). However, the longevity of siRNA-mediated gene silencing in mammalian cells appears to be greatly reduced compared to RNAi activity in other eukaryotes, such as C. elegans (McManus and Sharp 2002).
The observation that siRNAs can be employed to inhibit gene expression in mammalian cells in vitro (Elbashir et al. 2001) has led to much speculation about their potential utility as therapeutic agents. To administer siRNAs as therapeutic agents, two main strategies are being explored. A number of studies have focused upon designing gene delivery and expression systems for short hairpin RNAs (shRNAs; Lee et al. 2002; Miyagishi and Taira 2002; Paul et al. 2002; Sui et al. 2002). Using such a gene-therapy-based approach, shRNAs have been shown to inhibit hepatitis B virus (HBV) expression in animal models (McCaffrey et al. 2003). The second strategy focuses on the use of synthetic siRNAs that are chemically modified to confer resistance to nuclease degradation. Recently, a few modifications have been described that greatly increase the nuclease-resistance of siRNAs without compromising their activity in cell culture studies (Braasch et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003; Harborth et al. 2003). In particular, siRNAs that have been modified with 2′-flouro (2′-F) pyrimidines appear to have favorable properties in vitro (Chiu and Rana 2003; Harborth et al. 2003). Moreover, one report recently suggested 2′-F modified siRNAs have enhanced activity in cell culture as compared to 2′-OH containing siRNAs (Chiu and Rana 2003). In this study, we examine the utility of 2′-F containing siRNAs in a murine model to begin to determine if such nuclease-resistant siRNAs can inhibit gene expression in vivo and if such modifications enhance the activity of the modified siRNAs relative to unmodified siRNAs in mammals. Our results demonstrate that 2′-F modified siRNAs are functional in mice but that they do not necessarily have enhanced intracellular activity over 2′-OH siRNAs.
RESULTS AND DISCUSSION
Effect of 2′ modification on siRNA activity in vitro
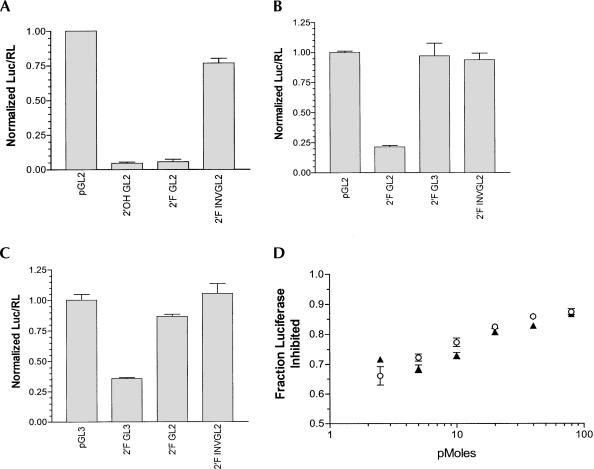
Increasing the stability of siRNA to nuclease degradation could potentially increase siRNA half-life and therefore therapeutic potential in cells and animals. In particular, 2′-F pyrimidines have been incorporated into RNA to reduce nuclease-mediated degradation of the RNA transcripts (Pieken et al. 1991; Rusconi et al. 2002). Moreover, Chiu and Rana (2003) recently demonstrated that siRNAs modified with 2′-flouro-uridine (2′-FU) and 2′-flouro-cytidine (2′-FC) have activities comparable to unmodified siRNAs in their ability to knock down expression of a green fluorescence protein (GFP) reporter gene in cell culture. We employed the siRNA-luciferase reporter system developed by Elbashir et al. in 2001 to test the effects of 2′-F pyrimidine modifications on siRNA activity in NIH/3T3 cells. As shown in Figure 1A ▶, both 2′-F and 2′-OH containing GL2 siRNAs suppressed GL2-luciferase expression by a similar amount, as did 2′-F-2′-OH hybrid siRNAs (data not shown). This suppression was approximately 30-fold compared to 2′-F or 2′-OH inverted control siRNAs and vector transfected cells. In addition, as illustrated in Figure 1 ▶, B and C, the siRNA-mediated inhibition was specific for its intended target sequence, as 2′-F GL2 siRNA did not inhibit pGL3 expression and vice versa. To determine if 2′-F siRNAs are more potent than 2′-OH siRNAs at lower concentrations, an siRNA titration experiment was performed. No significant differences in the interference of luciferase expression were observed between 2′-OH and 2′-F siRNA over a range of concentrations. (Fig. 1D ▶) These results demonstrate that modification of siRNA with 2′-F pyrimidines does not have an adverse effect on siRNA suppression of luciferase expression in this system and are consistent with the results reported on inhibition of GFP expression (Chiu and Rana 2003). Moreover, the experiments demonstrate that 2′-F modification of siRNA does not greatly affect target specificity as seen in its ability to distinguish between the GL2 and GL3 luciferase target RNAs.
