MITOCHONDRIAL DNA MUTATIONS IN HUMAN DISEASE (original) (raw)
. Author manuscript; available in PMC: 2007 Jan 4.
Published in final edited form as: Nat Rev Genet. 2005 May;6(5):389–402. doi: 10.1038/nrg1606
Abstract
The human mitochondrial genome is extremely small compared with the nuclear genome, and mitochondrial genetics presents unique clinical and experimental challenges. Despite the diminutive size of the mitochondrial genome, mitochondrial DNA (mtDNA) mutations are an important cause of inherited disease. Recent years have witnessed considerable progress in understanding basic mitochondrial genetics and the relationship between inherited mutations and disease phenotypes, and in identifying acquired mtDNA mutations in both ageing and cancer. However, many challenges remain, including the prevention and treatment of these diseases. This review explores the advances that have been made and the areas in which future progress is likely.
Until recently, mitochondrial genetics and diseases that are due to defects in mitochondrial DNA (mtDNA) were often considered to be curiosities, outside mainstream genetics. These views were not helped by the different genetic rules by which mtDNA is inherited compared with nuclear DNA, the presence of several mtDNA copies in individual cells and a belief that mtDNA disease is rare. In addition, the clinical syndromes caused by mtDNA mutations have variable phenotypes and are often described by instantly forgettable eponyms or acronyms1. These views perhaps ignore the fact that the mitochondrial genome is central to the study of evolutionary genetics, has an important role in forensic medicine2 and that the human, bovine and mouse mitochondrial genomes were the first mammalian genomes to be completely sequenced3–5.
The increasing ease with which the mitochondrial genome can be analysed, and the availability of a consensus human sequence6, have both helped to recognize mtDNA disorders as a frequent cause of genetic disease. It is difficult to estimate the true prevalence of mtDNA disease owing to its many clinical guises, presentations and the involvement of numerous causative mutations. Data on a single-point mutation (3243A>G) in Finland indicate that approximately 1 in 6,000 individuals are affected, whereas estimates from the British population intimate that about 1 in 3,500 people either have mtDNA disease or are at risk of developing it7,8. These estimates do not include the recent apparent association of mtDNA mutations and common clinical features (for example, hypertension)9, which indicate that the incidence could be higher still. In addition, there is increasing evidence from animal models10 and human studies11 that acquired mtDNA mutations and mitochondrial dysfunction are involved in ageing and age-related diseases such as diabetes12,13. Although our present knowledge already indicates that mtDNA mutations are an important cause of disease, the true impact of these mutations on human health remains to be determined.
This review focuses on abnormalities of the mitochondrial genome in relation to human disease. Mitochondrial disease can also arise from nuclear gene disorders because most proteins involved in mitochondrial metabolism and all those involved in mtDNA maintenance are nuclear-encoded. Mutations in these nuclear genes can mimic the features seen in patients with mtDNA defects, and indeed some nuclear genetic disorders result in secondary abnormalities of the mitochondrial genome. Excellent reviews describe the defects that are due to mutations in nuclear genes that are involved in mitochondrial oxidative metabolism1,14,15. Here we review our current understanding of the role of mtDNA in human disease and highlight areas that are either controversial or merit further studies. We first review the basic features of mitochondrial genetics. Much progress has been made in this area and a clear understanding is important to further our knowledge of the diagnosis, pathogenesis and treatment of mtDNA disease. We then turn to clinical syndromes that are due to mtDNA mutations, recognizing that mtDNA mutations might cause various clinical features, which often makes diagnosis a considerable challenge. Even when diagnosed, there is no curative treatment for patients who have mtDNA disease. Nonetheless, there are several experimental possibilities to either prevent transmission or to correct the genetic defect and these are discussed here. Although models of mtDNA disease exist, these are predominantly cell-based rather than transgenic mouse models. Some valuable models are available, but few mimic the characteristics of pathogenic mtDNA mutations. We conclude by considering the possible role of the mitochondrial genome in ageing, which, despite much speculation, has only recently been supported by firm evidence from an animal model.
