Prelamin A and lamin A appear to be dispensable in the nuclear lamina (original) (raw)
Abstract
Lamin A and lamin C, both products of Lmna, are key components of the nuclear lamina. In the mouse, a deficiency in both lamin A and lamin C leads to slow growth, muscle weakness, and death by 6 weeks of age. Fibroblasts deficient in lamins A and C contain misshapen and structurally weakened nuclei, and emerin is mislocalized away from the nuclear envelope. The physiologic rationale for the existence of the 2 different Lmna products lamin A and lamin C is unclear, although several reports have suggested that lamin A may have particularly important functions, for example in the targeting of emerin and lamin C to the nuclear envelope. Here we report the development of lamin C–only mice (Lmna+/+), which produce lamin C but no lamin A or prelamin A (the precursor to lamin A). Lmna+/+ mice were entirely healthy, and Lmna+/+ cells displayed normal emerin targeting and exhibited only very minimal alterations in nuclear shape and nuclear deformability. Thus, at least in the mouse, prelamin A and lamin A appear to be dispensable. Nevertheless, an accumulation of farnesyl–prelamin A (as occurs with a deficiency in the prelamin A processing enzyme Zmpste24) caused dramatically misshapen nuclei and progeria-like disease phenotypes. The apparent dispensability of prelamin A suggested that lamin A–related progeroid syndromes might be treated with impunity by reducing prelamin A synthesis. Remarkably, the presence of a single LmnaLCO allele eliminated the nuclear shape abnormalities and progeria-like disease phenotypes in Zmpste24–/– mice. Moreover, treating Zmpste24–/– cells with a prelamin A–specific antisense oligonucleotide reduced prelamin A levels and significantly reduced the frequency of misshapen nuclei. These studies suggest a new therapeutic strategy for treating progeria and other lamin A diseases.
Introduction
Lamin A and lamin C, alternatively spliced products of Lmna (1), are key structural components of the nuclear lamina, an intermediate filament meshwork lying beneath the inner nuclear membrane (2). Both lamin A and lamin C contain an aminoterminal globular domain, a central helical rod domain, and a carboxyterminal globular domain (2–4). Lamins A and C dimerize to form parallel coiled-coil homodimers, then associate head-to-tail to form strings, and ultimately form a higher-order filamentous meshwork that provides structural support for the nucleus (2, 3). Lamins A and C also interact with a number of proteins in the heterochromatin, including transcription factors, and bind to membrane proteins within the inner nuclear membrane (e.g., emerin) (5).
Lamins A and C are identical for 566 amino acids, but contain different carboxyterminal domains (1, 4, 6, 7). Lamin C (572 amino acids) contains 6 unique amino acids at its carboxyl terminus. Lamin A (646 amino acids) is actually generated from a precursor protein, prelamin A, by a series of 4 posttranslational processing steps (reviewed in ref. 8). Prelamin A (664 amino acids) terminates with a CaaX motif, which first triggers farnesylation of a carboxyterminal cysteine (the C of the CaaX motif) by protein farnesyltransferase. Second, the last 3 amino acids of the protein (i.e., the aaX) are released from the protein, likely a redundant function of a pair of prenyl protein–specific endoproteases of the ER, Rce1 and Zmpste24. Third, the newly exposed farnesylcysteine is methylated by Icmt, a membrane methyltransferase of the ER. Fourth, the carboxyterminal 15 amino acids of prelamin A, including the farnesylcysteine methyl ester, are clipped off by Zmpste24 and degraded, leaving behind mature lamin A. At first glance, all of the prelamin A processing could appear to be unnecessary, given that mature lamin A lacks all of these posttranslational modifications. However, several studies have suggested that the posttranslational processing steps play a role in the targeting of prelamin A to the nuclear envelope (9, 10). The hydrophobic farnesylcysteine methyl ester probably assists in the delivery of prelamin A to the inner nuclear membrane, where Zmpste24 cleaves the carboxyl terminus and releases mature lamin A (10).
The physiologic importance of lamins A and C has been dramatically illustrated by Lmna knockout mice (Lmna–/–) (11). Lmna is not needed for embryonic development; however, Lmna–/– mice grow slowly after birth and die by 5–6 weeks of age, with histologic evidence of muscular dystrophy (11). The absence of lamin A and lamin C causes grossly misshapen cell nuclei (11) and structurally weakened nuclear envelopes (12). Interestingly, emerin is mislocalized to the ER in Lmna–/– cells, indicating an essential role for lamins A and C in the targeting of emerin to the inner nuclear membrane (11). Mutations in the emerin gene cause X-linked Emery-Dreifuss muscular dystrophy, suggesting that emerin is critical for muscle physiology (7).
The complete processing of farnesyl–prelamin A to mature lamin A is critically important. In the setting of Zmpste24 deficiency, the release of the last 15 amino acids from prelamin A does not occur, resulting in an accumulation of farnesyl–prelamin A at the nuclear envelope (13, 14). This accumulation of farnesyl–prelamin A is toxic (15, 16) and results in strikingly misshapen nuclei at the cellular level and a host of progeria-like disease phenotypes at the “whole-animal” level (e.g., osteoporosis, osteolytic lesions in bones, alopecia, micrognathia, and dental abnormalities) (14, 15). Of note, the classic pediatric progeroid syndrome Hutchinson-Gilford progeria syndrome (HGPS) is caused by the production of a mutant form of farnesyl–prelamin A (frequently called progerin) that cannot be processed to mature lamin A (17). The HGPS mutation can be considered a toxic gain-of-function mutation.
Although a deficiency in lamins A and C and an accumulation of farnesyl–prelamin A both have disastrous consequences for mice (11, 13, 14), the physiological importance of mature lamin A relative to lamin C has remained obscure. Would the absence of lamin A alone be disastrous for mammalian cells? The answer is simply not known, although several studies have suggested that lamin A could be particularly vital for cells. One study suggested that lamin A, but not lamin C, was capable of assisting with the targeting of emerin to the nuclear envelope (18). Another study suggested that lamin A assists both emerin and lamin C in reaching the nuclear envelope (19).
Determining whether lamin A is truly essential for mammalian cells is an extremely important topic, for the simple reason that several human diseases — from restrictive dermopathy (20, 21) to progeria (17) to cardiomyopathy (22, 23) — are caused by genetic defects that affect prelamin A and lamin A but not lamin C. If lamin A were shown to be dispensable, one could begin to think about treating lamin A diseases with therapies designed to reduce lamin A expression. On the other hand, if lamin A played some crucial role in cellular biology or mammalian physiology, for example in the targeting and function of emerin, such an approach would probably be inappropriate.
In this study, we report the development and characterization of mice that synthesize lamin C but not lamin A (lamin C–only mice). Lamin C–only mice (Lmna+/+) were entirely healthy, with normal growth rates and no evidence of bone or muscle disease. The targeting of lamin C and the nuclear membrane protein emerin were normal in Lmna+/+ fibroblasts, and analysis of cell nuclei revealed only minor alterations in nuclear shape and nuclear deformability. Remarkably, a single LmnaLCO allele eliminated the nuclear shape abnormalities and progeria-like disease phenotypes in Zmpste24–/– mice. Our studies revealed that lamin A appears to be dispensable, opening up a new strategy for the treatment of progeria and other lamin A diseases.