FIGURE 1.
Comparison of 2′-OH and 2′-F siRNAs in cell culture. NIH-3T3 cells were transfected with pGL2 or pGL3 luciferase plasmids and 2′-OH or 2′-F siRNA. Normalized luciferase activity was calculated by dividing luciferase counts by Renilla counts and normalizing to the vector control. (A) Activity of 2′-OH and 2′-F GL2 siRNA against pGL2 luciferase target. Six replicates are shown (mean ± SEM). (B, C) 2′-F siRNA show specificity for their respective targets. (D) The inhibitory effects of 2′-OH siRNAs (open circles) and 2′-F siRNAs (closed triangles) titrate similarly. Three replicates are shown (mean ± SEM).
Effect of 2′-F modification on siRNA stability
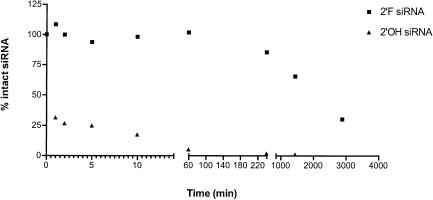
As there does not seem to be any decrease in activity of 2′-F modified siRNA compared to unmodified 2′-OH siRNA, we directly tested the stability of modified versus unmodified siRNAs in a biological fluid. We incubated 5′ 32P-labeled 2′-F and 2′-OH GL2 siRNAs in human plasma and measured the amount of intact RNA present following various incubation times. As shown in Figure 2 ▶, 2′-F modified siRNA showed a dramatic increase in stability over 2′-OH siRNA. Greater than 50% of the 2′-OH siRNA was degraded within the first minute of exposure to plasma and it was virtually all degraded by 4 h. By contrast, more than 50% of the 2′-F siRNA molecules remained full length after 24 h. In addition, more than 25% of the 2′-F siRNA remained intact after 48 h, a level comparable to the 1-min time point for 2′-OH. These results show that 2′-F modification of siRNA does indeed dramatically increase the molecule’s stability in human plasma compared to unmodified siRNA.
FIGURE 2.
siRNA stability in plasma. siRNA quantities at various time points were calculated by dividing the total counts of full-length siRNA by the input starting material. The stability of 2′-F (squares) and 2′-OH (triangles) siRNAs in human plasma are compared over a period of 48 h.
