The PET1-CMS Mitochondrial Mutation in Sunflower Is Associated with Premature Programmed Cell Death and Cytochrome c Release (original) (raw)
Abstract
In mammals, mitochondria have been shown to play a key intermediary role in apoptosis, a morphologically distinct form of programmed cell death (PCD), for example, through the release of cytochrome c, which activates a proteolytic enzyme cascade, resulting in specific nuclear DNA degradation and cell death. In plants, PCD is a feature of normal development, including the penultimate stage of anther development, leading to dehiscence and pollen release. However, there is little evidence that plant mitochondria are involved in PCD. In a wide range of plant species, anther and/or pollen development is disrupted in a class of mutants termed CMS (for cytoplasmic male sterility), which is associated with mutations in the mitochondrial genome. On the basis of the manifestation of a number of morphological and biochemical markers of apoptosis, we have shown that the PET1-CMS cytoplasm in sunflower causes premature PCD of the tapetal cells, which then extends to other anther tissues. These features included cell condensation, oligonucleosomal cleavage of nuclear DNA, separation of chromatin into delineated masses, and initial persistence of mitochondria. In addition, immunocytochemical analysis revealed that cytochrome c was released partially from the mitochondria into the cytosol of tapetal cells before the gross morphological changes associated with PCD. The decrease in cytochrome c content in mitochondria isolated from male sterile florets preceded a decrease in the integrity of the outer mitochondrial membrane and respiratory control ratio. Our data suggest that plant mitochondria, like mammalian mitochondria, play a key role in the induction of PCD. The tissue-specific nature of the CMS phenotype is discussed with regard to cellular respiratory demand and PCD during normal anther development.
INTRODUCTION
Mammalian mitochondria have been shown to play a pivotal role in the induction of apoptosis, a morphologically distinct form of programmed cell death (PCD) (reviewed in Cai et al., 1998; Green and Reed, 1998; Mignotte and Vayssiere, 1998). In mammals, apoptosis is characterized by condensation of the nucleus and the cytoplasm, cleavage of nuclear DNA into ∼180-bp oligonucleosomal units (DNA laddering), and packaging of the cell corpse in vesicles that are ingested by phagocytes. PCD is a genetically based pathway, in contrast to necrosis or “accidental” cell death, which is characterized by swelling of the cell, rupture of the plasma membrane, and spilling of the cell contents. It has been shown in mammalian systems that mitochondria can play a role in the induction of PCD by release of intermembrane space components into the cytosol, including cytochrome c, a key component of the electron transport chain (Liu et al., 1996). The mechanism by which cytochrome c release occurs has yet to be established unequivocally, but it seems to be regulated by antiapoptotic and proapoptotic proteins of the Bcl-2 family (Green and Reed, 1998). Once in the cytosol, cytochrome c activates a proteolytic cascade mediated by caspases (cysteinylaspartate proteases), leading to the oligonucleosomal cleavage of DNA (Enari et al., 1998) and the organized breakdown of the cell.
In plants, several tissues or whole organs undergo cell death as part of their normal development, for example, in xylogenesis, anther dehiscence, and senescence, or in response to pathogens and environmental stresses (reviewed in Greenberg, 1996; Jones and Dangl, 1996; Beers, 1997; Pennell and Lamb, 1997; Richberg et al., 1998; Lam et al., 1999). In contrast to mammalian apoptosis, the genetic basis underlying these developmental and (a)biotically induced cell deaths in plants remains to be discovered. However, many of the cytological and biochemical markers of mammalian apoptosis have been observed in plants, suggesting that gene-specific or “programmed” cell death does exist. By analogy with mammalian apoptosis, plant mitochondria have been suggested to play a pivotal role in the integration of environmental and developmental signals that trigger cell death (Jones, 2000, and references therein).
To further investigate the potential link between mitochondrial function and PCD in plants, we decided to reexamine cytoplasmic male sterility (CMS) in sunflower, which is characterized by the inability to produce viable pollen associated with a mutation in the mitochondrial genome. In humans, several pathologies associated with mutations in the mitochondrial genome have been linked to the role of mitochondria in apoptosis (Wallace, 1999), and it is possible that detailed investigations of mitochondrial mutations in plants may reveal a similar link.
CMS has been described in >150 plant species (Kaul, 1988), and, in combination with the associated nuclear restorer of fertility genes, it has been used extensively in plant breeding for the production of a range of F1 hybrid crops. In CMS plants, anther development may be severely affected, as is the case in homeotic-like mutants of tobacco and Brassica species (Kofer et al., 1991, 1992; Makaroff, 1995), but in most CMS mutants, anthers develop normally and microsporogenesis is arrested during or soon after meiosis (Kaul, 1988). Generally, the tapetum undergoes cellular degradation during meiosis of the meiocytes, followed soon after by the death of the immature microspores.
CMS is associated with the expression of novel open reading frames (ORFs) in the mitochondrial genome in most species investigated to date (Schnable and Wise, 1998) or with the deletion of mitochondrial genes in tobacco (Gutierres et al., 1997). The novel ORFs are thought to have originated by aberrant recombination events in the plant mitochondrial genome. The DNA sequences of the novel ORFs often contain partial sequences of other mitochondrial genes and may be cotranscribed with functional mitochondrial genes. PET1-CMS in sunflower was identified in an interspecific cross between Helianthus petiolaris and H. annuus (Leclercq, 1969) and is associated with the expression of a novel mitochondrial gene, orf522, which encodes a 15-kD polypeptide (Horn et al., 1991; Laver et al., 1991; Monéger et al., 1994). The orf522 gene was created by a recombination event involving an inversion/insertion rearrangement 3′ to the atp1 gene (Köhler et al., 1991; Laver et al., 1991). The first 18 amino acids of ORF522 are identical to ORFB, which may be the plant homolog of yeast and mammalian ATP8, a subunit of the F0F1-ATPase (Gray et al., 1998).
As part of the normal developmental sequence associated with pollen formation and release, several anther tissues undergo cell death in a precisely coordinated and temporal progression (Goldberg et al., 1993; Wu and Cheung, 2000). The tapetal cells lyse and release lipid components that coat the pollen exine (Piffanelli and Murphy, 1998). The stomium cells die and shear to allow release of the pollen from the anther locule. The endothecium and epidermal cell layers undergo cell death, and special cell wall structures in the endothecium cells provide the mechanism whereby the locules spring open at dehiscence. Recently, Papini et al. (1999) showed that the degradation of tapetal cells in two angiosperms (Lobivia rauschii and Tillandsia albida) shows cytological features characteristic of PCD when studied by electron microscopy. These include shrinkage of the cell, condensation of chromatin, swelling of the endoplasmic reticulum, and the persistence of mitochondria. In addition, Wang et al. (1999) observed oligonucleosome-sized DNA cleavage in barley anthers at the end of the unicellular stage of pollen development.
Sunflower is a suitable system in which to study mitochondrial biogenesis and function during flower development because the florets develop in a spatial and temporal sequence such that the older stages are found at the periphery of the capitulum. The stages of pollen development as described by Horner (1977) have been related to bud length (Smart et al., 1994). Therefore, whorls of florets at successive stages of development can be dissected easily, providing enough material for the isolation of mitochondria and associated in situ microscopic analysis. Because the male and female parts of the flower are clearly separated and pollen development takes place when the ovary is relatively undeveloped, florets enriched in anthers can be isolated easily.
To study the role of mitochondria in plant PCD, we have characterized anther development in fertile and PET1-CMS sunflower with respect to morphological and biochemical markers of PCD. We have shown that in plants carrying the PET1-CMS mitochondrial mutation, first the tapetal cells and then the young microspores lost their shape and condensed concomitant with oligonucleosomal cleavage of nuclear DNA. Initially, mitochondria persisted and, in tapetal cells, released cytochrome c into the cytosol. The release of cytochrome c preceded cellular condensation, DNA fragmentation, and a decrease in the integrity of the outer mitochondrial membrane and the respiratory control ratio. These data provide strong evidence that PET1-CMS cytoplasm causes premature induction of PCD in the tapetum, leading to death of the microspores, which results in the male sterile phenotype, and suggest that plant mitochondria are involved in the induction of PCD.
RESULTS
Cytological Features of Anther Development in the Fertile and Sterile Sunflower Lines
Several reports have compared anther development in PET1-CMS sunflower and the isonuclear fertile line at the resolution of light and electron microscopy (Horner, 1977; Laveau et al., 1989; Smart et al., 1994). However, these reports focus on the initial cytological abnormalities during anther development in the sterile line to provide an understanding of the underlying causes of the CMS phenotype. We performed a light microscopic study of the later stages of cellular degradation in PET1-CMS anthers compared with fertile anthers to determine whether the dying tissues exhibit features of necrosis or PCD.