Results
Lamin C–only mice.
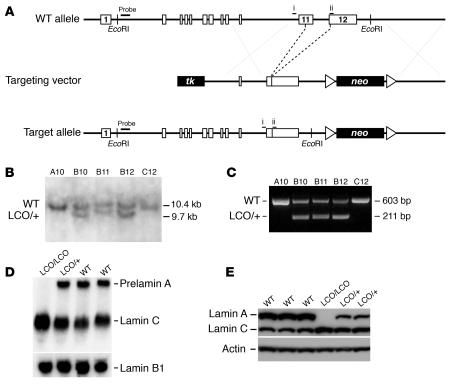
Lamin C–only mice were generated during an attempt to create a Lmna allele yielding wild-type lamin C and the mutant prelamin A produced in HGPS. We used gene targeting to create mice with a mutant Lmna allele lacking the last 150 nucleotides of exon 11 and all of intron 11 (Figure 1, A–C), a mutation that we anticipated would lead to the production of the truncated prelamin A found in HGPS (17). Unexpectedly, however, this deletion (which created an exon 11–12 fusion exon) prevented the formation of a prelamin A transcript because the exon 11–12 fusion was not recognized by the mRNA splicing machinery. Thus, a lamin C transcript was the sole product of this mutant Lmna allele (Figure 1D). We designated this the lamin C–only allele (LmnaLCO). In keeping with the absence of the prelamin A transcript, Western blots of Lmna+/+ fibroblasts with an antibody against lamins A and C revealed lamin C and absolutely no lamin A, even on overexposed Western blots (Figure 1E).
Figure 1.
Production of a mutant Lmna allele, LmnaLCO. (A) A sequence-replacement vector was used to remove intron 11 and the last 150 nucleotides of exon 11. Unexpectedly, this mutation eliminated the splicing event required to produce prelamin A. Exons are shown as boxes. Locations of the PCR primers (i and ii) and the 5′ flanking probe for Southern blots are shown. tk, thymidine kinase gene (for negative selection). (B) Southern blot detection of the LmnaLCO (LCO/+) allele in _Eco_RI-cleaved genomic DNA from ES cells. (C) PCR identification of wild-type and LmnaLCO alleles. Shown are results with wild-type (A10, C12) and LmnaLCO/+ (B10, B11, B12) ES cell clones from gene-targeting experiments. (D) Northern blot of total RNA from Lmna+/+ (+/+), LmnaLCO/+ (LCO/+), and Lmna+/+ (LCO/LCO) fibroblasts; the blot was hybridized with a mouse Lmna cDNA probe that detects both prelamin A and lamin C transcripts and a mouse Lmnb1 cDNA probe that detects lamin B1, a closely related lamin protein. (E) Western blots of 3 different wild-type, 2 heterozygous, and 1 homozygous primary fibroblast cell lines at the same passage number with a polyclonal antibody against lamin A/C and actin. A minor form of lamin A lacking exon 10 has been reported previously in some human cells (43). It is conceivable that this minor lamin could be synthesized, but this would require splicing from exon 9 to the exon 11–12 fusion. We have not identified this minor splice variant in the lamin C–only fibroblasts.
In wild-type fibroblasts, prelamin A and lamin C transcripts were present in similar amounts as determined by Northern blots (Figure 1D). In Lmna+/+ cells, all Lmna transcription was diverted to the production of lamin C, thus the levels of the lamin C transcript in Lmna+/+ cells were higher than in wild-type cells (Figure 1D). Similarly, the amount of lamin C protein in Lmna+/+ and LmnaLCO/+ fibroblasts was higher than in wild-type cells as judged by Western blots (Figure 1E).
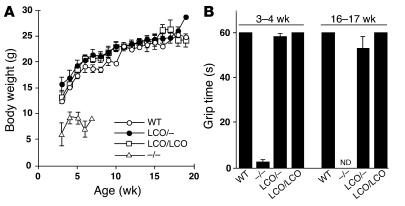
_Lmna–/–_mice exhibit growth retardation and muscle weakness, which was evidenced by an inability to hang upside-down from a grid (11). Neither abnormality was present in _Lmna+/+_mice. Indeed, Lmna+/+ mice were indistinguishable from wild-type control mice in terms of their growth and grip strength (Figure 2). The mice appeared normal over 2 years of observation, and necropsy and histological studies did not uncover any abnormalities in skeletal or cardiac muscle or any other tissue (data not shown).
Figure 2.
Weight gain and grip strength is normal in Lmna+/+ and LmnaLCO/ – mice. (A) Female LmnaLCO/ – (n = 9) and Lmna+/+ mice (n = 13) exhibit normal growth, indistinguishable from wild-type mice (n = 9). Similar results were obtained with male mice (not shown). The growth rate for female Lmna –/ – mice (n = 7) is shown for comparison. (B) Grip strength in 3- to 4-week-old and 16- to 17-week-old mice as judged by the length of time that they were able to hang upside-down from a grid (14). For 3- to 4-week-old mice, n = 7 (wild-type), 13 (Lmna –/ –), 9 (LmnaLCO/ –), and 11 (Lmna+/+). For 16-week-old mice, n = 3 (wild-type), 6 (LmnaLCO/ –), and 7 (Lmna+/+). Grip strength in Lmna –/ – mice at 16 weeks of age was not determined (ND) because the mice did not survive to this age. NS, wild type versus Lmna+/+.
We considered the possibility that the increased lamin C synthesis in Lmna+/+ mice somehow compensated for the loss of lamin A, thereby preventing the emergence of disease phenotypes. However, we also bred mice with 1 Lmna knockout allele and 1 LmnaLCO allele (LmnaLCO/+ mice); these mice synthesized lamin C exclusively in approximately the same amount as in wild-type mice. The LmnaLCO/+ mice (n = 9) appeared entirely normal, and their growth rate and grip strength were indistinguishable from wild-type and Lmna+/+ mice (Figure 2).
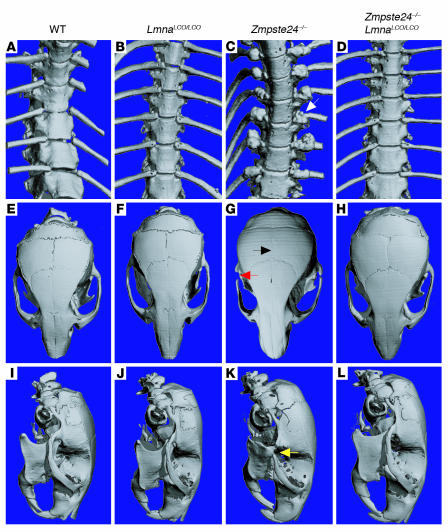
Zmpste24–/– mice (which produce lamin C and prelamin A but no mature lamin A) invariably develop rib fractures and osteolytic lesions in multiple bones (14). We reasoned that _Lmna+/+_mice might also be susceptible to such bone abnormalities, inasmuch as _Lmna+/+_mice, like _Zmpste24–/–_mice, have a complete inability to produce mature lamin A. To test this hypothesis, we performed μCT scans on Lmna+/+ and Zmpste24–/– mice. The Zmpste24–/– mice exhibited the hallmark broken ribs and osteolytic lesions in the posterior zygomatic arch (Figure 3) described previously (14). In contrast, the bones in Lmna+/+ mice were normal, strongly suggesting that the bone disease in Zmpste24–/– mice is caused by the accumulation of farnesyl–prelamin A rather than an absence of mature lamin A.