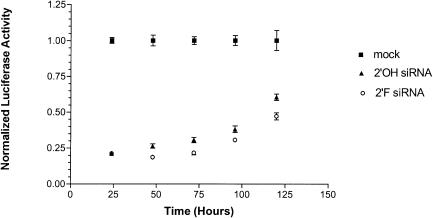
Given that 2′-F modification of siRNA results in a significant increase in stability in vitro, we hypothesized that 2′-F modified siRNA would have longer lasting effects in cell culture. We transfected 2′-OH and 2′-F GL2 siRNA into the HeLa R19-Luc cell line that stably expresses the GL2 luciferase gene. We assayed suppression of luciferase over a period of 5 d (Fig. 3 ▶). Surprisingly, we saw little difference in the duration or magnitude of suppression between 2′-F and 2′-OH siRNAs. Whereas the amount of inhibition of luciferase activity was similar for both siRNAs, 2′-F siRNA induced only a slightly increased level of inhibition compared to 2′-OH over time. This observation contrasts with the results obtained by Chiu and Rana (2003), who reported that in their system the effects of 2′-F modified siRNA were more persistent than that of unmodified siRNA. The difference in results may be due to the system used. Whereas we used a cell line that was stably transfected with a reporter vector, Chiu and Rana used transient transfection of their reporter vector, which could have been diluted out of the cells over time as they divided. In addition, in our system we are constitutively expressing luciferase; as the cells divide the siRNA is being diluted but the reporter is not. This latter approach may more closely approximate the conditions found in vivo. Finally, our results are consistent with those of Song et al. (2003), who recently demonstrated that 2′-OH siRNA activity is limited by dilution in cell culture, not intracellular stability.
FIGURE 3.
2′-F and 2′-OH siRNA effects over a 120-h time course. The luciferase activities of 2′-OH (triangles) and 2′-F (open circles) siRNAs were compared over a 120-h time course in a HeLa cell line constitutively expressing luciferase (squares). The luciferase activities of nine replicates were normalized to mock transfected cells.
Effect of 2′-F modification on siRNA activity in vivo
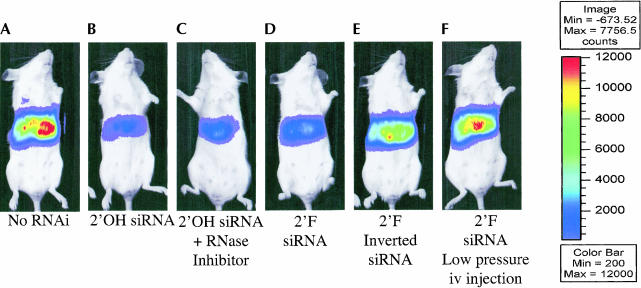
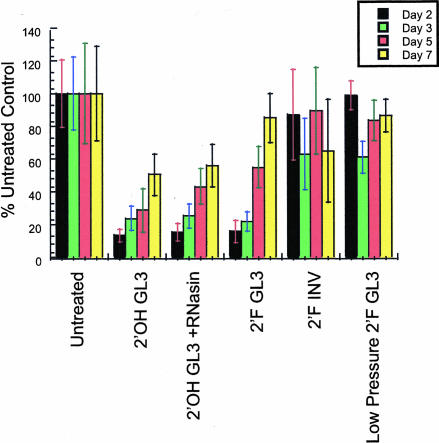
To test the effects of 2′-F modifications of siRNA activity in vivo, we utilized a hydrodynamic transfection method to codeliver pGL3 luciferase reporter plasmids and synthetic 2′-OH and 2′-F GL3 siRNAs. High-pressure injection of naked DNA into the tail vein of rodents leads to efficient transgene expression in the liver (Liu et al. 1999; Zhang et al. 1999). Recently, we modified the hydrodynamic trans-fection method described above in order to codeliver siRNAs (McCaffrey et al. 2002a, 2003) and luciferase expression plasmids to the livers of mice. Luciferase expression was monitored in living animals using quantitative whole-body imaging (Contag et al. 1997). As seen in Figure 4 ▶, both 2′-OH and 2′-F siRNA were able to reduce lucifer-ase expression in vivo by greater than 85%. However, this suppression decreased over time (Fig. 5 ▶). Moreover, 2′-F modification of the siRNA did not grant an advantage to either the magnitude or duration of response, in agreement with the response observed in the cell culture experiments. A change in delivery method of siRNA from high pressure to low pressure resulted in abolishing siRNA activity, presumably because not as much siRNA was taken up by the liver (Fig. 5 ▶). It is also relevant to note that the addition of RNase inhibitors to the injection cocktail did not yield any obvious enhancement of 2′-OH siRNA effect and stability. In contrast, delivery of functional luciferase mRNAs to the livers of mice by hydrodynamic transfection requires formulation with RNasin (McCaffrey et al. 2002b). Thus double-stranded siRNAs may be more resistant to nuclease degradation during delivery compared to long single-stranded RNAs. Hydrodynamic transfection results in rapid cytoplasmic delivery of nucleic acids. Although 2′-F modified siRNAs have much greater stability in plasma than 2′-OH siRNA, it is possible that when this delivery system is used, the molecules are delivered and induce a response before significant degradation can occur. Furthermore, these data suggest that once siRNAs are localized within the cell, modifications that enhance nuclease stability do not influence activity or persistence. Nevertheless, these studies clearly demonstrate that nuclease-resistant siRNAs are functional in mammals. Thus it will be interesting to determine if such stability translates into improved siRNA activity in other animal models, disease settings, or in the context of other delivery modalities.