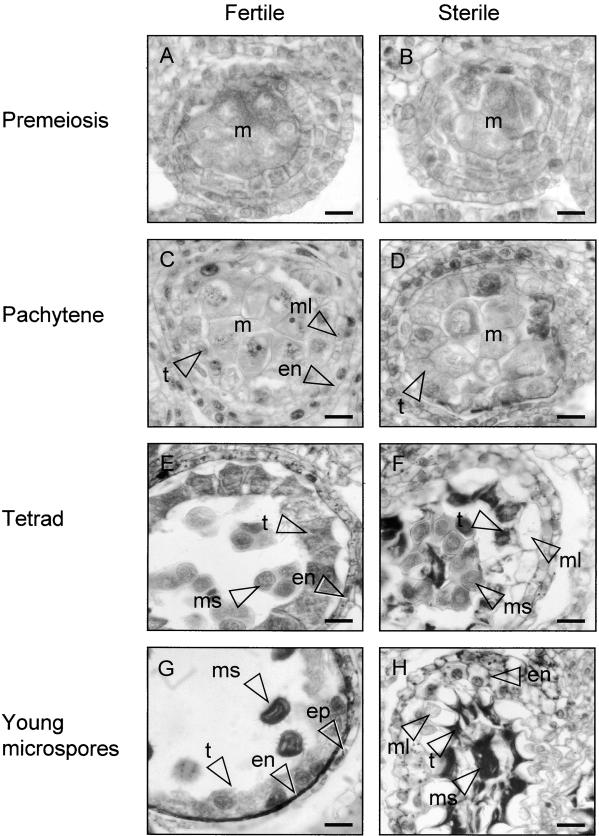
In histologically stained anther sections observed by light microscopy, the first sign of abnormal development in PET1-CMS sunflower became evident at pachytene, when the tapetal cells began to enlarge (Figures 1C and 1D; see also Horner, 1977). This was followed by condensation of the tapetal cytoplasm at the tetrad stage (Figure 1F), loss of regular cell shape, and separation of the chromatin into delineated masses (cf. Figures 5K and 5L).
Figure 1.
Anther Development in Florets of the Fertile and Sterile Sunflower Lines.
Four stages of anther development in the fertile line and the corresponding stages of development in the sterile line were compared using histological staining. The images are of cross-sections through single locules.
(A) Fertile line, premeiosis.
(B) Sterile line, premeiosis.
(C) Fertile line, pachytene.
(D) Sterile line, pachytene.
(E) Fertile line, tetrad stage.
(F) Sterile line, tetrad stage.
(G) Fertile line, stage 6 according to Horner (1977): young microspores.
(H) Sterile line, stage 6 as in (G).
en, endothecium cell(s); ep, epidermal cell(s); m, meiocytes; ml, middle layer; ms, microspore(s); t, tapetal cell(s). Bars = 20 μm.
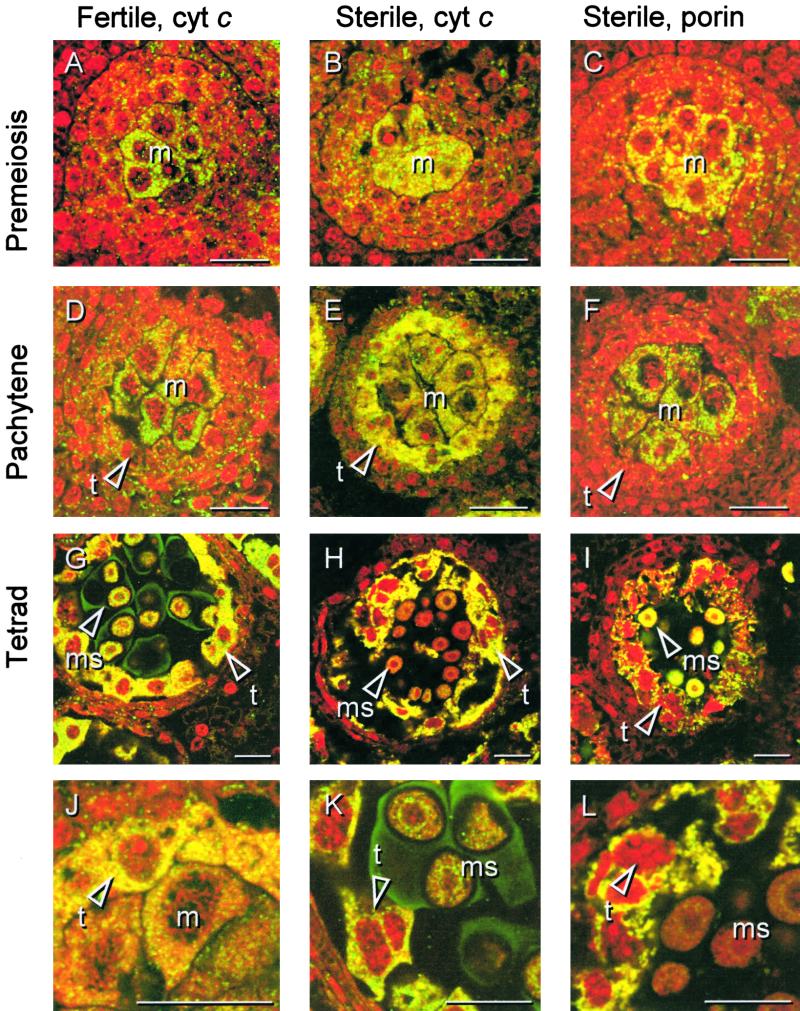
Figure 5.
Immunolocalization of Cytochrome c in Florets of the Fertile and Sterile Sunflower Lines.
Three stages of anther development in the fertile line were compared with the corresponding stages in the sterile line with respect to the in situ localization of cytochrome c. Thin sections from PEG-embedded plant material were labeled with monoclonal antibodies against rat cytochrome c or, as depicted for the sterile line only, with monoclonal antibodies against porin, an integral outer mitochondrial membrane protein. The sections were secondarily labeled with a FITC-conjugated antibody (yellow/green), and nucleic acids were stained with propidium iodide (red) to facilitate identification of the developmental stage.
(A), (B), and (C) Premeiosis.
(D), (E), and (F) Pachytene.
(G), (H), and (I) Tetrad stage.
(A), (D), and (G) show localization of cytochrome c during anther development in the fertile line. (B), (E), and (H) show localization of cytochrome c during anther development in the sterile line. (C), (F), and (I) show localization of porin during anther development in the sterile line.
(J) Subcellular localization of cytochrome c in the sterile line at pachytene shown at higher magnification.
(K) Cytochrome c localization and propidium iodide staining in the fertile line at the tetrad stage shown at higher magnification. The arrowhead points to the chromatin of a tapetal cell.
(L) Cytochrome c localization and propidium iodide staining in the sterile line at the tetrad stage shown at higher magnification. The arrowhead points to the chromatin of a dying tapetal cell.
cyt c, cytochrome c; m, meiocyte(s); t, tapetal cell(s); ms, microspore(s). Bars = 20 μm.
At the tetrad stage, the microspores in the sterile line appeared normal, although they were compressed in the center of the locule (Figure 1F). The middle layer in the sterile line was enlarged, whereas this cell layer had disappeared from the fertile anthers at this stage (Figure 1E). When the florets of the PET1-CMS line had reached a length that corresponds to stage 6 (young microspores) during fertile development (Horner, 1977; Smart et al., 1994), the tapetal cells were further condensed and the microspores were degraded into a lump of condensed material, crushed by the enlarged middle layer (Figure 1H).
In the fertile line, the tapetal cytoplasm stained darker at the tetrad stage (cf. Figures 1C and 1E), the cell walls started to disappear (Horner, 1977), and the cells became plasmodial. When the microspores had developed a spiky exine (Figure 1G), the tapetal cytoplasm stained less intensely and the cells became irregular in shape, with large, diffuse nuclei. The cells and nuclei of the endothecium and epidermal cells condensed from the tetrad stage onward.
Premature Death of Anther Tissues in the Sterile Line Is Associated with Oligonucleosomal Cleavage of DNA
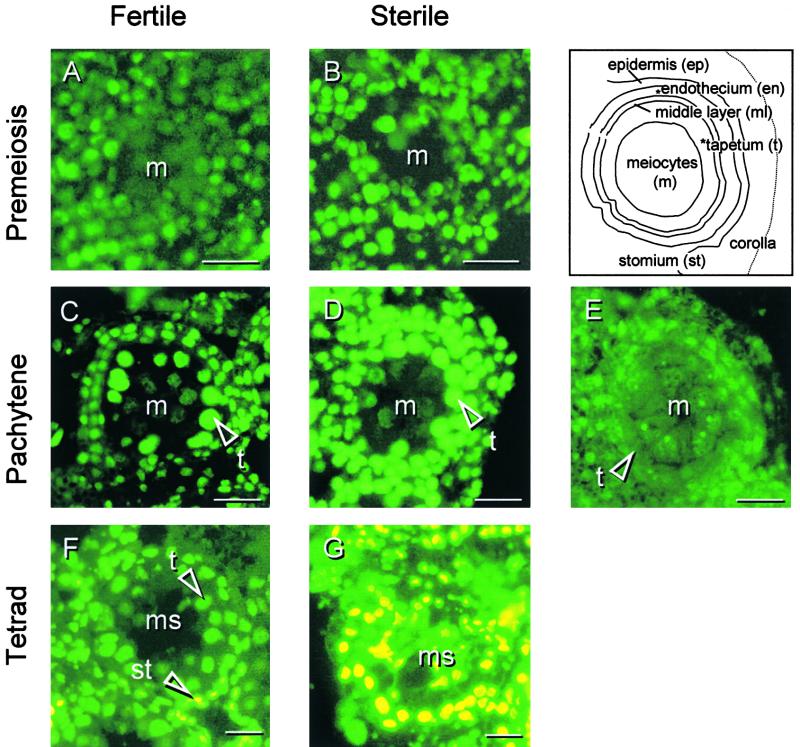
A distinct feature of PCD is the cleavage of nuclear DNA into oligonucleosome-sized fragments (∼180 bp). To determine at which stage of anther development and in which tissues the fragmentation of nuclear DNA occurs, fertile and sterile anthers were compared using the TUNEL assay (TdT-mediated dUTP nick-end labeling). This is an in situ technique that measures the fragmentation of DNA by incorporating fluorescein 12–dUTP at the free 3′-OH ends (Figure 2, bright green to yellow). The stages of development and cytology were identified by differential interference contrast microscopy (data not shown) to avoid the use of fluorescent DNA dyes that have partially overlapping emission spectra with fluorescein.