Figure 3.
Bone abnormalities are absent in Lmna+/+ and Zmpste24 –/ –Lmna+/+ mice. Surface renderings of μCT scans of the spine and skull are shown for 28-week-old wild-type (A, E, and I), Lmna+/+ (B, F, and J), Zmpste24 –/ – (C, G, and K) and Zmpste24 –/ –Lmna+/+ mice (D, H, and L). (A–D) Thoracic spine. In the Zmpste24 –/ – mouse (C), callus around a rib fracture is indicated by a white arrow. Rib fractures were absent in Lmna+/+ and Zmpste24 –/ –Lmna+/+ mice. (E–H) Top view of skulls. Zmpste24 –/ – mice showed a loss of the zigzag appearance of the cranial sutures (black arrow) and an osteolytic lesion of the zygomatic arch (red arrow). Wild-type, Lmna+/+, and Zmpste24 –/ –Lmna+/+ skulls were normal. (I–L) Lateral view of skulls. Zmpste24 –/ – mice exhibited a small mandible and an osteolytic lesion in the posterior portion of the zygomatic arch (yellow arrow). These abnormalities were absent in wild-type, Lmna+/+, and Zmpste24 –/ –Lmna+/+ skulls.
Analysis of fibroblasts from lamin C–only mice.
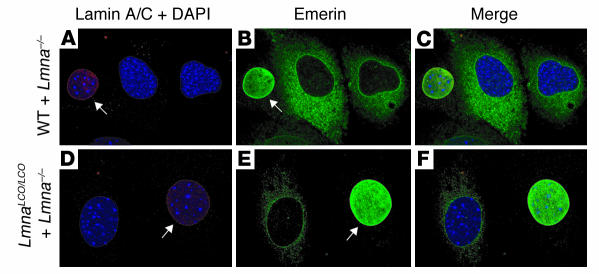
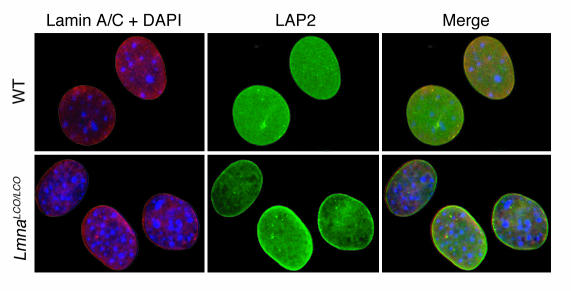
In Lmna–/– cells, emerin is mislocalized to the ER and does not reach its proper location in the inner nuclear membrane (14). We suspected that emerin localization in Lmna+/+ cells might be abnormal, in view of earlier reports suggesting that lamin A, and not lamin C, directs emerin to the inner nuclear membrane (18, 19). To explore this issue, we examined emerin localization in Lmna–/– and Lmna+/+ cells with confocal immunofluorescence microscopy (Figure 4). These studies revealed unequivocally that emerin targeting to the nuclear envelope was normal in Lmna+/+ cells. In addition, both lamina-associated polypeptide 2 (LAP2) and lamin C were normally positioned at the nuclear envelope in Lmna+/+ cells (Figure 5). Previous studies have shown that emerin is normally positioned at the nuclear rim in Zmpste24–/– cells (13).
Figure 4.
Emerin localization is normal in Lmna+/+ primary embryonic fibroblasts. The localization of lamins A and C (red) and emerin (green) in wild-type, Lmna –/ –, and Lmna+/+ fibroblasts was determined by confocal fluorescence microscopy. DAPI was used to visualize DNA. To obtain side-by-side comparisons with Lmna –/ – cells, wild-type and Lmna+/+ fibroblasts were mixed together with Lmna –/ – cells in equal proportions and plated on the same coverslip. (A–C) Wild-type cells plus Lmna –/ – cells. Cells were stained with an anti–lamin A/C antibody, and wild-type cells (arrows) were identified by red fluorescence. As expected based on the studies of Sullivan and coworkers (11), emerin was normally located within the nuclei of wild-type cells, but was mislocalized to the ER in Lmna –/ – cells. (D–F) Lmna+/+ cells plus Lmna –/ – cells. Cells were plated and stained as described for wild-type cells. Lmna+/+ cells (arrows) were identified by red fluorescence. Emerin was located in the nucleus of Lmna+/+ cells with a pattern indistinguishable from that of wild-type cells.
Figure 5.
Lamin C is normally targeted to the nuclear envelope in Lmna+/+ primary embryonic fibroblasts. Wild-type and Lmna+/+ cells were incubated with antibodies against lamin A/C (red) and LAP2 (green) or DAPI to visualize DNA (blue), and the cells were examined by epifluorescence microscopy. The red fluorescence in wild-type cells identifies both lamin A and lamin C, whereas the red fluorescence in Lmna+/+ cells represents only lamin C staining. Both lamin C and LAP2 in Lmna+/+ cells were located in the nucleus and nuclear rim, and their distribution was indistinguishable from that of wild-type cells.
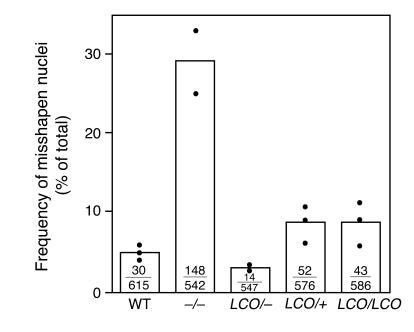
A large fraction of Lmna–/– cells contain misshapen nuclei (11). To determine whether the absence of lamin A caused similar abnormalities, we examined the percentage of misshapen nuclei in LmnaLCO/+, Lmna+/+, LmnaLCO/+, Lmna–/–, and Lmna+/+ primary embryonic fibroblasts. In 3 independent experiments, each involving several independent cell lines and scored by 2 genotype-blinded observers, the percentage of misshapen nuclei in LmnaLCO/+ and Lmna+/+ cells was only slightly greater than in wild-type cells (P = 0.007, χ2 test; Figure 6). No increase in the frequency of misshapen nuclei, however, was observed in LmnaLCO/+ cells (Figure 6).
Figure 6.
Analysis of nuclear shape in wild-type, Lmna –/ –, LmnaLCO/ –, LmnaLCO/+, and Lmna+/+ fibroblasts. The nuclear envelope of cells was visualized with antibodies against lamin B or LAP2, and the number of cells with misshapen nuclei was scored by 2 trained observers blinded to genotype. Bars show the mean frequency of misshapen nuclei; the number of misshapen nuclei and the total number of nuclei examined are shown within each bar. The graph summarizes data for 3 independent experiments (filled circles). Both LmnaLCO/+ and Lmna+/+ cells had a slight but statistically significant increase in number of misshapen nuclei compared with wild-type cells (P < 0.007, χ2 test).