FIGURE 4.
2′-F modified siRNA inhibits gene expression in living mice. Representative bioluminescence images of light emitted from living mice transfected, 2 d earlier, with the luciferase expression plasmids pGL3-control. Mice (N = 4–5 mice per group) received pGL3-control alone (A) or were cotransfected with (B) 2′-OH GL3 siRNA, (C) 2′-OH GL3 siRNA and the RNase inhibitor, RNasin, (D) 2′-F GL3 siRNA, (E) 2′-F inverted GL2 siRNA. The mouse in panel F received a low pressure i.v. injection of 2′-F GL3 siRNA on day 1 to test if stabilized siRNAs could be taken up by the liver without hydrodynamic transfection. Mice in panels B, C, and D show significant reductions in emitted light as a result of RNA. Hydrodynamic transfection with 2′-F inverted GL2 siRNA and low pressure i.v. transfection of 2′-F GL3 siRNA did not result in significant RNAi.
FIGURE 5.
Inhibition of luciferase activity over time. Compared to mice that received pGL3-control alone, the amount of light emitted from mice as a result of luciferase activity is reduced upon hydrodynamic transfection with siRNAs and 2′-F siRNAs but not with control 2′-F siRNAs. Maximal RNAi was observed at 48 h post-transfection and the degree of inhibition declined over time. Administration of 2′-F siRNAs by low pressure i.v. transfection did not result in substantial gene silencing, presumably because these RNAs are not efficiently taken up by hepatocytes in vivo.
MATERIALS AND METHODS
RNA preparation
Single-stranded 21-nt RNAs were chemically synthesized by Dhar-macon Research, Inc. Synthetic oligonucleotides were desalted, deprotected, and gel purified. The sequences for GL2, GL3, and the inverted GL2 siRNAs have been previously reported (Elbashir et al. 2001). All pyrimidines in both sense and antisense strands were modified with 2′-F pyrimidines. To anneal the siRNAs, 20 μM single strands were incubated in annealing buffer (100 mM potassium acetate, 30 mM HEPES at pH 7.4, 2 mM magnesium acetate) at 95°C for 1 min, then at 37°C for 1 h.
Cell culture and transfections
NIH/3T3 cells were maintained at 37°C, 5% CO2 in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco). HeLa R19-Luc cells were maintained at 37°C, 5% CO2 in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 5 μg/mL blasticidin (Invitrogen). Cells were regularly passaged to maintain exponential growth.