Figure 2.
DNA Fragmentation in Florets of the Fertile and Sterile Sunflower Lines.
Three stages of anther development in the fertile line and the corresponding stages of development in the sterile line were compared with respect to nuclear DNA fragmentation using the TUNEL assay (bright green to yellow is positive stain).
(A) Fertile line, premeiosis.
(B) Sterile line, premeiosis.
(C) Fertile line, pachytene.
(D) Sterile line, pachytene.
(E) As a negative control, the terminal deoxynucleotidyl transferase enzyme was omitted from the labeling mixture. Sterile line, pachytene.
(F) Fertile line, tetrad stage.
(G) Sterile line, tetrad stage.
m, meiocyte(s); ms, microspores; st, stomium cells; t, tapetal cell(s). Bars = 20 μm.
At premeiosis, there was no difference in TUNEL staining between fertile and sterile anthers (Figures 2A and 2B). However, in the sterile anthers at pachytene (Figure 2D), the nuclei of all anther tissues except for the meiocytes stained more intensely than in their fertile equivalents, suggesting that DNA fragmentation had been initiated. When the sterile florets reached the tetrad stage (Figure 2G), the nuclei of all anther tissues, including the microspores (the meiotic products of the meiocytes), showed clear DNA fragmentation.
At the tetrad stage in fertile anthers, the cells forming the stomium contained condensed nuclei and fragmented DNA (Figure 2F). Nuclear DNA fragmentation in fertile anthers was not investigated beyond the tetrad stage because fertile anther development was used as a reference to anther development in the sterile line. However, Wang et al. (1999) have reported that toward the end of the unicellular stage of pollen development in barley, the nuclei of tapetal cells and the tissues of the anther wall were TUNEL positive and oligonucleosomal DNA cleavage was observed.
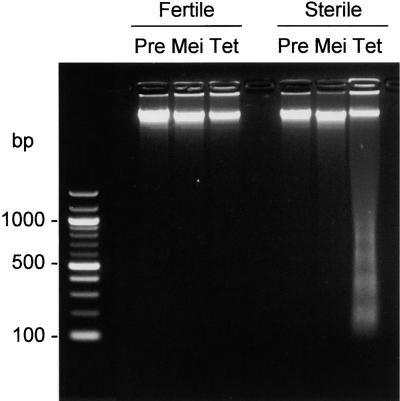
The in situ TUNEL assay does not discriminate between random fragmentation and oligonucleosome-sized cleavage of DNA. To discriminate between the former, which is associated with necrosis, and the latter, which is characteristic of PCD, total DNA was extracted from fertile and sterile florets at the stages of anther development approximating those investigated using the TUNEL assay and analyzed by agarose gel electrophoresis with ethidium bromide staining. The results shown in Figure 3 indicate PCD-specific laddering of DNA in sterile florets at the tetrad stage, correlating with the progressive indication of DNA fragmentation in sterile anthers observed using the TUNEL assay. The lowest fragment corresponded approximately to the size of DNA contained within nucleosomes (z180 bp). No induction of DNA cleavage was observed in the fertile florets at the tetrad stage, despite the appearance of TUNEL-positive nuclei in the stomia, which apparently were too few in number to detect by agarose gel electrophoresis and ethidium bromide staining.
Figure 3.
Oligonucleosomal Cleavage of Total DNA in Florets of the Fertile and Sterile Sunflower Lines.
Agarose gel electrophoresis of total DNA stained with ethidium bromide. The developmental stages correspond to those stages used for the TUNEL assay (Figure 2). Pre, premeiosis; Mei, meiosis; Tet, tetrad stage.
First Signs of Abnormal Development in the Tapetum of Sterile Anthers Are Associated with the Release of Cytochrome c from the Mitochondria
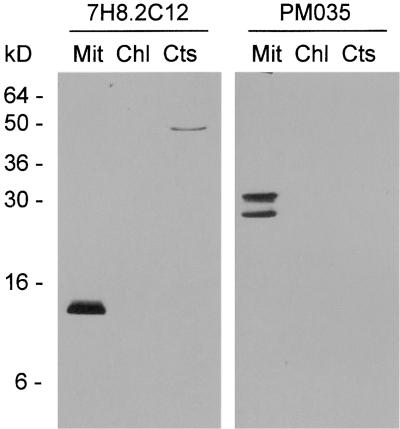
Several reports have demonstrated the translocation of cytochrome c from the mitochondria into the cytosol upon induction of PCD (Balk et al., 1999; Stein and Hansen, 1999; Sun et al., 1999). Therefore, the localization of cytochrome c during anther development was investigated in the fertile and sterile sunflower lines using a monoclonal antibody against rat cytochrome c (7H8.2C12). The epitope recognized by the antibody has been mapped (Jemmerson et al., 1991), and the amino acid sequence of the epitope is identical in rat and sunflower cytochrome c. Cross-reactivity was confirmed by protein gel blot analysis of cytochrome c purified from sunflower seedlings (data not shown). The identity of purified cytochrome c was confirmed by its spectrum (Margoliash and Walasek, 1967) and internal sequence data (see Methods). The 7H8.2C12 monoclonal antibody was tested for use in immunocytochemistry by protein gel blot analysis of subcellular fractions of sunflower tissues. The antibody labeled an ∼12-kD protein in sunflower mitochondria (Figure 4). The antibody cross-reacted weakly with a 50-kD protein in the cytosolic fraction when used for protein gel blot analysis, but in situ, a mitochondria-like, punctate labeling pattern was observed. No cross-reaction with chloroplast proteins was detected.
Figure 4.
Characterization of the Monoclonal Antibodies Used for in Situ Labeling.
Protein gel blot analysis using monoclonal antibodies against rat cytochrome c (7H8.2C12) and maize porin (PM035) on 30 μg of mitochondrial (Mit), chloroplast (Chl), and cytosolic (Cts) proteins from florets (mitochondria and cytosol) or leaves (chloroplast) of the fertile sunflower line.
Localization of porin, an integral outer mitochondrial membrane protein, was used as a control for mitochondrial density and distribution. A monoclonal antibody against maize porin (PM035) recognized three proteins of 28, 30, and 31 kD in sunflower mitochondria (Figure 4). These proteins are likely to be isoforms, as have been reported to exist in wheat (Elkeles et al., 1995). Thin sections of anthers from successive stages of development were labeled with the antibodies as indicated in Figure 5 and visualized with a fluorescein isothiocyanate (FITC)–conjugated secondary antibody (Figure 5, yellow-green). Nucleic acids were stained with propidium iodide (red) to confirm the stage of anther development.
In both fertile and sterile anthers at premeiosis (Figures 5A to 5C), cytochrome c and porin accumulated in the meiocytes, corresponding to the high mitochondrial numbers reported previously (Smart et al., 1994). At pachytene, the localization pattern of porin in sterile anthers (Figure 5F) was similar to the localization pattern of cytochrome c in fertile anthers (Figure 5D), most likely reflecting a high mitochondrial density in the meiocytes. However, when sections of sterile anthers at pachytene were labeled for cytochrome c (Figure 5E), in addition to the punctate labeling in the meiocytes the tapetal cell layer was found to be brightly stained. At higher magnification (Figure 5J), the labeling pattern of cytochrome c in the tapetal cells of sterile anthers at pachytene appeared both punctate and diffuse, in contrast to the entirely diffuse pattern observed in PCD-induced HeLa cells (Bossy-Wetzel et al., 1998). This finding suggests that only part of the cytochrome c is released into the cytosol.
The total intensity of the fluorescence associated with cytochrome c in the tapetum of sterile anthers at pachytene appeared much greater than that for the comparable sections of fertile anthers (Figure 5D), which is likely to be attributable to the increased accessibility of the antigen rather than to a sudden increase in the amount of cytochrome c. Similarly, Varkey et al. (1999) described “display” of the otherwise hidden cytochrome c epitope in Drosophila melanogaster nurse cells undergoing PCD. In D. melanogaster, however, cytochrome c is thought to remain bound to the outer mitochondrial membrane upon release from the intermembrane space, whereas our data suggest that at least part of the cytochrome c is translocated into the cytosol.
At the tetrad stage, the cytological degradation of tapetal cells in the sterile line became apparent (Figures 5H and 5L). In the tapetal cells of both fertile and sterile anthers, the fluorescence associated with cytochrome c was increased and appeared granular/diffuse (Figures 5G and 5H). In contrast, the localization pattern of porin was still punctate, suggesting that mitochondria were not yet degraded completely (Figure 5I; data shown for sterile anthers only). In fertile anthers, cytochrome c was localized mostly around the nucleus of the young microspores (Figures 5G and 5K), corresponding to the localization of mitochondria (Dickinson and Li, 1988; Smart et al., 1994). A similar localization pattern could be seen for porin in sterile anthers (Figure 5I), whereas in sterile anthers the fluorescence associated with cytochrome c was much reduced in the microspores (Figures 5H and 5L).