Mitochondrial genetics: the basics
Mitochondria are found in all nucleated cells and are the principal generators of cellular ATP by oxidative phosphorylation (OXPHOS), incorporating the electron-transferring respiratory chain (complexes I–IV) and the ATP synthase (complex V). Mitochondria are the only location of extra-chromosomal DNA within the cell (except in plant chloroplasts), and they are under the dual genetic control of both nuclear DNA and the mitochondrial genome. The mitochondrial genome consists of a multicopy, circular dsDNA molecule (16.6 kb in humans), which encodes 13 essential polypep-tides of the OXPHOS system and the necessary RNA machinery (2 rRNAs and 22 tRNAs) for their translation within the organelle (FIG. 1). The remaining protein subunits that make up the respiratory-chain complexes, together with those required for mtDNA maintenance, are nuclear-encoded, synthesized on cytoplasmic ribosomes, and are specifically targeted and sorted to their correct location within the organelle.
Figure 1. The role of the mitochondrial genome in energy generation.
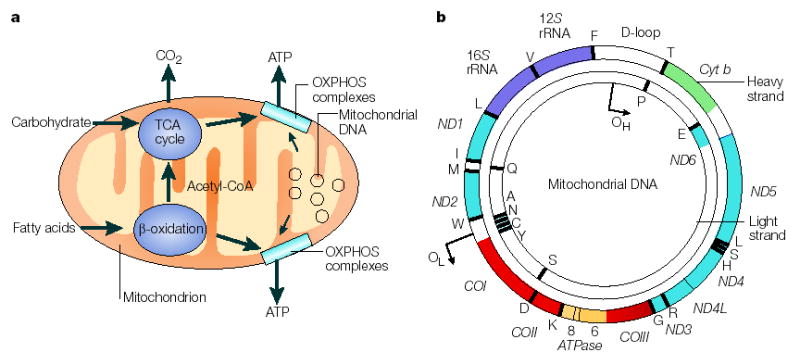
a This highlights the importance of the mitochondrial genome in contributing polypeptide subunits to the five enzyme complexes that comprise the oxidative phosphorylation (OXPHOS) system within the inner mitochondrial membrane — the site of ATP synthesis. The reoxidation of reducing equivalents (NADH (reduced flavin adenine dinucleotide)) that are produced by the oxidation (reduced nicotinamide adenine dinucleotide) and FADH2 of carbohydrates (the tricarboxylic acid (TCA) cycle) and fatty acids (β-oxidation) is coupled to the generation of an electrochemical gradient across the inner mitochondrial membrane, which is harnessed by the ATP synthase to drive the formation of ATP. b A map of the human mitochondrial genome. The genes that encode the subunits of complex I (ND1–ND6 and ND4L) are shown in blue; cytochrome c oxidase (COI_–_COIII) is shown in red; cytochrome b of complex III is shown in green; and the subunits of the ATP synthase (ATPase 6 and 8) are shown in yellow. The two ribosomal RNAs (rRNAs; 12_S_ and 16_S_, shown in purple) and 22 tRNAs, indicated by black lines and denoted by their single letter code, which are required for mitochondrial protein synthesis are also shown. The displacement loop (D-loop), or non-coding control region, contains sequences that are vital for the initiation of both mtDNA replication and transcription, including the proposed origin of heavy-strand replication (shown as OH). The origin of light-strand replication is shown as OL.
Mitochondrial genetics is different from Mendelian genetics in almost every aspect, from the uniparental inheritance of disease mutations, to the presence of many copies of the genome within a single cell and the basic mechanisms that underlie replication and control of transcription (TABLE 1). The differences between the two genetic systems that are in human cells are probably a relic of evolution16, but lead to some fascinating biology that dictates the functional consequences of mtDNA mutations.
Table 1.