Sixteen hours before all transfections, cells were trypsinized, diluted with fresh media, and plated 5 × 104 cells per well in 24-well plates. Cotransfection of siRNA and reporter plasmids into NIH/3T3 cells was carried out using Superfect (Qiagen) following the protocol provided by the manufacturer for transient transfection of adherent cells. Per well, 1 μg of pGL2-control or pGL3-control (Promega), 1 ng pRL-TK (Promega), and 0.26 μg siRNA duplex were used with a final volume of 360 μL. Each experiment was repeated in triplicate. Cells were incubated at 37°C, 5% CO2 for 24 h, and luciferase and Renilla expression was monitored using the Dual-Luciferase Reporter 1000 Assay from Promega. Luciferase and Renilla readings were taken on the EG&G Berthold Lumat LB 9507 using 100 μL reagent and a 28-sec reading time per sample. For the time course experiments, HeLa/R19-Luc cells were transfected with 20 pmoles of siRNA per well using GeneSilencer siRNA Transfection Reagent (Gene Therapy Systems) according to the protocol provided by the manufacturer for transfection of adherent cells. Cells were incubated at 37°C, 5% CO2 for desired times. At 24 h, one sample set was read and a duplicate plate was trypsinized and split 1:5 into new 24-well plates. Luciferase expression was monitored using the Dual-Luciferase Reporter 1000 Assay from Promega.
Reporter plasmids were amplified in XL-1 Blue (Stratagene) and purified using Clonetech’s NucleoBond Plasmid Maxi kit, both per manufacturer’s instructions.
RNA stability
siRNA was 5′-end labeled using T4 Polynucleotide Kinase (New England Biolabs) and [γ-32P]ATP (Amersham Biosciences). Ten picomoles of labeled siRNA was then incubated at 37°C in 500 μL pooled human plasma (George King BioMedical Inc.) containing 5 U heparin (Elkins-Sinn, Inc.) and 5 mM calcium chloride. At each time point, 2 μL plasma was removed and added to 8 μL TE and 10 μL 2× loading dye (0.5× TBE, 6% Ficoll, 0.1% bromophenol blue, 0.1% xylene cyanol FF), frozen on dry ice, and stored at −20°C. An additional 5 units of heparin were added after the 24-h time point was taken. A starting time point was taken by calculating how many counts were in each 2-μL sample and using that number of counts of labeled siRNA. When all time points had been taken, the samples were thawed and run on a 15% nondenaturing TBE acrylamide gel at 10 W for approximately 2 h. The gel was then exposed to a phosphorimager screen for 2 h and quantified on a Molecular Dynamics Storm 840 Phosphorimager. The total counts of the full-length siRNA band at each time point were divided by the total counts of the starting material to determine the percentage full length siRNA remaining.
Hydrodynamic transfections and bioluminescence imaging
Hydrodynamic transfections of plasmids in PBS were carried out as described (Liu et al. 1999; Zhang et al. 1999) with some modifications (McCaffrey et al. 2002a). Briefly, 1.8 mL of DPBS (with no MgCl2 or CaCl2) was mixed with 3 μg of pGL3-Control (Promega), 3 μg of pThAAT (Yant et al. 2000), 10 μg of indicated siRNA, and if indicated, 20 μL of recombinant RNasin (Promega). This mixture was introduced by tail vein injection over a period of 5 sec. Mice that did not receive the injection in one smooth bolus were excluded without analysis. The mice in the “low pressure” group received 3 μg of pGL3-Control on day 0 followed by a slow infusion of GL3 siRNA in 200 μL of DPBS the following day to assess uptake of siRNAs in the absence of hydrodynamic transfection. Light emitted as a result of luciferase reporter expression was monitored using whole-body imaging of in vivo luciferase activity (Contag et al. 1997). At indicated times, 80 μL of 30 mg/mL luciferin (Biosynth International) was injected i.p. Ten minutes post-injection, live anesthetized mice were imaged for 10 sec to 1 min using an intensified CCD camera (IVIS Imaging System, Xenogen). A representative mouse whose emitted light was closest to the average for that group (4–5 mice per group) is shown (Fig. 4 ▶). This image is comprised of a pseudocolor image representing intensity of emitted light (red most intense and blue least intense) superimposed on a grayscale reference image (for orientation). Serum hAAT levels were measured by ELISA at day 3 as described (Yant et al. 2000) and were similar in all groups, indicating that transfection efficiencies were similar in each group (data not shown). Eighteen- to 22-g female BALB/c mice were obtained from Jackson Laboratory. Animals were treated according to NIH Guidelines for Animal Care and the Guidelines of Stanford University.