To semiquantify the amount of cytochrome c associated with mitochondria during fertile and sterile anther development, mitochondria were prepared from the upper, mostly anther-containing part of florets at premeiosis, meiosis, and tetrad stage. Small samples of the mitochondrial fraction were fixed and processed for electron microscopy to confirm that the population of mitochondria isolated displayed the ultrastructural characteristics described by Laveau et al. (1989) and Smart (1994) (data not shown). Mitochondrial proteins were separated by SDS-PAGE, and the relative amount of cytochrome c was determined by protein gel blot analysis and densitometry. Figure 6 shows the relative amount of cytochrome c per milligram of mitochondrial protein averaged over four experiments. The same protein gel blots were stripped and relabeled with a monoclonal antibody against maize E1α, which is a subunit of pyruvate dehydrogenase, an enzyme complex in the mitochondrial matrix. In contrast to the monoclonal antibody against porin (Figure 4), the E1α antibody recognized a single polypeptide and therefore is more suitable for quantification. When the relative amount of cytochrome c was normalized for E1α levels, similar results were obtained, as shown in Figure 6, indicating that any changes found are not caused by differing impurities of the mitochondrial preparations (data not shown).
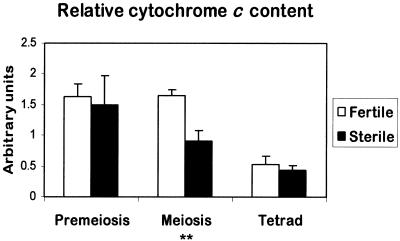
Figure 6.
Relative Cytochrome c Content in Isolated Mitochondria from Florets of the Fertile and Sterile Sunflower Lines.
The relative amount of cytochrome c was determined by protein blot analysis and densitometry, working within a linear range of 20 to 40 μg of mitochondrial proteins as determined experimentally. The data are the average values of four independent experiments (±se), for each of which mitochondrial preparations were derived from four to eight sunflower heads. Statistical analysis was performed using Student's t test on sample means (** 0.001 < P ≤ 0.01).
During fertile anther development, the levels of cytochrome c associated with mitochondria were similar at premeiosis and meiosis but decreased at the tetrad stage (Figure 6). In sterile florets, the mitochondrial levels of cytochrome c at premeiosis were similar to the levels in fertile florets, whereas at meiosis, the relative amount of cytochrome c was significantly lower (0.001 < P ≤ 0.01) than in fertile florets. At the tetrad stage, the mitochondrial cytochrome c levels in sterile florets decreased further to a level that was similar to that found in fertile florets.
To confirm that cytochrome c was translocated from the mitochondria into the cytosol, cytosolic extracts were analyzed by protein gel blot analysis. However, because it was estimated based on specific enzyme activity assays that 1 part of mitochondria (mg of protein) is proportional to more than 10 parts of cytosolic protein (mg) extract, the bulk of protein was removed by ammonium sulfate precipitation to enable the detection of cytochrome c by protein gel blot analysis. Low amounts of cytochrome c then could be detected in the 60 to 80% ammonium sulfate precipitate of the cytosolic protein fractions at the later stages of anther development in sterile florets but not at any stage of anther development in fertile florets (data not shown).
Decrease in Mitochondrial Cytochrome c in Sterile Anthers Precedes a Decline in Outer Membrane Integrity and Respiratory Control Ratio
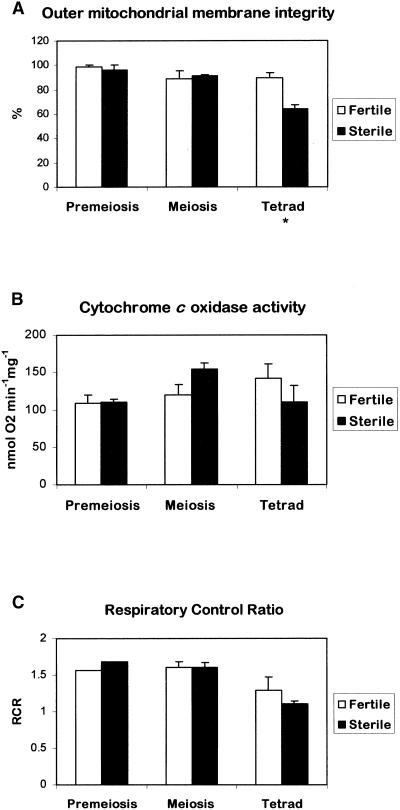
Mitochondria from successive stages of anther development also were used to investigate whether the significant decrease in mitochondrial cytochrome c in sterile anthers at meiosis was the result of a reduction in the integrity of the outer mitochondrial membrane and whether the reduced levels of cytochrome c had an effect on respiration. The respiratory assays were conducted on mitochondrial preparations from two independent experiments that were representative of the four experiments used to quantify the cytochrome c content (Figure 6). The outer mitochondrial membrane integrity assay is based on the accessibility of complex IV (cytochrome c oxidase) to exogenous cytochrome c that is reduced by ascorbate. The outer mitochondrial membrane integrity was similar in mitochondria from fertile and sterile anthers at premeiosis and meiosis but was reduced significantly (0.01 < P ≤ 0.05) in mitochondria from sterile anthers at the tetrad stage (Figure 7A).
Figure 7.
Respiratory Properties of Isolated Mitochondria from Florets of the Fertile and Sterile Sunflower Lines.
Standard oxygen electrode assays were used to determine outer membrane integrity (A), cytochrome c oxidase activity (B), and respiratory control ratio (C). The data are averages of two independent experiments (±se) of those used for Figure 6. Statistical analysis was performed using Student's t test on sample means (* 0.01 < P ≤ 0.05).
To investigate whether the decrease in mitochondrial cytochrome c is associated with downregulation of the amount and/or activity of cytochrome c oxidase and not with a sudden, PCD-associated release, cytochrome c oxidase activity was measured in isolated mitochondria. No significant decrease in cytochrome c oxidase activity was observed during anther development in either the fertile or the sterile line, suggesting that the decrease in cytochrome c is not caused by downregulation of the cytochrome c oxidase pathway (Figure 7B). In contrast, Bino et al. (1986) observed a lower activity of cytochrome c oxidase in CMS petunia just after the onset of cellular degradation and in CMS-S maize before degeneration of the microspores compared with their respective isonuclear fertile lines. However, the enzyme activity measured reflects the maximum activity in the presence of excess cytochrome c substrate, which may not reflect the situation in vivo. Unfortunately, it was not possible to estimate the absolute amounts of cytochrome c oxidase by protein gel blot analysis because there are no antibodies available that cross-react with sunflower cytochrome c oxidase subunits.
The release of cytochrome c from the mitochondria is likely to affect the activity of the respiratory chain and, consequently, ATP synthesis. To obtain a measure of this effect, the oxygen consumption rate of isolated mitochondria was determined with and without ADP. The ratio is known as the respiratory control ratio (RCR) and reflects the degree of coupling of ATP synthesis to electron transport. Figure 7C shows that the RCR was similar in mitochondria isolated from fertile and sterile anthers at premeiosis and meiosis but that there was a significant reduction in RCR at the tetrad stage compared with meiosis in the sterile sunflower line. The RCR also was reduced in mitochondria from fertile anthers at the tetrad stage, but not significantly.
Mitochondrial Biogenesis in Anthers
In PET1-CMS sunflower, the CMS-specific mitochondrial protein ORF522 is expressed in all tissues, but the deleterious phenotype associated with its expression is limited to the anthers. Because this is true for many types of CMS investigated to date, two hypotheses are commonly proposed that could explain the tissue specificity of the CMS phenotype: (1) there exists an anther-specific compound that interacts with the CMS-associated polypeptide, and (2) the respiratory demands are highest in the developing tapetum and/or meiocytes. We have shown a dramatic increase in mitochondrial gene expression during anther development in sunflower (Smart et al., 1994), which was consistent with the observation by Lee and Warmke (1979), who showed, using electron microscopy and quantitative analysis, that during meiosis in maize the mitochondrial number per cell increases 20- and 40-fold in meiocytes and tapetal cells, respectively.
However, while mitochondrial numbers increase rapidly during pollen development, their volumes decrease associated with a loss of internal membrane structures (Lee and Warmke, 1979; Dickinson and Li, 1988). Therefore, mitochondrial number may not be a good measure of mitochondrial biogenesis on a per cell basis. Thus, we performed a quantitative analysis of mitochondrial protein using a fluorescent label and optical sectioning by confocal microscopy in meiocytes and tapetal cells at premeiosis and at pachytene. The data were compared with data from root periderm cells, a vegetative tissue with high respiratory activity. Thick tissue sections (30 to 70 μm) were labeled using an antibody against mitochondrial CPN60 (Robertson et al., 1995), a mitochondrial chaperonin involved in protein import and assembly and hence mitochondrial biogenesis (Prasad et al., 1990). CPN60 labeling was visualized with a FITC-conjugated secondary antibody. The integrated CPN60-specific fluorescence (signal) was expressed per cell (Figure 8A) and per unit volume (Figure 8B).