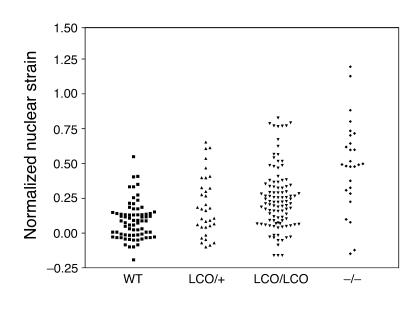
To further explore the possibility that the nuclear lamina in Lmna+/+ cells might be abnormal, we examined the extent of nuclear deformation in response to the application of biaxial strain to cells. Uniform biaxial strain was applied to wild-type and Lmna+/+ cells grown on fibronectin-coated silicon membranes, and the extent of nuclear deformation was measured and expressed as a ratio of nuclear strain to membrane strain. Measurements of normalized nuclear strain overlapped substantially in Lmna+/+ and wild-type cells; when all measurements were included, however, we noted a small but significant increase in normalized nuclear strain in Lmna+/+ cells compared with wild-type cells (wild-type, 0.091 ± 0.016, n = 72, versus Lmna+/+, 0.250 ± 0.022, n = 103; P < 0.0001; Figure 7). In the same series of experiments, the mean normalized nuclear strain in Lmna–/– cells was far higher (0.629 ± 0.055; P < 0.001 versus wild type).
Figure 7.
_Lmna+/+_and _LmnaLCO/+_cells exhibit greater nuclear deformation by biaxial strain than do wild-type cells. Biaxial strain was applied to wild-type, LmnaLCO/+, Lmna+/+, and Lmna –/ – cells, and the extent of nuclear deformation was measured and expressed as a ratio of nuclear strain to membrane strain (Normalized nuclear strain). LmnaLCO/+ and Lmna+/+ cells exhibited a small but statistically significant increase in normalized nuclear strain compared with wild-type cells (P < 0.001, unpaired t test with Welch’s correction).
Correcting cellular and disease phenotypes in Zmpste24–/– mice by interfering with the synthesis of prelamin A.
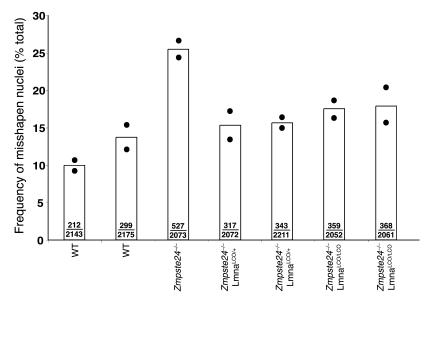
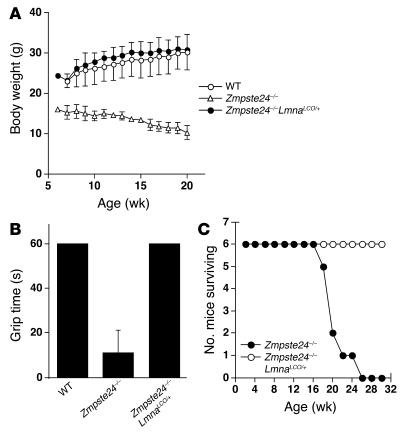
Zmpste24–/– mice manifest an accumulation of farnesyl–prelamin A at the nuclear envelope, develop misshapen nuclei, and exhibit a host of progeria-like disease phenotypes (e.g., osteolytic lesions in bones, alopecia, and reduced subcutaneous fat) (14). We suspected that all of the abnormalities were due to the accumulation of farnesyl–prelamin A rather than to the absence of lamin A. We predicted that we might be able to “cure” Zmpste24–/– mice, at both the cellular and whole-animal levels, by introducing a LmnaLCO allele, as this intervention would reduce the accumulation of prelamin A in cells. Indeed, this prediction was upheld. At the cellular level, Zmpste24–/–LmnaLCO/+ and Zmpste24–/–Lmna+/+ primary embryonic fibroblasts had far fewer misshapen nuclei than fibroblasts from littermate Zmpste24–/– embryos (P < 0.0001, χ2 test; Figure 8). At the whole-animal level, the impact of the LmnaLCO allele was even more dramatic. Zmpste24–/–LmnaLCO/+ and Zmpste24–/–Lmna+/+ mice were phenotypically normal, with normal growth rates and grip strengths indistinguishable from wild-type mice (Figure 9). In contrast, all Zmpste24–/– mice exhibited severely retarded growth and impaired grip strength by 4 months of age. Without exception, the Zmpste24–/– mice died or required euthanasia by 28 weeks of age, whereas all of the Zmpste24–/–LmnaLCO/+ mice were alive, fertile, and healthy at that age (Figure 9). As judged by a μCT scan, a Zmpste24–/–Lmna+/+ mouse was entirely free of bone abnormalities (Figure 3).
Figure 8.
A single LmnaLCO allele reduces the number of misshapen nuclei in primary Zmpste24 –/ – mouse embryonic fibroblasts. Primary embryonic fibroblasts were isolated from embryos generated from Zmpste24+/ –LmnaLCO/+ intercrosses. The cells were stained with lamin A/C and LAP2 antibodies, and the number of cells with abnormal shaped nuclei was determined by fluorescence microscopy. Bars show the mean frequency of misshapen nuclei; the number of misshapen nuclei and the total number of nuclei examined are recorded within each bar. The bar graph summarizes data from 2 independent experiments (filled circles). P < 0.001, Zmpste24 –/ – versus Zmpste24 –/ –LmnaLCO/+ or Zmpste24 –/ –Lmna+/+.
Figure 9.
Growth rates, grip strength, and survival of Zmpste24 –/ –LmnaLCO/+ and Zmpste24 –/ – mice. Compound heterozygotes (Zmpste24+/ –LmnaLCO/+) were intercrossed to generate littermate Zmpste24 –/ –LmnaLCO/+ and Zmpste24 –/ –Lmna+/+ mice. (A) The growth rate of male Zmpste24 –/ – mice (n = 4) was retarded, whereas Zmpste24 –/ –LmnaLCO/+ mice (n = 5) exhibited normal growth indistinguishable from that of wild-type mice (n = 7). Similar results were obtained with male Zmpste24 –/ –Lmna+/+ mice (not shown). (B) Grip strength in 17-week-old wild-type (n = 7), Zmpste24 –/ – (n = 4), and Zmpste24 –/ –LmnaLCO/+ mice (n = 5), as judged by the length of time that they were able to hang upside-down from a grid (14). P < 0.001, t test, Zmpste24 –/ – versus Zmpste24 –/ –LmnaLCO/+. (C) The survival rates were followed over a 30-week period. By 26 weeks of age, all of the Zmpste24 –/ – mice had died (or were euthanized at the request of the veterinary staff). In contrast, all of the Zmpste24 –/ – mice with a single LmnaLCO allele were alive and exhibiting normal health. n = 6 mice per group.