Acknowledgments
This work was supported by funds from the National Institutes of Health (NIH) to B.A.S. HeLa R19-Luc cell line was generously provided by Mariano Garcia-Blanco (Duke University Medical Center).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409**:** 363–366. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., and Corey, D.R. 2003. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42**:** 7967–7975. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L. and Rana, T.M. 2003. siRNA function in RNAi: A chemical modification analysis. RNA 9**:** 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag, C.H., Spilman, S.D., Contag, P.R., Oshiro, M., Eames, B., Dennery, P., Stevenson, D.K., and Benaron, D. A. 1997. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol. 66**:** 523–531. [DOI] [PubMed] [Google Scholar]
- Czauderna, F., Fechtner, M., Dames, S., Aygun, H., Klippel, A., Pronk, G.J., Giese, K., and Kaufmann, J. 2003. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31**:** 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., and Sharp, P.A. 2003. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell. Biol. 4**:** 457–467. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411**:** 494–498. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391**:** 806–811. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286**:** 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404**:** 293–296. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418**:** 244–251. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Vandenburgh, K., Manninga, H., Scaringe, S.A., Weber, K., and Tuschl, T. 2003. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 13**:** 83–105. [DOI] [PubMed] [Google Scholar]
- Lee, N.S., Dohjima, T., Bauer, G., Li, H., Li, M.J., Ehsani, A., Salvaterra, P., and Rossi, J. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20**:** 500–505. [DOI] [PubMed] [Google Scholar]
- Liu, F., Song, Y., and Liu, D. 1999. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6**:** 1258–1266. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P., Meuse, L., Pham, T.T., Conklin, D.S., Hannon, G.J., and Kay, M.A. 2002a. RNA interference in adult mice. Nature 418**:** 38–39. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P., Ohashi, K., Meuse, L., Shen, S., Lancaster, A.M., Lukavsky, P.J., Sarnow, P., and Kay, M.A. 2002b. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5**:** 676–684. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P., Nakai, H., Pandey, K., Huang, Z., Salazar, F.H., Xu, H., Wieland, S.F., Marion, P.L., and Kay, M.A. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21**:** 639–644. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3**:** 737–747. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M. and Taira, K. 2002. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20**:** 497–500. [DOI] [PubMed] [Google Scholar]
- Paul, C.P., Good, P.D., Winer, I., and Engelke, D.R. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20**:** 505–508. [DOI] [PubMed] [Google Scholar]
- Pieken, W.A., Olsen, D.B., Benseler, F., Aurup, H., and Eckstein, F. 1991. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253**:** 314–317. [DOI] [PubMed] [Google Scholar]
- Rusconi, C.P., Scardino, E., Layzer, J., Pitoc, G.A., Ortel, T.L., Monroe, D., and Sullenger, B.A. 2002. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419**:** 90–94. [DOI] [PubMed] [Google Scholar]
- Song, E., Lee, S.K., Dykxhoorn, D.M., Novina, C., Zhang, D., Craw-ford, K., Cerny, J., Sharp, P.A., Lieberman, J., Manjunath, N., et al. 2003. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77**:** 7174–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y., and Forrester, W.C. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 99**:** 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant, S.R., Meuse, L., Chiu, W., Ivics, Z., Izsvak, Z., and Kay, M.A. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 25**:** 35–41. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101**:** 25–33. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Budker, V., and Wolff, J.A. 1999. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 10**:** 1735–1737. [DOI] [PubMed] [Google Scholar]