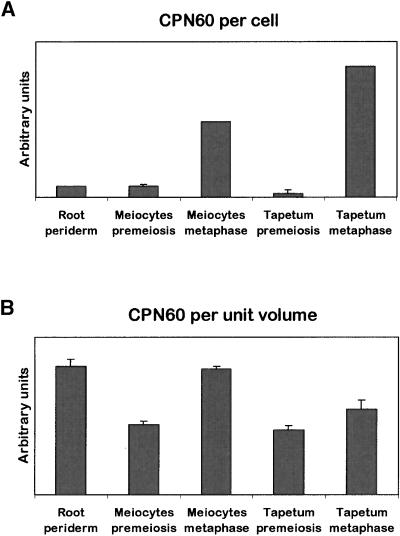
Figure 8.
Quantification of Mitochondrial CPN60 in Several Sunflower Tissues.
Thick sections of several sunflower tissues were labeled with an antibody against CPN60, and the fluorescent signal of the secondary antibody was quantified using confocal microscopy and optical sectioning. The relative CPN60 signal is expressed per cell (A) and per unit volume (B). The data are average values of at least three cells (±se).
The CPN60 signal increased approximately sevenfold in meiocytes and 20-fold in tapetal cells from premeiosis to the meiotic metaphase and was highest in tapetal cells at metaphase. These increases were comparable to the approximately eightfold and ∼12-fold increases, respectively, found by Lee and Warmke (1979) in maize during the same developmental period. The CPN60 signal per cell in root periderm cells was similar to that found in premeiotic meiocytes. Because of the dramatic difference in cell sizes, the CPN60 signal per unit volume in root periderm cells actually was comparable to that in meiocytes at pachytene. Because of an increase in cell size, the increase in CPN60 signal per unit volume during meiosis was far less dramatic than the increase per cell (approximately twofold and 1.5-fold, respectively) for meiocytes and tapetal cells, respectively. In comparison, Lee and Warmke (1979) found a 4.5-fold and threefold increase, respectively, in mitochondrial number per unit volume in maize.
DISCUSSION
PET1-CMS Mutation Is Associated with Premature PCD in Anther Tissues
To study the role of mitochondria in PCD in plants, we have characterized anther development in PET1-CMS sunflower and compared that with anther development in the fertile line. Plants carrying the PET1-CMS cytoplasm are male sterile because of the death of the tapetum and meiocytes near the end of meiosis (Leclercq, 1969; Horner, 1977).
Anther tissues undergo cell death as part of their normal developmental program leading to dehiscence. Recently, Papini et al. (1999) showed that the degradation of tapetal cells in two angiosperms (L. rauschii and T. albida) shows cytological features characteristic of PCD when studied by electron microscopy. In addition, Wang et al. (1999) observed oligonucleosomal DNA cleavage in barley anthers at the end of the unicellular stage of pollen development. In sunflower, we observed the onset of PCD leading to anther dehiscence at the tetrad stage, when the nuclei of the stomium condensed and the nuclear DNA was fragmented. Also, the endothecium and epidermal cells started to condense. In contrast to fertile development, in PET1-CMS sunflower, first the tapetum and then the microspores lost their regular cell shape and condensed starting at the pachytene stage. The cells of the middle layer and later the endothecium and epidermis enlarged.
The TUNEL assay showed that the nuclear DNA of all anther tissues was fragmented at the tetrad stage and, as shown by agarose gel electrophoresis and ethidium bromide staining, that this fragmentation was in fact oligonucleosomal cleavage. The chromatin of the tapetal cells at the tetrad stage separated into delineated masses. Despite the cell collapse and fragmentation of DNA, the mitochondria were not degraded completely, as shown by in situ localization of porin. The latter finding confirms the electron microscopic observation by Horner (1977) that “most organelles seem to be intact, even though the mitochondria are somewhat pleomorphic with some evidently degenerating.” Similarly, Laveau et al. (1989) observed that “the mitochondria and plastids seem normal at the first signs of abnormal development, but soon after the mitochondria show a loss of cristae and a lighter matrix both in tapetal cells and meiocytes.”
Cellular condensation, oligonucleosomal cleavage, the specific appearance of the chromatin, and the initial persistence of the mitochondria all are features of mammalian apoptosis, a morphologically distinct form of PCD. In contrast to what is seen in apoptosis, the nuclei of the dying cells did not condense and the cells were not broken down into apoptotic bodies that could be phagocytosed by neighboring cells. The latter effect, however, is observed rarely in plants, although apoptotic body-like structures have been observed during the PCD induced by mycotoxins (Wang et al., 1996).
The PET1-CMS phenotype is associated with a mutation in the mitochondrial genome leading to the expression of a novel polypeptide, ORF522. Although there is no direct evidence that ORF522 causes the death of the male reproductive tissues, overexpression of ORF522 has been shown to be lethal to Escherichia coli (Nakai et al., 1995). Therefore, our demonstration that the premature death of tapetal cells in the PET1-CMS line exhibited many features of apoptosis strongly suggests that mitochondria are involved in plant PCD.
Cytochrome c Release
A link between plant mitochondria and PCD is supported by our observation that in tapetal cells, cytochrome c was partially released from the mitochondria into the cytosol during both fertile anther development and anther development in the sterile line. In fertile anthers, the release of cytochrome c was observed at the tetrad stage, whereas in the sterile line, cytochrome c release took place at pachytene coincident with the first cytological signs of abnormal development, in agreement with our hypothesis that the PET1-CMS cytoplasm induces premature PCD. However, cytochrome c release into the cytosol was not observed in all tissues with characteristics of apoptosis (e.g., the stomium during fertile development and the microspores in the sterile line). In the latter tissues, the mitochondrial level of cytochrome c was reduced without a corresponding immunolabeling of the cytosol, which could account for the difficulty of detecting cytochrome c in the cytosolic protein fraction of florets. This suggests that in these cells, cytochrome c was degraded rapidly either inside the mitochondria or upon translocation into the cytosol. Complete degradation of the mitochondria could be ruled out because immunolabeling with anti-porin antibodies showed a mitochondria-like, punctate pattern in the microspores.
In mitochondria isolated from successive stages of sterile anther development in sunflower, cytochrome c release preceded a decrease in outer mitochondrial membrane integrity and respiratory control ratio, which is in agreement with our previous results obtained from mitochondria isolated from cucumber cotyledons undergoing heat-induced PCD (Balk et al., 1999). These data raise the question of how cytochrome c is released from the mitochondria. The mechanism of cytochrome c release during mammalian apoptosis is a topic of ongoing debate; however, two mechanistic principles have been proposed (Green and Reed, 1998). One is that the mitochondrial matrix swells as a result of either hyperpolarization or permeability transition of the inner mitochondrial membrane and that, because the inner membrane is folded, the increased volume of the matrix ruptures the outer mitochondrial membrane. The other possibility is that a transient pore is formed in the outer mitochondrial membrane, either by proapoptotic proteins on their own or by interaction with porin.
Our assay for outer mitochondrial membrane integrity is more likely to measure the rupture of the outer membrane, because the transient pores might close upon isolation of the mitochondria and resuspension in isoosmotic buffer. For example, Kluck et al. (1999) reported that two proapoptotic proteins, Bid and Bax, cause complete release of cytochrome c but only a limited permeabilization of the outer membrane in isolated Xenopus mitochondria using an assay similar to that used in our studies. Therefore, our data suggest that during anther development in PET1-CMS sunflower, cytochrome c is released from the tapetal mitochondria via transient pores rather than by swelling of the matrix and rupture of the outer mitochondrial membrane. This is in agreement with the normal appearance of mitochondria as observed by electron microscopy at the onset of tapetum degradation (Horner, 1977; Laveau et al., 1989). The mitochondria isolated from sterile florets at the tetrad stage, however, did show a reduction in outer mitochondrial membrane integrity as well as a reduced respiratory control ratio. These data suggest that at the tetrad stage, both the outer and inner mitochondrial membranes in part of the mitochondrial population have lost their integrity, which is not surprising given the state of cellular degradation visible in histologically stained sections.
Tissue-Specific Phenotype of CMS
How does the PET1-CMS cytoplasm cause the premature death of anther tissues? The male sterile phenotype of the PET1-CMS mutants is associated with the expression of ORF522, a 15-kD hydrophobic protein that is bound to mitochondrial membranes (Horn et al., 1996). Any physiological differences between the PET1-CMS line and the isonuclear fertile line remain to be demonstrated (Hustedt, 1995), but based on its mitochondrial localization, ORF522 is assumed to affect respiration. Although ORF522 is expressed in all tissues, it may cause a relatively minor degree of mitochondrial dysfunction that results in premature cell death in anther tissues only. One explanation for this tissue and developmental specificity is that respiratory demands are highest in the developing tapetum and/or meiocytes, which is supported by the observation that mitochondrial numbers per cell increased dramatically during meiosis in maize (Lee and Warmke, 1979) and Cosmos bipinnatus (Dickinson and Li, 1988) and by the anther-specific accumulation of transcripts of respiratory genes (Smart et al., 1994; Zabaleta et al., 1998). By quantifying the fluorescent signal associated with a biogenesis-associated mitochondrial protein (CPN60), we have confirmed this dramatic increase during anther development in sunflower. However, if the CPN60 signal per unit volume is considered, the values for root tissues are equal to or higher than those found for anther tissues. Therefore, high respiratory demands might not be the only reason for the anther-specific phenotype of CMS.