Comparison between the human nuclear and mitochondrial genomes*
| Characteristic | Nuclear genome | Mitochondrial genome |
|---|---|---|
| Size | ~3.3 x 109 bp | 16,569 bp |
| Number of DNA molecules per cell | 23 in haploid cells; 46 in diploid cells | Several thousand copies per cell (polyploidy) |
| Number of genes encoded | ~20,000–30,000 | 37 (13 polypeptides, 22 tRNAs and 2 rRNAs) |
| Gene density | ~1 per 40,000 bp | 1 per 450 bp |
| Introns | Frequently found in most genes | Absent |
| Percentage of coding DNA | ~3% | ~93% |
| Codon usage | The universal genetic code | AUA codes for methionine; TGA codes for tryptophan; AGA and AGG specify stop codons |
| Associated proteins | Nucleosome-associated histone proteins and non-histone proteins | No histones; but associated with several proteins(for example, TFAM) that form nucleoids |
| Mode of inheritance | Mendelian inheritance for autosomes and the X chromosome; paternal inheritance for the Y chromosome | Exclusively maternal |
| Replication | Strand-coupled mechanism that uses DNA polymerases α and δ | Strand-coupled and strand-displacement models; only uses DNA polymerase γ |
| Transcription | Most genes are transcribed individually | All genes on both strands are transcribed as large polycistrons |
| Recombination | Each pair of homologues recombines during the prophase of meiosis | There is evidence that recombination occurs at a cellular level but little evidence that it occurs at a population level |
Replication, transcription and translation of mitochondrial DNA
Both replication and transcription are important in terms of understanding the development of mtDNA mutations and their biochemical consequences. There is now good evidence to indicate that the minimal mitochondrial replisome17 consists of several proteins. These include: the heterodimeric mtDNA polymerase γ, which consists of a catalytic subunit with proof-reading ability (PolgA) and a processivity subunit (PolgB)18, Twinkle, which has 5′–3′ DNA helicase activity19, and a mitochondrial single-stranded binding protein. Together with mitochondrial transcription factor A (TFAM), these proteins associate with mtDNA to form structures that are termed NUCLEOIDS, which are believed to be the units of mtDNA transmission and inheritance.
NUCLEOID
A dynamic complex that consists of several copies of mitochondrial DNA and key maintenance proteins within the organelle.
POLYCISTRONIC
A form of gene organization that results in transcription of an mRNA that codes for multiple gene products, each of which is independently translated from the mRNA.
The mode of mtDNA replication has recently become the subject of much debate. On the basis of ultrastructural and biochemical analyses, mtDNA replication had been considered to occur by a strand-displacement model20, in which replication of the leading (heavy (H)) strand occurs first, initiating at a specific site (called OH) in the non-coding control region (FIG. 1). When leading-strand replication has reached two-thirds around the genome, the origin of replication (OL) on the lagging (light (L)) strand is exposed, allowing L-strand synthesis to occur in the opposite direction. Recent studies using two-dimensional gel electrophoresis challenge this model by providing evidence, in human, mouse and avian tissues, of a more conventional replication mechanism in which leading-lagging strand DNA replication is coordinated21–23. The two modes of replication have been hotly debated24–26 and the recent description of another D-loop origin of replication adds further interest to this debate27. Nascent chains that start at this origin do not terminate prematurely and seem to be mainly responsible for mtDNA maintenance under steady-state conditions.
Our understanding of the transcriptional machinery in mammalian mitochondria has improved, predominantly owing to the identification of specific proteins and the development of an in vitro system to dissect out the regulatory features28. In human mtDNA, transcriptional initiation can occur on both strands. Transcription from the mitochondrial promoters produces a POLYCISTRONIC precursor RNA that is then processed to produce individual tRNA and mRNA molecules29,30. To initiate transcription, the dedicated mitochondrial RNA polymerase (POLRMT) requires TFAM, and either mitochondrial transcription factor B1 (TFB1M) or B2 (TFB2M)31,32. Recent evidence shows that TFAM induces a structural change of the light-strand promoter that is required for POLRMT-dependent promoter recognition33. The importance of mitochondrial transcription to cellular dysfunction as a result of pathogenic mtDNA mutations is a neglected area of research that might give important insights into some of the tissue-specific or mutation-specific effects.