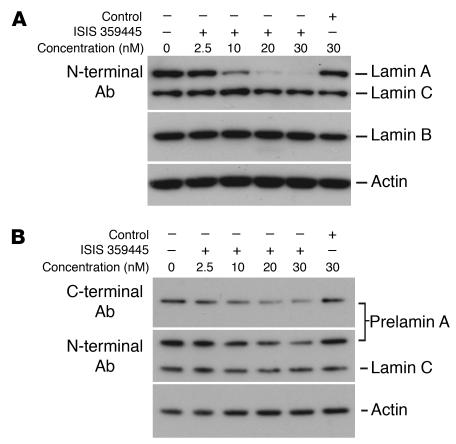
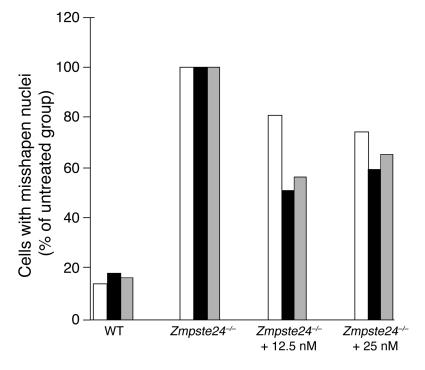
The studies with Zmpste24–/–LmnaLCO/+ mice showed that adding a single LmnaLCO allele to Zmpste24–/– mice resulted in a parallel improvement in nuclear shape and disease phenotypes. Those data, along with the absence of discernible phenotypes in Lmna+/+ mice, suggested that it might be possible to treat progeroid syndromes effectively and with impunity by blocking the synthesis of prelamin A. To begin to test this idea, we screened 78 mouse prelamin A–specific antisense oligonucleotides for their capacity to reduce prelamin A synthesis in mouse fibroblasts. At a concentration of 10 nM, 11 of the oligonucleotides reduced prelamin A mRNA levels by more than 75%, but one (ISIS 359445) reduced prelamin A mRNA levels by more than 95%. ISIS 359445 also reduced lamin A levels in wild-type fibroblasts (Figure 10A) and farnesyl–prelamin A levels in Zmpste24–/– fibroblasts (Figure 10B) without a significant effect on lamin C. We predicted that ISIS 359445 would reduce the frequency of misshapen nuclei in Zmpste24–/– cells. Indeed, this was the case: Zmpste24–/– cells treated with either 12.5 or 25 nM ISIS 359445 had significantly fewer misshapen nuclei than untreated Zmpste24–/– cells (P < 0.0001; Figure 11).
Figure 10.
Western blots of extracts from immortalized wild-type and Zmpste24 –/ – embryonic fibroblasts after treatment with ISIS 359445, a prelamin A–specific antisense oligonucleotide. Cells were transfected with the antisense oligonucleotide, and after 48 hours lamin A (in wild-type cells) or prelamin A (in Zmpste24 –/ – cells) levels were measured by Western blot. (A) Wild-type cells. ISIS 359445 inhibited lamin A protein levels in a concentration-dependent manner, whereas a nonspecific control oligonucleotide had no effect. (B) Zmpste24 –/ – cells. ISIS 359445 inhibited prelamin A protein levels as measured using a prelamin A antibody, whereas the control oligonucleotide had no effect.
Figure 11.
Improved nuclear shape in immortalized Zmpste24 –/ – fibroblasts treated with an antisense oligonucleotide specific for mouse prelamin A (ISIS 359445). Zmpste24 –/ – cells were treated with ISIS 359445; after 72 hours, the cells were stained with antibodies against lamin A/C and LAP2, and the number of cells with misshapen nuclei was scored by 3 trained observers blinded to genotype and treatment. Bars show the frequency of misshapen nuclei as a percentage of untreated Zmpste24 –/ – cells for each individual observer (observer 1, white bars; observer 2, black bars; observer 3, gray bars). There was a statistically significant decrease in the number of misshapen nuclei for cells treated with 12.5 and 25 nM oligonucleotide compared with untreated Zmpste24 –/ – cells for each observer (P < 0.001, χ2 test). The average number of cells with misshapen nuclei in the untreated Zmpste24 –/ – cells was 78.5 cells. The higher baseline level of misshapen nuclei in the Zmpste24 –/ – fibroblasts in this experiment compared with Figure 8 is likely due to the fact that the cells were immortalized. We have previously noted much higher levels of misshapen nuclei in immortalized Zmpste24 –/ – fibroblasts than in primary Zmpste24 –/ – fibroblasts (44).
Discussion
The molecular basis for the synthesis of lamin A and lamin C from a single gene was initially defined in 1993 by the laboratory of Howard Worman at Columbia University (1). Since that discovery, mutations in LMNA have been linked to a potpourri of human genetic diseases (e.g., muscular dystrophy, partial lipodystrophy, cardiomyopathy, progeria, and neuropathy), prompting considerable interest in the nuclear lamina (2, 4, 7). Studies of Lmna knockout mice have underscored the importance of lamin A and lamin C for the integrity of the nuclear envelope, for the targeting of emerin to the nuclear envelope, and for the function of vital tissues such as skeletal and cardiac muscle (11). However, quite surprisingly, we knew very little about the relative importance of lamin A and lamin C, although the prevailing view is that lamin A could be particularly important, with a special role in the targeting of important proteins to the nuclear envelope (18, 19). The existence of a complex and highly conserved pathway for lamin A biogenesis (8, 24) has tended to add weight to the idea that lamin A might be particularly vital. In the current study, we created and analyzed lamin C–only mice, and these studies have yielded fresh insights into lamin A/C biology. The lamin C–only mice (both LmnaLCO/+ and Lmna+/+) were fertile, grew normally, had normal grip strengths and a normal gait, and were entirely healthy over a 2-year period of observation. Thus, at least in the mouse, lamin A appears to be dispensable. At a cellular level, we identified a slightly increased frequency of misshapen nuclei in Lmna+/+ cells compared with wild-type cells along with increased nuclear deformability in response to biaxial strain. However, the differences between wild-type cells and Lmna+/+ cells were small, and we were unable to identify an increase in misshapen nuclei in LmnaLCO/+ cells.
Our studies revealed that lamin A is not required for the targeting of either lamin C or emerin to the nuclear envelope in mouse fibroblasts. The fact that lamin C reaches the nuclear envelope without lamin A probably helps to explain why there were only minimal abnormalities in nuclear shape and deformability in Lmna+/+ cells. The normal targeting of emerin and lamin C to the nuclear envelope in Lmna+/+ cells was somewhat surprising. Raharjo et al. (18) studied emerin targeting in Lmna–/– fibroblasts and showed that a prelamin A cDNA, but not a lamin C cDNA, restored proper targeting of emerin to the inner nuclear membrane. Similarly, Vaughan et al. (19) also concluded that prelamin A plays a special role in the targeting of emerin (and lamin C) to the nuclear envelope. On the other hand, our findings are in keeping with the fact that lamin C is capable of binding emerin (25), although perhaps not as strongly as lamin A (26). Also, our results are consistent with recent studies by Muchir et al. (27), who studied emerin localization in fibroblasts harvested from a human fetus homozygous for a Y259X mutation in LMNA. They found that both emerin and nesprin-1α were mislocalized to the ER in the mutant fibroblasts and that the mislocalization could be normalized by expressing either a lamin C or a prelamin A cDNA.