The alternative hypothesis of an anther-specific compound proposed by Flavell (1974), could play a role in the form of the “ready-to-be-activated” PCD machinery involved in anther dehiscence. Although the genetic basis of PCD remains to be determined, anther-specific protease activities have been characterized in lily (DeGuzman and Riggs, 2000), and a cysteine endopeptidase is expressed in the circular cell cluster, connective and stomium, in tobacco (Koltunow et al., 1990). In addition, tapetum-specific serine proteases have been cloned from lily (Taylor et al., 1997) and pine (Walden et al., 1999), and a tapetum-specific gene (A9) with similarity to cysteine protease inhibitors has been identified in several plant species (Paul et al., 1992; Walden et al., 1999). Therefore, the combination of high respiratory demands and the tissue-specific expression of PCD-associated proteins—activated prematurely by mitochondrial components in the cytosol—could explain the tissue-specific phenotype of CMS. Also, the timing of cell death in the tapetum is critical for the development of functional pollen.
The ORF522 protein shares 18 amino acids at its N terminus with ORFB. Recently, ORFB, which is encoded in all plant mitochondrial genomes studied to date, was found to share a low degree of sequence and structural homology with ATP8 (Gray et al., 1998), a subunit of the FoF1-ATP synthases in Reclinomonas, fungi, and mammals. Because the N terminus of ORFB is important for assembly (Hadikusumo et al., 1988), the presence of ORF522 might affect the assembly of FoF1-ATPase. Interestingly, FoF1-ATPase is thought to play a role in apoptosis (reviewed in Matsuyama and Reed, 2000) in mammals and possibly yeast, although the precise mechanism remains to be determined. Interesting in this respect is a report by Hernould et al. (1998) describing the death of the tapetum followed by the meiocytes in tobacco transformed with an unedited copy of mitochondrion-targeted ATP9.
The many CMS-associated mitochondrial mutations in a range of plant species have proved a very useful tool for the study of mitochondrial function and the role of nuclear genes therein. We have shown that CMS in sunflower causes premature PCD associated with the release of cytochrome c during anther development. CMS, therefore, could be used to identify mitochondrial components that are able to induce cell death and cytosolic components that are involved in the breakdown of the cell (e.g., proteases and endonucleases).
METHODS
Materials
Sunflower (Helianthus annuus) seed were provided by Rhône Poulenc Agrochimie (France). Fertile (RPA842B) and sterile (RPA842A) lines are isonuclear containing the sunflower nuclear genotype. The fertile (maintainer) line carries the sunflower cytoplasm and the sterile line carries the H. petiolaris cytoplasm. Plants were grown in nonheated greenhouses during the summer. Male florets were harvested after ∼40 days. A male floret corresponds to the upper part of the sunflower floret, which contains the anthers. Flowering occurred after ∼50 days.
The monoclonal antibody against rat cytochrome c (clone 7H8.2C12) was purchased from Pharmingen (San Diego, CA). The monoclonal antibodies against porin (PM035) and the E1α subunit of the pyruvate dehydrogenase complex (PM030), both from maize, were purchased from GT Monoclonal Antibodies (Lincoln, NE).
Fixation, Embedding, and Sectioning
Plant material was fixed in 50% (v/v) ethanol, 4% (v/v) formaldehyde, and 5% (v/w) glacial acetic acid for 30 min at room temperature and washed twice with 70% (v/v) ethanol for 15 min. The tissue was stored in 70% (v/v) ethanol at 4°C until further use. Fixed plant material was dehydrated through an ethanol series of 75, 85, 95, and 100% (v/v) and embedded in polyethylene glycol (PEG), 1500 grade (Fisher Scientific), at 56°C. The PEG blocks were sectioned into 7-μm sections, spread on Superfrost Plus slides (Merck), and baked on a hot plate at 40°C overnight. The slides were placed in MilliQ water for 30 sec to remove the PEG and air dried. The sections were then rehydrated through an ethanol series of 100, 90, 70, and 50% (v/v) and MilliQ water for 2 min each before further procedures (see below) or histological staining with Safranin O and Fast Green according to Langdale (1994). All in situ data were obtained from serial sections of florets of the same sunflower line and developmental stage in two independent experiments (including plant material).
TdT-Mediated dUTP Nick-End Labeling
Seven-micrometer sections were washed in PBS (160 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 1.5 mM KH2PO4) for 5 min and incubated in 20 μg/mL proteinase K in 100 mM Tris-HCl, pH 8.0, and 50 mM Na2EDTA (100 μL per slide in a humid chamber). The sections were washed in PBS for 5 min and fixed in 4% (w/v) paraformaldehyde in PBS for 10 min. The sections were washed again in PBS for 5 min, and the 3′-OH ends of DNA were labeled with fluorescein 12–dUTP using the apoptosis detection system fluorescein (Promega), according to the supplier's instructions. The fluorescent signal was viewed using an Axiophot microscope (Zeiss, Jena, Germany) and photographed using the camera supplied (Zeiss) and Provia Fujichrome 400 film (Fuji, Tokyo, Japan).
DNA Isolation and Agarose Gel Electrophoresis
Male florets (0.5 g) were ground in liquid nitrogen. The frozen powder was added to 15 mL of extraction buffer (100 mM Tris-HCl, pH 8.0, 50 mM EDTA, 500 mM NaCl, and 10 mM β-mercaptoethanol) followed by the addition of 1 mL of 20% (w/v) SDS. After incubation at 65°C for 10 min, 5 mL of potassium acetate was added and the sample was incubated on ice for 20 min. The proteins were pelleted by centrifugation at 17,400_g_ at 4°C for 20 min. The supernatant was filtered through Miracloth (Calbiochem) into a tube containing 10 mL of cold isopropanol and incubated on ice for 30 min. The nucleic acids were pelleted by centrifugation at 17,400_g_ for 15 min. The pellet was air dried and resuspended in 0.7 mL of 50 mM Tris-HCl, pH 8.0, and 10 mM EDTA. The resuspension was centrifuged at maximum speed in a microcentrifuge to remove nondissolved debris. Then, one-tenth volume of 3 M sodium acetate and two-thirds volume of cold isopropanol were added to the supernatant, and the samples were incubated on ice for 15 min. The nucleic acids were pelleted at maximum speed in a microfuge for 10 min at 4°C, dried, and resuspended in 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA. RNA was digested in the presence of 0.2 mg/mL RNase A for 30 min at 37°C. DNA (10 μg/lane) was separated on 1.5% agarose gels in 1 × Tris-acetate-EDTA containing 0.1 μg/mL ethidium bromide.
Immunocytochemistry
Seven-micrometer sections were washed in PBS for 2 min and covered in primary antibody diluted 1:100 in PBS and 0.5% (w/v) BSA and incubated at 4°C in a humid chamber overnight. The slides were washed three times, first in PBS and 0.5% (w/v) BSA, then in PBS and 0.01% (v/v) Tween 20, and finally in PBS. The sections were incubated with fluorescein isothiocyanate (FITC)–conjugated rabbit anti-mouse IgG in PBS (Sigma; dilution according to the supplier's instructions) at room temperature in a humid chamber for 1 hr. The slides were washed as described above. The nucleic acids were stained with 10 μg/mL propidium iodide for 5 min. The sections were mounted in citifluor/PBS and sealed with nail polish. The fluorescent signal was viewed using an MRC 1024 confocal system (Bio-Rad) and a BX50WI fixed-stage upright microscope (Olympus, Tokyo, Japan) with a ×60 1.2–numerical aperture oil immersion lens.
Isolation of Mitochondria
Male florets were ground using a mortar and pestle on ice in cold grinding buffer containing 0.4 M mannitol, 25 mM 3-(N_-morpholino)-propanesulfonic acid–KOH, pH 7.8, 1 mM EGTA, 0.1% (w/v) BSA, and 40 mM β-mercaptoethanol. All further procedures were performed at 4°C. Cell debris was pelleted by a quick centrifugation step during which the rotor was stopped as soon as it reached 6000_g. The supernatant was removed and recentrifuged at 12,000_g_ for 15 min to pellet the mitochondria. The supernatant (cytosol) was stored at −80°C for further analysis. The crude mitochondrial pellet was resuspended in wash buffer containing 0.4 M mannitol, 5 mM 3-(N_-morpholino)-propanesulfonic acid–KOH, pH 7.5, 1 mM EGTA, and 0.1% (w/v) BSA and loaded onto step gradients of 13.5 and 27% (v/v) Percoll (Amersham Pharmacia Biotech) in resuspension buffer containing 0.4 M mannitol, 10 mM Tricine-KOH, pH 7.2, and 1 mM EGTA. The gradients were spun at 30,000_g for 45 min in a swinging-bucket rotor. The buff-colored fraction at the 13.5 to 27% interface was collected and washed in resuspension buffer by centrifugation at 12,000_g_ for 15 min. The pellet was resuspended in resuspension buffer, and the protein concentration was determined using the Coomassie Plus protein assay reagent (Pierce Chemical Co.) according to the supplier's instructions.
Purification of Cytochrome c
Cytochrome c was purified from mitochondria isolated from etiolated sunflower seedlings according to the method of Diano and Martinez (1971). Amino acid sequences were generated from two tryptic peptides, AVIWEENTLYD and MVFPGLK, that match the published sequence (Ramshaw et al., 1970).