The mRNAs for the 13 mtDNA-encoded OXPHOS proteins are translated on mitochondrial ribosomes. Mutations in mitochondrial tRNA or rRNA genes affect pathology by disrupting mitochondrial translation34 and recent research has concentrated on identifying nuclear factors, including mitochondrial ribosomal proteins (MRPs), that are essential for this process. The first mutations in these nuclear genes, including MRPS16 (mitochondrial ribosomal protein S16)35 and EFG1 (officially known as GFM1; G elongation factor, mitochondrial 1)36 have recently been described in consanguineous families that have generalized mitochondrial translation defects.
Homoplasmy and heteroplasmy
The polyploid nature of the mitochondrial genome — up to several thousand copies per cell — gives rise to an important feature of mitochondrial genetics, homoplasmy and heteroplasmy. In simple terms, homoplasmy is when all copies of the mitochondrial genome are identical; heteroplasmy is when there is a mixture of two or more mitochondrial genotypes. The value of these terms is apparent when we consider mtDNA mutations that lead to disease. Some mutations affect all copies of the mitochondrial genome (homoplasmic mutation), whereas others are only present in some copies of the mitochondrial genome (heteroplasmic mutation). In the presence of heteroplasmy, there is a threshold level of mutation that is important for both the clinical expression of the disease and for biochemical defects, as routinely demonstrated by the cytochemical assessment of cytochrome c oxidase (COX; complex IV) activity in an individual cell (FIG. 2). Numerous single-cell and _TRANS_MITOCHONDRIAL CYBRID CELL studies have shown that the mutated form is functionally recessive and that a biochemical phenotype is associated with high levels of mutation above a crucial threshold37.
Figure 2. Cytochrome c oxidase deficiency in mitochondrial DNA-associated disease and ageing.
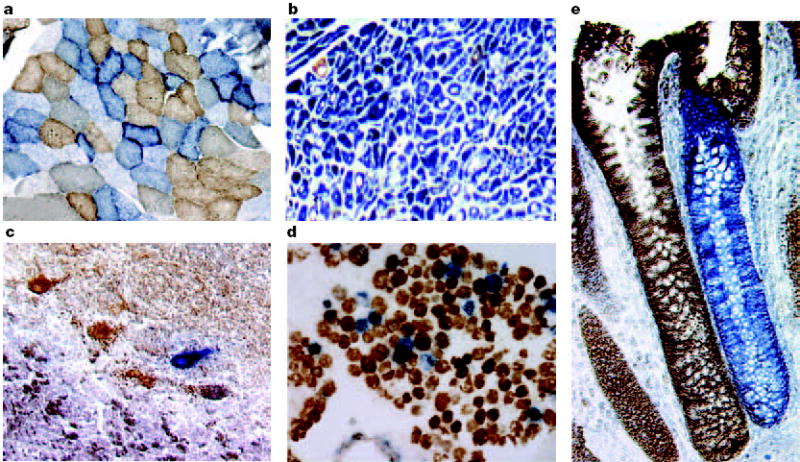
Transverse tissue sections that are reacted for both cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activities sequentially, with COX-positive cells shown in brown and COX-deficient cells shown in blue. a Skeletal muscle from a patient with a heteroplasmic mitochondrial tRNA point mutation. The section shows a typical ‘mosaic’ pattern of COX activity, with many muscle fibres harbouring levels of mutated mtDNA that are above the crucial threshold to produce a functional enzyme complex. b Cardiac tissue (left ventricle) from a patient with a homoplasmic tRNA mutation that causes hypertrophic cardiomyopathy, which demonstrates an absence of COX in most cells. c A section of cerebellum from a patient with an mtDNA rearrangement that highlights the presence of COX-deficient neurons. d,e Tissues that show COX deficiency that is due to clonal expansion of somatic mtDNA mutations within single cells — a phenomenon that is seen in both post-mitotic cells (d; extraocular muscles) and rapidly dividing cells (e; colonic crypt) in ageing humans.
_TRANS_MITOCHONDRIAL CYBRID
Derived from the term ‘cytoplasmic hybrid’. A cell line that is used to study the pathophysiological consequences of any given mitochondrial DNA mutation in a control nuclear background. Cybrids are generated by fusing patient cytoplasts (enucleated cells that contain mitochondria) with a cell line that lacks mitochondrial DNA.