Our suggestion that lamin A and prelamin A are dispensable means that mice lacking these proteins appear healthy and that Lmna+/+ cells have only subtle abnormalities in shape and mechanics; dispensable does not mean that we believe that prelamin A and lamin A have no purpose. Prelamin A and lamin A have survived millions of years of mammalian evolution, and it remains possible that lamin A has unique and important functions that we have not detected. We simply want to suggest that the physiologic function of prelamin A, whatever it might be, could be relatively unimportant compared to the severe disease phenotypes elicited by an accumulation of farnesyl–prelamin A. Thus, from the perspective of a patient with progeria, living without prelamin A might be quite preferable to living with it.
In Zmpste24–/– mice, farnesyl–prelamin A cannot be cleaved to produce mature lamin A (13, 14). The absence of the final endoproteolytic processing step results in the accumulation of farnesyl–prelamin A at the nuclear envelope, misshapen cell nuclei, and a host of disease phenotypes resembling those occurring in humans with HGPS (e.g., retarded growth, osteolytic lesions in bone, and micrognathia) (13, 14). In the current study, we demonstrate that heterozygosity (or homozygosity) for the LmnaLCO allele eliminated all disease phenotypes in Zmpste24–/– mice. Also, even a single LmnaLCO allele reduced the frequency of misshapen nuclei in Zmpste24–/– fibroblasts. These genetic experiments reinforce the notion that all of the cellular and tissue abnormalities in Zmpste24–/– mice are due to the toxic effects of farnesyl–prelamin A.
The fact that reduced prelamin A synthesis improved the misshapen nuclei and disease phenotypes associated with Zmpste24 deficiency immediately suggested a therapeutic opportunity: that lamin A diseases such as HGPS might be ameliorated by treatment with a prelamin A–specific antisense oligonucleotide. To explore this possibility, we screened for and identified an antisense oligonucleotide, ISIS 359445, that reduced prelamin A expression in mouse cells. This oligonucleotide reduced prelamin A levels in Zmpste24–/– cells, as judged by Western blots, and simultaneously reduced the frequency of misshapen nuclei, as judged by immunofluorescence microscopy.
The fact that the antisense oligonucleotide reduced prelamin A levels in cells and improved nuclear shape is exciting, raising the possibility that systemic administration of these antisense compounds might ameliorate disease phenotypes in Zmpste24–/– mice (14) or newly created mice with a targeted “Hutchinson-Gilford” mutation in Lmna (28). We are cautiously optimistic that ISIS 359445 might be effective in vivo. One reason for optimism is that chimeric phosphorothioate oligodeoxynucleotides containing 2′-O-methoxyethyl (2′-MOE) modifications are extremely resistant to nucleases, have long half-lives (up to a week in mice), and support the RNAse H–mediated removal of the target transcript (29, 30). Oligonucleotides specific for mouse (or human) apoB (employing the same chemistry) have previously been found to be very effective, lowering the levels of both apoB100 and LDL cholesterol in the plasma (31). In that study, therapeutic success was aided by the fact that antisense compounds reach high concentrations in the liver, where virtually all of the apoB100 and LDL cholesterol originate. In the case of the progeria, the antisense reagents would probably need to reduce prelamin A levels in bone, muscle, and fat — tissues where the concentrations of oligonucleotide tend to be lower (30). However, even if oligonucleotide concentrations in these tissues were relatively low, it is still conceivable that disease phenotypes could be partially ameliorated. In this regard, it was heartening that a 50% lowering of prelamin A expression (i.e., with a single LmnaLCO allele) completely eliminated disease phenotypes. Thus, we would not be surprised if a mere 10–20% reduction in prelamin A expression favorably affected the course of the disease. Clearly, experimental studies to test the efficacy of the prelamin A antisense reagents are needed.
We urge caution about the implications of the lamin C–only mice, as it is possible that they may not be entirely relevant to humans. It is possible that the elimination of lamin A could cause disease phenotypes in humans, but not in mice. It is sobering to remember that mice appear to be “tougher” than humans when it comes to mutations in lamins A and C (28, 32, 33). For example, humans carrying a single LMNA H222P allele develop muscular dystrophy, whereas 2 copies of this allele are required to elicit disease in mice (32). Similarly, humans carrying 1 LMNA nonsense allele develop muscular dystrophy (34), whereas mice with a single Lmna knockout allele are entirely normal (11).
Recently, we showed that farnesyltransferase inhibitor (FTI) drugs mislocalized the mutant prelamin A away from the nuclear envelope and largely eliminated the nuclear shape abnormalities in mouse fibroblasts carrying a targeted HGPS mutation (28). Later, we showed that the drugs worked to reduce misshapen nuclei in mouse Zmpste24–/– fibroblasts as well as fibroblasts from humans with HGPS and restrictive dermopathy (35). Currently we are faced with the challenge of determining whether FTIs work to ameliorate disease phenotypes in Zmpste24–/– or HGPS mice. No answers are yet available; nevertheless, we have several concerns about the FTI approach. The first is whether it will be possible to deliver FTIs in sufficiently high doses to block a large fraction of prelamin A farnesylation, and to do so without causing unacceptable side effects. The second concern is that the blockade of farnesylation will leave cells with an abnormal lamin protein (i.e., nonfarnesylated prelamin A) that conceivably could elicit its own disease phenotypes (28, 35). Thus it may be possible that FTIs could improve HGPS disease phenotypes but produce other disease phenotypes as a result of the accumulation of nonfarnesylated prelamin A. Obviously, a prelamin A–specific antisense drug approach would not be burdened by the latter problem. Depending on the outcome of future experiments in mice, we can imagine that combinations of FTIs and antisense reagents might be useful in treating HGPS.
Scaffidi and Misteli (36) reported studies with an antisense oligonucleotide in HGPS fibroblasts. They found that a morpholino oligonucleotide specific for the novel, aberrantly spliced prelamin A transcript in HGPS reduced expression levels for the mutant transcript, leading to reduced numbers of misshapen nuclei. This work was promising, although the approach is only applicable to a single LMNA mutation, and there are obstacles to using conventional morpholino oligonucleotides in humans. The strategy proposed here, simply reducing prelamin A expression, is distinct and would be relevant to the treatment of all lamin A diseases, even those caused by missense mutations (e.g., an R644C mutation associated with cardiomyopathy and progeroid phenotypes; refs. 22, 36). In addition, as noted earlier, our strategy uses an antisense chemistry that has been tested extensively, with considerable success, in both animals and humans (29–31, 37, 38).
Methods
Generation of a lamin C–only allele.