Isolation of Chloroplasts
All procedures were performed at 4°C. Young sunflower leaves were homogenized with a Polytron (Kinematica AG, Lucerne, Switzerland) in a minimal amount of SRM (0.165 M sorbitol and 25 mM Hepes-KOH, pH 8.0). The homogenate was filtered through two layers of Miracloth (Calbiochem) and centrifuged at 3200_g_ for 2 min. The pellet was resuspended in SRM and loaded onto 35% (v/v) Percoll (Amersham Pharmacia Biotech) pads in SRM. After centrifugation at 1400_g_ for 7.5 min, the pellet was resuspended in SRM and washed by centrifugation at 3200_g_ for 2 min. The pellet was resuspended in a small volume of SRM. The purity of the chloroplast sample was assessed under the light microscope, and the protein concentration was measured using the Coomassie Plus protein assay reagent (Pierce Chemical Co.). The chloroplasts were frozen at −80°C until further use.
Protein Gel Blot Analysis
Protein samples stored at −80°C were separated by SDS-PAGE [15% (w/v) monomer and 2.6% (w/w) cross-linking agent] and transferred to nitrocellulose (0.2 μm; Schleicher and Schuell) by semidry electroblotting. The membrane was labeled with the 7H8.2C12 monoclonal antibody against rat cytochrome c as described by Bossy-Wetzel et al. (1998). For labeling with the porin and E1α monoclonal antibodies, the membrane was blocked against nonspecific binding by incubation in PBS and 0.1% (v/v) Tween 20 (PBS-T) plus 1% (w/v) BSA for 1 hr. The membrane was then incubated in the primary antibody diluted 1:500 in block buffer at 4°C overnight. The membrane was washed three times in PBS-T, incubated with horseradish peroxidase–conjugated sheep anti-mouse Ig (Amersham Life Science), and diluted 1:10,000 in PBS-T for 1 hr. After four washes, labeling was detected by chemiluminescence (DuPont–New England Nuclear Life Science Products) and exposure to Kodak BioMax ML film.
The signal corresponding to cytochrome c was quantified using densitometry. The linear range of mitochondrial protein per lane (20 to 40 μg) and film exposure time (1 to 5 min) were determined experimentally. The films were scanned and analyzed using a Fluor-S Multi-Imager and Multi-Analyst software (both from Bio-Rad).
Oxygen Electrode Assays
All respiratory studies were performed by polarographic measurement of oxygen in the aqueous phase using a Clark-type oxygen electrode (Hansatech, King's Lynn, Norfolk, UK) linked to a chart recorder (Rikadenki, Tokyo, Japan). Freshly isolated mitochondria (50 to 100 μg) were assayed in 1 mL of 0.3 M mannitol, 10 mM Tes-KOH, pH 7.5, 3 mM MgSO4, 10 mM NaCl, 5 mM KH2PO4, and 0.1% (w/v) BSA. Outer mitochondrial membrane integrity was measured as the ratio of oxygen consumption in the presence of 50 μg/mL cytochrome c and 5 mM ascorbate with or without the addition of 0.05% (w/v) Triton X-100. Maximal cytochrome c oxidase activity was considered to be the rate of oxygen consumption in the presence of Triton X-100 in the outer membrane integrity assay. In a separate assay, 10 mM succinate and 0.2 mM ATP were added to the mitochondria, followed by the addition of 0.1 mM ADP to measure the rate of oxygen consumption in state 3. The oxygen consumption in state 4 was measured when the ADP was depleted as judged by a reduction in oxygen consumption rate. The respiratory control ratio was calculated as the ratio of state 3 to state 4 oxygen consumption rate.
Quantitative Analysis of CPN60
Fixed tissue was sectioned into 30- to 70-μm sections using a vibratome (series 1000; General Scientific, Redhill, Surrey, UK) in MilliQ water using Wilkinson razor blades. The sections were transferred to eight-well slides coated with 2% (v/v) 3-aminopropyltriethoxysilane in acetone and subsequently with 2.5% (v/v) glutaraldehyde in PBS. The sections were air dried and then digested with 2% (w/v) cellulase and 1% (w/v) driselase in 25 mM Tris-HCl, pH 7.5, 140 mM NaCl, and 3 mM KCl (TBS) for 1 to 1.5 hr. The slides were washed in TBS for 5 min and incubated with CPN60 antibody (Robertson et al., 1995) diluted 1:1000 in TBS plus 1% (w/v) BSA and 0.01% (w/v) Tween 20 overnight at 4°C. The slides were washed in TBS for 5 min and incubated with goat anti-rabbit FITC-conjugated antibody for 1 hr at room temperature in the dark. The slides were washed again and incubated with propidium iodide (Molecular Probes, Eugene, OR) at a concentration of 10 μg/mL for 20 min to stain nucleic acids, facilitating the determination of the developmental stage.
After washing for 5 min, the slides were mounted in citifluor in glycerol/PBS. The labeling was viewed using an MRC 600 confocal system (Bio-Rad) and a Diaphot inverted microscope (Nikon, Tokyo, Japan) with a ×60 1.4–numerical aperture oil immersion lens (laser, 488 nm; filters, DE1 and R1). All images used for quantitative analysis were made under identical settings (ND0, PMT1 3, PMT2 4, Kalman 3, Zoom 1.6, normal scanning speed). Z-series of 1-μm steps were made for each tissue. The FITC signal was integrated using the software provided (COMOS; Bio-Rad). Bleaching of the signal was not detected during the time used for scanning. No significant levels of labeling above background were detected in the control, in which the primary antibody was omitted. To avoid differences in signal attributable to attenuation, cells from just under the cut surface were selected for analysis.
The value for the total CPN60 signal per cell was obtained with the following equation: Σ(_P_x × S_x) − (Σ_P × BG), where _P_x is the number of pixels per optical section x, _S_x is the average signal per optical section x, and BG is the average background signal (measured in the nucleus; n = 3). The CPN60 signal per unit volume was calculated as the total CPN60 signal per cell divided by Σ_P_x.
Acknowledgments
We thank Paul McCabe for encouraging this project, Lee Sweetlove for critical reading of the manuscript, Tony C. Willis for protein sequencing, and Cledwyn Merriman for processing of the samples for electron microscopy. This work was supported by the Gatsby Charitable Foundation, the Biotechnology and Biological Science Re-search Council, and Lincoln College, Oxford (J.B.).
References
- Balk, J., Leaver, C.J., and McCabe, P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463**,** 151–154. [DOI] [PubMed] [Google Scholar]
- Beers, E. (1997). Programmed cell death during plant growth and development. Cell Death Differ. 4**,** 649–661. [DOI] [PubMed] [Google Scholar]
- Bino, R., Suurs, L.C.J.M., De Hoop, S.J., Van der Neut, A., Van Went, J.L., and Van Marrewijk, G.A.M. (1986). Characterization of cytoplasmic male sterility in Petunia hybrida and Zea mays: Localization and activity of cytochrome c oxidase. Euphytica 35**,** 905–918. [Google Scholar]
- Bossy-Wetzel, E., Newmeyer, D.D., and Green, D.R. (1998). Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J. 17**,** 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, J., Yang, J., and Jones, D.P. (1998). Mitochondrial control of apoptosis: The role of cytochrome c. Biochim. Biophys. Acta 1366**,** 139–149. [DOI] [PubMed] [Google Scholar]
- DeGuzman, R., and Riggs, C. (2000). A survey of proteinases active during meiotic development. Planta 210**,** 921–924. [DOI] [PubMed] [Google Scholar]
- Diano, M., and Martinez, G. (1971). Le cytochrome c des mitochondries du tubercule de pomme de terre. C. R. Acad. Sci. Paris 272**,** 2692–2694. [Google Scholar]
- Dickinson, H.G., and Li, F.L. (1988). Organelle behaviour during higher plant gametogenesis. In The Division and Segregation of Organelles, S. Boffey and D. Lloyd, eds (Cambridge, U.K.: Cambridge University Press), pp. 131–148.
- Elkeles, A., Devos, K.M., Graur, D., Zizi, M., and Breiman, A. (1995). Multiple cDNAs of wheat voltage–dependent anion channels (VDAC): Isolation, differential expression, mapping and evolution. Plant Mol. Biol. 29**,** 109–124. [DOI] [PubMed] [Google Scholar]
- Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., and Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391**,** 43–50. [DOI] [PubMed] [Google Scholar]
- Flavell, R. (1974). A model for the mechanism of cytoplasmic male sterility in plants, with special reference to maize. Plant Sci. Lett. 3**,** 259–263. [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5**,** 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W., et al. (1998). Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 26**,** 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281**,** 1309–1312. [DOI] [PubMed] [Google Scholar]
- Greenberg, J. (1996). Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 93**,** 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres, S., Sabar, M., Lelandais, G., Chetrit, P., Diolez, P., Degand, H., Boutry, M., Vedel, F., de Kouchkovsky, Y., and DePaepe, R. (1997). Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 94**,** 3436–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadikusumo, R., Meltzer, S., Choo, W.M., Jean-François, M.J.B., Linnane, A.W., and Marzuki, S. (1988). The definition of mitochondrial H+ ATPase assembly defects in _mit_− mutants of _Saccharomyces cerevisia_e with a monoclonal antibody to the enzyme complex as an assembly probe. Biochim. Biophys. Acta 933**,** 212–222. [DOI] [PubMed] [Google Scholar]
- Hernould, M., Suharsono, S., Zabaleta, E., Carde, J.P., Litvak, S., Araya, A., and Mouras, A. (1998). Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Mol. Biol. 36**,** 499–508. [DOI] [PubMed] [Google Scholar]
- Horn, R., Köhler, R.N., and Zetsche, K. (1991). A mitochondrial 16-kDa protein is associated with cytoplasmic male sterility in sunflower. Plant Mol. Biol. 17**,** 29–36. [DOI] [PubMed] [Google Scholar]
- Horn, R., Hustedt, J.E.G., Horstmeyer, A., Hahnen, J., Zetsche, K., and Friedt, W. (1996). The CMS-associated 16 kDa protein encoded by orfH522 in the PET1 cytoplasm is also present in other male-sterile cytoplasms of sunflower. Plant Mol. Biol. 30**,** 523–538. [DOI] [PubMed] [Google Scholar]
- Horner, H. (1977). A comparative light- and electron-microscopic study of microsporogenesis in male-fertile and cytoplasmic male-sterile sunflower (Helianthus annuus). Am. J. Bot. 64**,** 745–759. [Google Scholar]
- Hustedt, J.E.G. (1995). Physiologische und Molekularbiologische Aspekte der Cytoplasmatischen Männlichen Sterilität bei der Sonnenblume (Helianthus annuus L). Inaugural Dissertation (Giessen, Germany: Justus-Liebig-Universität).