The concept of homoplasmy is more apparent than real. In most individuals there is no evidence of heteroplasmy, but all available evidence indicates that mtDNA is constantly undergoing mutation, with clonal expansion or loss of either point mutations or deletions38,39. Because these acquired mutations occur at random, all acquired mutations will be present at a low level and therefore might not be detected in a tissue homogenate or blood sample. Perhaps we should only use ‘homoplasmy’ to describe a state in which we cannot detect these acquired mutations, although it would be naive to think that all mitochondrial genomes within an individual, tissue or even a cell are identical.
Inheritance of mitochondrial DNA
The standard model of mtDNA inheritance is that it is transmitted strictly through the maternal line40 and that mtDNA lineages are therefore clonal. However, this model has recently been challenged. Low levels of paternal transmission of mtDNA have been observed in crosses between mouse species, but not within species41, although further studies showed that this paternal mtDNA was not transmitted to the subsequent generation42. In addition, there is evidence that recombination has contributed to the distribution of mtDNA polymorphisms within the human population43. Although recombination might occur at the cellular level44,45, the occurrence of recombination within the population is contentious46. Studies of a patient with mitochondrial myopathy — who carried a novel, 2-bp pathogenic deletion in the NADH (reduced nicotinamide adenine dinucleotide) dehydrogenase subunit 2 (ND2) gene in his muscle mtDNA — have also challenged the maternal inheritance of mtDNA47. Using haplotype analysis, this sporadic mutation was present in paternally derived mtDNA, although the deletion itself was not evident in the father’s mtDNA. With the exception of skeletal muscle, all other tissues contained a single, maternally derived mtDNA haplotype. Subsequent studies of other patients with mitochondrial myopathies have not shown any evidence of paternal transmission48–50, even when assisted reproduction techniques were applied51,52.
LEBER HEREDITARY OPTIC NEUROPATHY
A mitochondrial disease that is characterized by optic nerve dysfunction, which leads to bilateral visual failure in young adults.
GENETIC BOTTLENECK
A temporary reduction in population size that causes the loss of genetic variation.
Mitochondrial DNA transmission
The current genetic advice is that fathers with mtDNA mutations are at no risk of transmitting the defect to their offspring. Maternal transmission of mutated mtDNA occurs, but the risk depends on the type of mutation and possibly the segregation of the mutation within maternal tissues. Identifying specific mtDNA mutations and investigating family members for evidence of transmission will give guidance to the likelihood of transmission through the germline. Detailed studies of large patient cohorts provide invaluable information on the risk of transmission. A recent analysis of a single, large-scale mtDNA deletion in 226 families, showed that the risk of recurrence in the offspring of an affected mother was 4.11% (REF. 53). If the mother was unaffected, then there was no record of affected siblings, which indicates that the risk of recurrence is negligible.
Homoplasmic mtDNA mutations are transmitted to all maternal offspring. However, the genetic advice given to these families is not straightforward. Most patients with LEBER HEREDITARY OPTIC NEUROPATHY (LHON; see also Online links box) have homoplasmic mtDNA mutations. Although all offspring inherit the mutation, only some will develop the disease. Approximately 50% of males, but only 10% of females, develop impaired vision54, which certainly indicates that nuclear genetic factors are important in the expression of the disease. Another homoplasmic mtDNA mutation, the 1555A>G 12_S_ ribosomal RNA (RNR1) mutation55, is an important cause of post-lingual deafness. Its clinical expression depends on the administration of amino-glycoside antibiotics, which highlights the importance of environmental factors in the expression of homoplasmic mtDNA disease. Although it is obvious that genetic counselling has to be tailored to specific mutations, further information is required to determine the risk for individual defects.
The transmission of heteroplasmic mtDNA point mutations is even more complex. Nuclear genetic56 and environmental factors almost certainly affect expression of the disease phenotype. In addition, there is a GENETIC BOTTLENECK during development, and the amount of mutated mtDNA that is transmitted to the offspring is variable57 (FIG. 3). Because many of the clinical features depend on the relative proportions of mutated versus wild-type mtDNA, the outcome for each pregnancy remains difficult to predict58.