The arms for the gene-targeting vector were generated by long-range PCR of genomic DNA from 129/SvJae ES cells. A 2.5-kb 5′ arm, which spanned from the middle of intron 9 to sequences downstream of the 3′ untranslated region of exon 12, was amplified with forward primer 5′-TTACAGAGCACCGAGCTCTCAGTTGTGTCC-3′ and reverse primer 5′-ATGTGTGGCACCTCTGTGTGCATGTGTCTG-3′. The 5′ arm was cloned into pCR2.1-TOPO and was subjected to 1 round of site-directed mutagenesis for the purpose of deleting the last 150 nucleotides of exon 11 and all of intron 11. This mutagenesis reaction was carried out with the QuikChange kit (Stratagene) with primer 5′-GACAAGGCTGCCGGTGGAGCGGGAGCCCAGAGCTCCCAGAACTGCAGCATCATGTAATCT-3′ and the reverse complemented primer. This deletion creates an in-frame fusion between codon 608 in exon 11 and codon 659 from exon 12. The mutation was verified by DNA sequencing. The mutagenized 5′ arm was removed from the pCR2.1-TOPO vector by restriction with _Eco_RI and cloned into the _Eco_RI site of pKS_loxP_NTmod (39). The 3′ arm of the vector was amplified with forward primer 5′-TTCTAGAAAGGTCAGCATATGTGGGGTGAGCGAGGGG-3′ and reverse primer 5′-TCCACAAACCCTGGCATCTCCATTGGAGATGTGG-3′ and cloned into pCR2.1-TOPO XL. The arm was then removed from the vector and cloned into the _Not_I and _Kpn_I sites of pKS_loxP_NTmod. The integrity of the completed vector was verified by restriction analyses and DNA sequencing. The vector was linearized with _Kpn_I and electroporated into 129/SvJae ES cells. Targeted clones were identified by Southern blotting of _Eco_RI-digested genomic DNA with a 5′ flanking probe. The probe, which is 348 bp in length, was amplified from genomic DNA with forward primer 5′-CAAGGAGCTCGGATTCTGTC-3′ and reverse primer 5′-GTCAGGGAAGAGTGCAGAGG-3′. The probe detected a 10.4-kb band in the wild-type Lmna allele and a 9.7-kb band from the targeted allele. The same probe detects a 7.2-kb band in the Lmna knockout allele (11). Genotyping was also performed by PCR across the deleted DNA segment with forward primer 5′-AGGTGCCGGTCCTAAGAAGT-3′ and reverse primer 5′-ACATGATGCTGCAGTTCTGG-3′. The PCR reaction yielded a 603-bp fragment from the wild-type allele and a 211-bp fragment from the targeted allele. Targeted ES cells were microinjected into blastocysts, and chimeric offspring were bred with C57BL/6 females to produce heterozygotes. Heterozygous mice (LmnaLCO/+) were bred with β-actin–Cre transgenic mice (40) to delete the floxed neo cassette.
Genetically modified mice.
Zmpste24–/– mice (14, 41) were mated with LmnaLCO/+ mice, and the offspring from Zmpste24–/–LmnaLCO/+ intercrosses were analyzed. In addition, LmnaLCO/+ mice were mated with Lmna+/– mice to create LmnaLCO/+ mice. Lmna genotyping was performed both by Southern blotting (11) and by PCR. Zmpste24 genotyping was performed by PCR with oligonucleotides 5′-TCACATGGAGTGAATGCTCTG-3′ and 5′-AGTGAACACCAGGCCAGTTT-3′; the mutant PCR product was 240 bp, and the wild-type product was 320 bp. All mice had a mixed genetic background (∼90% C57BL/6 and ∼10% 129/SvJae). The mice were fed a chow diet and housed in a virus-free barrier facility with a 12-hour light/dark cycle. Mice were weighed weekly, and their ability to hang upside-down from a grid was assessed as described previously (13, 14). All procedures were approved by UCLA’s Institutional Animal Care and Use Committee.
Cell culture and immunofluorescence microscopy.
Primary mouse embryonic fibroblasts were prepared from E13.5 embryos (14). For immunocytochemistry experiments, fibroblasts matched for passage number were grown on coverslips, fixed with 3% paraformaldehyde, and stained as described previously (15, 28, 35). Primary antibodies used were mouse LAP2 monoclonal (1:400 dilution; BD Biosciences), mouse emerin monoclonal (1:200 dilution; Novocastra Laboratories Ltd.), rabbit prelamin A antiserum (1:5,000 dilution), and rabbit lamin A/C polyclonal (1:50 dilution; Santa Cruz Biotechnology Inc.). Secondary antibodies used were anti-rabbit Cy3 (1:800 dilution; Jackson ImmunoResearch Laboratories Inc.) and anti-mouse Alexa Fluor 488 (1:600 dilution; Invitrogen Corp.). DNA was detected with DAPI (Sigma-Aldrich). Cell images were obtained on an Axiovert 40CFL microscope (Zeiss) with a 63×/1.25 oil immersion objective and processed with AxoVision software (version 4.2; Zeiss) or on a TCS SP2 Acusto-Optical beam splitter laser scanning confocal microscope (Leica Microsystems) with a 63×/1.44 oil immersion objective. Numbers of cells with normal-shaped nuclei (smooth oval shape) and abnormally shaped nuclei (blebs, grossly irregular shape, or multiple folds) were counted by 2 observers blinded to genotype. Statistical differences were calculated with the χ2 test.
Prelamin A antisense oligonucleotides.
To identify an active RNAse H–dependent antisense oligonucleotide for prelamin A, a series of 78 chimeric 2′-MOE–modified antisense oligonucleotides were evaluated in mouse brain endothelioma (bEND) cells for the ability to reduce the lamin A mRNA. The efficacy in lowering prelamin A levels was determined by TaqMan (Applied Biosystems) with forward primer 5′-CCTGGTGGGCTCTGTCTCAA-3′, reverse primer 5′-CCTTCGTCCCCCACTCTTC-3′, and probe 5′-ATGGAGGGCAATGTCAAGTGGGATACAG-3′. The most active antisense oligonucleotide was ISIS 359445 (5′-GGGTTCCTTAGTGTTTGCTG-3′; 2′-MOE–modified residues are underlined, other residues are 2′-deoxyribose; all linkages are thioate ester). ISIS 359445 or a control oligonucleotide were transfected into wild-type or Zmpste24–/– mice as described previously (40). Briefly, immortalized mouse fibroblasts were plated in 6-well tissue culture dishes (55,000 cells/well) or 24-well dishes (35,000 cells/well) and allowed to adhere overnight. The cells were washed with reduced serum Opti-MEM I medium (Invitrogen Corp.) and transfected with 2′-MOE–modified oligonucleotides (2.5–30 nM) using 3 μl/ml Lipofectin (Invitrogen Corp.) at 37°C for 5–7 hours. The medium was aspirated and replaced with fresh culture medium (DMEM plus 10% FCS). The cells were cultured for an additional 48–72 hours and processed for Western blotting (6-well) or immunofluorescence microscopy (24-well).
Western blot analyses.
Urea-soluble mouse embryonic fibroblast extracts were prepared as described previously (42), and aliquots were separated on 4–12% gradient polyacrylamide Bis-Tris gels with the NuPage System (Invitrogen Corp.). The size-separated proteins were electrophoretically transferred to nitrocellulose membranes for Western blotting. The antibody dilutions were 1:5,000 rabbit anti–prelamin A antiserum, 1:400 goat anti–lamin A/C antibody (Santa Cruz Biotechnology Inc.), 1:400 goat anti–lamin B antibody (Santa Cruz Biotechnology Inc.), 1:2,000 goat anti-actin IgG (Santa Cruz Biotechnology Inc.), 1:6,000 HRP-labeled anti-rabbit IgG (Amersham Biosciences), 1:6,000 HRP-labeled anti-goat IgG (Santa Cruz Biotechnology Inc.), and 1:4,000 HRP-labeled anti-mouse IgG (Amersham Biosciences). Antibody binding was detected with the ECL Plus chemiluminescence system (Amersham Biosciences) with subsequent exposure to X-ray film.