- Jemmerson, R., Johnson, J.G., Burrell, E., Taylor, P.S., and Jenkins, M.K. (1991). A monoclonal antibody specific for a cytochrome-c T-cell stimulatory peptide inhibits T-cell responses and affects the way the peptide associates with antigen-presenting cells. Eur. J. Immunol. 21**,** 143–151. [DOI] [PubMed] [Google Scholar]
- Jones, A. (2000). Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci. 5**,** 225–230. [DOI] [PubMed] [Google Scholar]
- Jones, A., and Dangl, J.L. (1996). Logjam at the Styx: Programmed cell death in plants. Trends Plant Sci. 1**,** 114–119. [Google Scholar]
- Kaul, M. (1988). Male Sterility in Higher Plants, Vol 10. (Heidelberg, Germany: Springer-Verlag).
- Kluck, R.M., Degli Esposti, M.D., Perkins, G., Renken, C., Kuwana, T., Bossy-Wetzel, E., Goldberg, M., Allen, T., Barber, M.J., Green, D.R., and Newmeyer, D.D. (1999). The pro-apoptotic proteins, Bid and Bax, cause a limited permeabilization of the mitochondrial outer membrane that is enhanced by cytosol. J. Cell Biol. 147**,** 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer, W., Glimelius, K., and Bonnett, H.T. (1991). Modifications of mitochondrial DNA cause changes in floral development in homeotic-like mutants of tobacco. Plant Cell 3**,** 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer, W., Glimelius, K., and Bonnett, H.T. (1992). Fusion of male-sterile tobacco causes modifications of mtDNA leading to changes in floral morphology and restoration of fertility in hybrid plants. Physiol. Plant. 85**,** 334–338. [Google Scholar]
- Köhler, R., Horn, R., Lossl, A., and Zetsche, K. (1991). Cytoplasmic male sterility in sunflower is correlated with the co-transcription of a new open reading frame with the atpA gene. Mol. Gen. Genet. 227**,** 369–376. [DOI] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Wallroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2**,** 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and del Pozo, O. (1999). Die and let live: Programmed cell death in plants. Curr. Opin. Plant Biol. 2**,** 502–507. [DOI] [PubMed] [Google Scholar]
- Langdale, J.A. (1994). In situ hybridization. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 165–180.
- Laveau, J.H., Schneider, C., and Berville, A. (1989). Microsporogenesis abortion in cytoplasmic male sterile plants from H. petiolaris or H. petiolaris fallax crossed by sunflower (Helianthus annuus). Ann. Bot. 64**,** 137–148. [Google Scholar]
- Laver, H., Reynolds, S.J., Monéger, F., and Leaver, C.J. (1991). Mitochondrial genome organisation and expression associated with cytoplasmic male sterility in sunflower (Helianthus annuus). Plant J. 1**,** 185–193. [DOI] [PubMed] [Google Scholar]
- Leclercq, P. (1969). Une sterilité male cytoplasmic chez le tournesol. Ann. Amelior. Plantes 19**,** 99–106. [Google Scholar]
- Lee, S.J., and Warmke, H.E. (1979). Organelle size and number in fertile and T-cytoplasmic male-sterile corn. Am. J. Bot. 66**,** 141–148. [Google Scholar]
- Liu, X.S., Kim, C.N., Yang, J., Jemmerson, R., and Wang, X.D. (1996). Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 86**,** 147–157. [DOI] [PubMed] [Google Scholar]
- Makaroff, C. (1995). Cytoplasmic male sterility in Brassica species. In The Molecular Biology of Plant Mitochondria, C. Levings III and I.K. Vasil, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 515–555.
- Margoliash, E., and Walasek, O.F. (1967). Cytochrome c from vertebrate and invertebrate sources. Methods Enzymol. 10**,** 339–348. [Google Scholar]
- Matsuyama, S., and Reed, J.C. (2000). Mitochondria-dependent apoptosis and cellular pH regulation. Cell Death Differ. 7**,** 1155–1165. [DOI] [PubMed] [Google Scholar]
- Mignotte, B., and Vayssiere, J.L. (1998). Mitochondria and apoptosis. Eur. J. Biochem. 252**,** 1–15. [DOI] [PubMed] [Google Scholar]
- Monéger, F., Smart, C.J., and Leaver, C.J. (1994). Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J. 13**,** 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, S., Noda, D., Kondo, M., and Terachi, T. (1995). High-level expression of a mitochondrial ORF-522 gene from the male-sterile sunflower is lethal to Escherichia coli. Breed. Sci. 45**,** 233–236. [Google Scholar]
- Papini, A., Mosti, S., and Brighigna, L. (1999). Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 207**,** 213–221. [Google Scholar]
- Paul, W., Hodge, R., Smartt, S., Draper, J., and Scott, R. (1992). The isolation and characterization of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol. Biol. 19**,** 611–622. [DOI] [PubMed] [Google Scholar]
- Pennell, R., and Lamb, C. (1997). Programmed cell death in plants. Plant Cell 9**,** 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P., and Murphy, D.J. (1998). Novel organelles and targeting mechanisms in the anther tapetum. Trends Plant Sci. 3**,** 250–253. [Google Scholar]
- Prasad, T., Hack, E., and Hallberg, R.L. (1990). Function of the maize mitochondrial chaperonin HSP60: Specific association between HSP60 and newly synthesized F1-ATPase alpha-subunits. Mol. Cell. Biol. 10**,** 3979–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramshaw, J.A.M., Thompson, E.W., and Boulter, D. (1970). The amino acid sequence of Helianthus annuus L. (sunflower) cytochrome c deduced from chymotryptic peptides. Biochem. J. 119**,** 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richberg, M., Aviv, D.H., and Dangl, J.L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1**,** 480–485. [DOI] [PubMed] [Google Scholar]
- Robertson, E.J., Williams, M., Harwood., J.L., Linsay, J.G., Leaver, C.J., and Leech, R.M. (1995). Mitochondria increase 3-fold and mitochondrial proteins and lipid change dramatically in postmeristematic cells in young wheat leaves grown in elevated CO2. Plant Physiol. 108**,** 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3**,** 175–180. [Google Scholar]
- Smart, C.J. (1994). A Molecular Investigation of Cytoplasmic Male Sterility during Male Flower Development in Sunflower (Helianthus annuus). PhD Dissertation (Oxford, UK: Oxford University).
- Smart, C.J., Monéger, F., and Leaver, C.J. (1994). Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6**,** 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J.C., and Hansen, G. (1999). Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol. 121**,** 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.-L., Zhao, Y., Hong, X., and Zhai, Z.-H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462**,** 317–321. [DOI] [PubMed] [Google Scholar]
- Taylor, A., Horsch, A., Rzepczyk, A., Hasenkampt, C., and Riggs, C. (1997). Maturation and secretion of a serine proteinase is associated with events of late microsporogenesis. Plant J. 12**,** 1261–1271. [DOI] [PubMed] [Google Scholar]
- Varkey, J., Chen, P., Jemmerson, R., and Abrams, J. (1999). Altered cytochrome c display precedes apoptotic cell death in Drosophila. J. Cell Biol. 144**,** 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden, A.R., Walter, C., and Gardner, R.C. (1999). Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol. 121**,** 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D.C. (1999). Mitochondrial diseases in man and mouse. Science 283**,** 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wang, H., Li, J., Bostock, R.M., and Gilchrist, D.G. (1996). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8**,** 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Hoekstra, S., Van Bergen, S., Lamers, G.E.M., Oppedijk, B.J., Van der Heijden, M.W., De Priester, W., and Schilperoort, R.A. (1999). Apoptosis in developing anthers and the role of ABA in this process during androgenesis in Hordeum vulgare L. Plant Mol. Biol. 39**,** 489–501. [DOI] [PubMed] [Google Scholar]
- Wu, H.-M., and Cheung, A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44**,** 267–281. [DOI] [PubMed] [Google Scholar]
- Zabaleta, E., Heiser, V., Grohmann, L., and Brennicke, A. (1998). Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 15**,** 49–59. [DOI] [PubMed] [Google Scholar]