Figure 3. The mitochondrial genetic bottleneck.
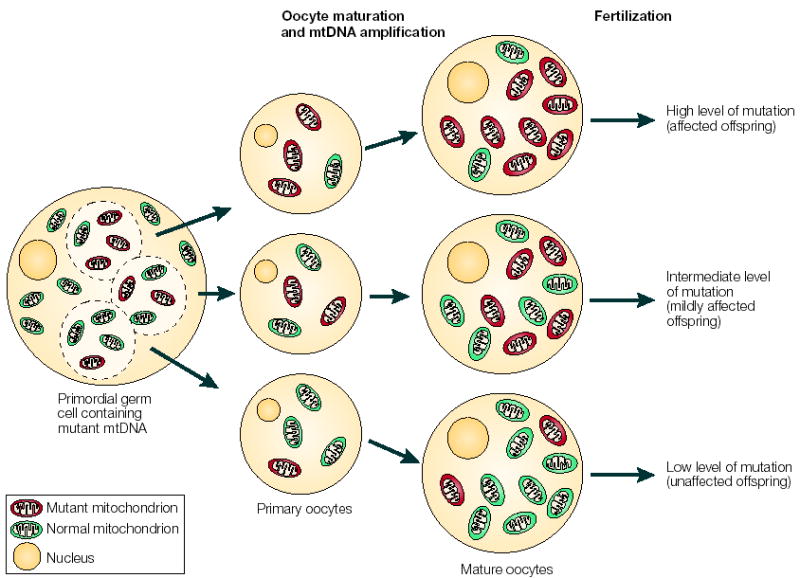
During the production of primary oocytes, a selected number of mitochondrial DNA (mtDNA) molecules are transferred into each oocyte. Oocyte maturation is associated with the rapid replication of this mtDNA population. This restriction-amplification event can lead to a random shift of mtDNA mutational load between generations and is responsible for the variable levels of mutated mtDNA observed in affected offspring from mothers with pathogenic mtDNA mutations. Mitochondria that contain mutated mtDNA are shown in red, those with normal mtDNA are shown in green.
Clinical features of human mtDNA disease
Mitochondria are vital components of all nucleated cells. Therefore, it is not surprising that mtDNA diseases affect many tissues and that the clinical features are so variable (BOX 1). Such diversity makes it difficult to define the impact of mtDNA mutations on human health, however, stratifying the clinical features into the following groups defines the extent of the problem: classic mtDNA syndromes, clinical syndromes with a high risk of mtDNA mutations, involvement in common disease phenotypes and mtDNA as a predisposition for common disease and ageing.
Box 1 | Common features of mitochondrial DNA-associated diseases
Adult
• Neurological: migraine | strokes | epilepsy | dementia | myopathy | peripheral neuropathy | DIPLOPIA | ATAXIA | speech disturbances | sensorineural deafness
• Gastrointestinal: constipation | irritable bowel | DYSPHAGIA
• Cardiac: heart failure | heart block | cardiomyopathy
• Respiratory: respiratory failure | nocturnal hypoventilation | recurrent aspiration | pneumonia
• Endocrinal: diabetes | thyroid disease | parathyroid disease | ovarian failure
• Ophthalmological: optic atrophy | cataract | ophthalmoplegia | PTOSIS
Paediatric
• Neurological: epilepsy | myopathy | psychomotor retardation | ataxia | spasticity | DYSTONIA | sensorineural deafness
• Gastrointestinal: vomiting | failure to thrive | dysphagia
• Cardiac: biventricular hypertrophic cardiomyopathy | rhythm abnormalities
• Respiratory: central hypoventilation | apnoea
• Haematological: anaemia | PANCYTOPAENIA
• Renal: renal tubular defects
• Liver: hepatic failure
• Endocrinal: diabetes | adrenal failure
• Ophthalmological: optic atrophy
DIPLOPIA
Double vision; derived from the Greek diplous, meaning double, and ops, meaning eye.