μ_CT scans_. Wild-type, Zmpste24–/–, Lmna+/+, and Zmpste24–/–Lmna+/+ mice were examined (n = 2 per genotype, 28–30 weeks of age) by compact cone-beam type tomography (MicroCT 40 scanner; Scanco Medical) (14). Whole-body scans were performed in the axial plane mounted in a cylindrical sample holder with a current of 0.16 mA and a voltage of 70 kV at an isotropic voxel size of 20.5 μm for the skulls and 30.7 μm for the remainder of the skeleton. The system was calibrated with a hydroxyapatite phantom of known density. μCT images were constructed in 1024 × 1024 pixel matrices with a standard convolution-back projection procedure with a Shepp-Logan filter. Three-dimensional images of whole bones reconstructed from individual μCT slices were used to qualitatively evaluate whole-bone structure and morphology. A constrained 3-dimensional Gaussian filter was used to partially suppress noise. Bone tissue was segmented from marrow and soft tissue with a thresholding procedure.
Nuclear strain experiments.
Experiments were performed as described previously by Lammerding et al. (12). In brief, cells were plated at subconfluent cell density on fibronectin-coated silicone membranes in DMEM supplemented with 10% FCS, followed by serum starvation for 48 hours in DMEM containing ITS (insulin, transferrin, selenite) supplement (Sigma-Aldrich). Before the strain experiments, cells were incubated with Hoechst 33342 nuclear stain (final concentration, 1 μg/ml; Invitrogen Corp.) in DMEM with ITS for 15 minutes. Subsequently, medium was changed to HBSS with calcium and magnesium (Invitrogen Corp.). Membranes were placed on a custom-made strain device, and uniform biaxial strain was applied to the silicone membrane. Membrane and nuclear strains were computed based on bright-field and fluorescence images acquired before, during, and after strain application using a custom-written image analysis algorithm (12). Normalized nuclear strain was defined as the ratio of nuclear strain to membrane strain to compensate for small variations in applied membrane strain (range 4.2–6.2 %).
Statistics.
The χ2 test was used for the analysis of nuclear shape, and the 2-tailed Student’s t test was used for the analysis of weight and grip strength. Data are expressed as mean ± SD. For nuclear strain studies, statistical analysis was performed using PRISM software (version 3.0; GraphPad). The data were tested to fit a Gaussian distribution and subsequently analyzed by unpaired t test with Welch’s correction to allow for unequal variances. All experiments were performed at least 3 independent times on cell lines independently derived from lamin C–only embryos and wild-type littermates.
Acknowledgments
This work was supported by NIH grants CA099506, AR050200, and AI054384 and grants from the Progeria Research Foundation. Confocal microscopy was performed at the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Facility at UCLA (Laurent Bentolila, director). We thank Brian Young for assistance with artwork.
Footnotes
See the related beginning on page .
Nonstandard abbreviations used: FTI, farnesyltransferase inhibitor; HGPS, Hutchinson-Gilford progeria syndrome; LAP2, lamina-associated polypeptide 2; LCO, lamin C–only; 2′-MOE, 2′-O-methoxyethyl.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 2.Muchir A, Worman HJ. The nuclear envelope and human disease. Physiology (Bethesda). 2004;19:309–314. doi: 10.1152/physiol.00022.2004. [DOI] [PubMed] [Google Scholar]
- 3.Hutchison CJ, Worman HJ. A-type lamins: guardians of the soma? Nat. Cell Biol. 2004;6:1062–1067. doi: 10.1038/ncb1104-1062. [DOI] [PubMed] [Google Scholar]
- 4.Mounkes LC, Burke B, Stewart CL. The A-type lamins. Nuclear structural proteins as a focus for muscular dystrophy and cardiovascular diseases. Trends Cardiovasc. Med. 2001;11:280–285. doi: 10.1016/s1050-1738(01)00126-8. [DOI] [PubMed] [Google Scholar]
- 5.Wilson KL. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- 6.Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. U. S. A. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 8.Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Hennekes H, Nigg EA. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- 10.Lutz RJ, Trujillo MA, Denham KS, Wenger L, Sinensky M. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi:10.1172/JCI200419670. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendás AM, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 14.Bergo MO, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong LG, et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18111–18116. doi: 10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raharjo WH, Enarson P, Sullivan T, Stewart CL, Burke B. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 2001;114:4447–4457. doi: 10.1242/jcs.114.24.4447. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan OA, et al. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- 20.Moulson CL, et al. Homozygous and compound heterozygous mutations in ZMPSTE24 cause the laminopathy restrictive dermopathy. J. Invest. Dermatol. 2005;125:913–919. doi: 10.1111/j.0022-202X.2005.23846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro CL, et al. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum. Mol. Genet. 2005;14:1503–1513. doi: 10.1093/hmg/ddi159. [DOI] [PubMed] [Google Scholar]
- 22.Genschel J, et al. A R644C mutation within lamin A extends the mutations causing dilated cardiomyopathy. Hum. Mutat. 2001;17:154. doi: 10.1002/1098-1004(200102)17:2<154::AID-HUMU11>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Csoka AB, et al. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J. Med. Genet. 2004;41:304–308. doi: 10.1136/jmg.2003.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck LA, Hosick TJ, Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J. Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- 26.Sakaki M, et al. Interaction between emerin and nuclear lamins. J. Biochem. (Tokyo). 2001;129:321–327. doi: 10.1093/oxfordjournals.jbchem.a002860. [DOI] [PubMed] [Google Scholar]
- 27.Muchir A, et al. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res. 2003;291:352–362. doi: 10.1016/j.yexcr.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Yang SH, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts containing a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean NM, Bennett CF. Antisense oligonucleotide-based therapeutics for cancer. Oncogene. 2003;22:9087–9096. doi: 10.1038/sj.onc.1207231. [DOI] [PubMed] [Google Scholar]
- 30.Geary RS, et al. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab. Dispos. 2003;31:1419–1428. doi: 10.1124/dmd.31.11.1419. [DOI] [PubMed] [Google Scholar]
- 31.Crooke RM, et al. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Arimura T, et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 33.Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum. Mol. Genet. 2005;14:2167–2180. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- 34.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 35.Toth JI, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu RZ, et al. Pharmacokinetics and pharmacodynamics of an antisense phosphorothioate oligonucleotide targeting Fas mRNA in mice. J. Pharmacol. Exp. Ther. 2001;296:388–395. [PubMed] [Google Scholar]
- 38.Zinker BA, et al. PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose, and improves insulin sensitivity in diabetic mice. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11357–11362. doi: 10.1073/pnas.142298199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 40.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 41.Leung GK, et al. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- 42.Steinert P, Zackroff R, Aynardi-Whitman M, Goldman RD. Isolation and characterization of intermediate filaments. Methods Cell Biol. 1982;24:399–419. doi: 10.1016/s0091-679x(08)60667-6. [DOI] [PubMed] [Google Scholar]
- 43.Machiels BM, et al. An alternative splicing product of the lamin A/C gene lacks exon 10. J. Biol. Chem. 1996;271:9249–9253. doi: 10.1074/jbc.271.16.9249. [DOI] [PubMed] [Google Scholar]
- 44.Young SG, Fong LF, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]