ATAXIA
The loss of the ability to coordinate muscular movement.
DYSPHAGIA
A difficulty in swallowing.
PTOSIS
The abnormal lowering or drooping of the upper eyelid that is caused by muscle weakness.
DYSTONIA
A neurological movement disorder that is characterized by involuntary muscle contractions that might cause twisting or jerking movements of the body.
PANCYTOPAENIA
A deficiency of all blood cells including red cells, white cells and platelets.
OPHTHALMOPLEGIA
Weakness of one or more of the muscles that control eye movement.
PEARSON SYNDROME
A severe disease during infancy that affects bone marrow and pancreas function owing to large-scale rearrangements of the mitochondrial genome.
Clinical features are specific and clinical diagnosis is straightforward for patients with the classic syndromes. However, in many patients, the clinical symptoms might be non-specific and mtDNA disease is only one of several possible diagnoses. Important clues to a possible mitochondrial origin for the symptoms include a maternal family history and the involvement of several tissues (BOX 2). For a large number of patients, diagnosis depends on the results of several lines of investigation. This includes histochemical and biochemical studies to determine the precise nature of the respiratory chain defect, analysis of the mitochondrial genome for common mutations and, finally, complete genome sequencing to look for rare or novel mutations59. The mitochondrial genome is highly variable and the ease of whole-genome sequencing does not resolve some diagnostic dilemmas because the interpretation of a novel sequence change can be difficult in relation to potential pathogenicity60. Accurate diagnosis of an mtDNA defect is essential because of the consequences for both the patient and the family members.
Classic syndromes
Genetic defects of the human mitochondrial genome were first described in 1988 (REFS 61,62) and arose from the investigation of two syndromes — Kearns–Sayre syndrome (KSS) and LHON. In KSS, single, large-scale mtDNA deletions (usually sporadic) were detected in muscle biopsies, and mitochondrial ultrastructural and cytochemical abnormalities in the muscle were apparent. For LHON, a strict maternal pattern of inheritance was evident and point mutations involving the ND family of genes that encode subunits of complex I were identified. Since 1988, several mutations63 of the mitochondrial genome have been identified and associated with disease (TABLE 2).
Table 2.
Clinical disorders that are caused by mutations in mitochondrial DNA
| Mitochondrial DNA Disorder | Clinical phenotype | mtDNA genotype | Gene | Status | Inheritance | Reference |
|---|---|---|---|---|---|---|
| Kearns–Sayre syndrome | Progressive myopathy, ophthalmoplegia, cardiomyopathy | A single, large-scale deletion | Several deleted genes | Heteroplasmic | Usually sporadic | 61,158 |
| CPEO | Ophthalmoplegia | A single, large-scale deletion | Several deleted genes | Heteroplasmic | Usually sporadic | 61, 64 |
| Pearson syndrome | Pancytopoenia, lactic acidosis | A single, large-scale deletion | Several deleted genes | Heteroplasmic | Usually sporadic | 65 |
| MELAS | Myopathy, encephalopathy lactic acidosis, stroke-like episodes | 3243A>G; 3271T>C | TRNL1 | Heteroplasmic | Maternal | 159 |
| Individual mutations | ND1 and ND5 | Heteroplasmic | Maternal | 160, 161 | ||
| MERRF | Myoclonic epilepsy, myopathy | 8344A>G; 8356T>C | TRNK | Heteroplasmic | Maternal | 162 |
| NARP | Neuropathy, ataxia, retinitis pigmentosa | 8993T>G | ATP6 | Heteroplasmic | Maternal | 163 |
| MILS | Progressive brain-stem disorder | 8993T>C | ATP6 | Heteroplasmic | Maternal | 67 |
| MIDD | Diabetes, deafness | 3243A>G | TRNL1 | Heteroplasmic | Maternal | 164 |
| LHON | Optic neuropathy | 3460G>A | ND1 | Hetero- or homoplasmic | Maternal | 165 |
| 11778G>A | ND4 | Hetero- or homoplasmic | Maternal | 62 | ||
| 14484T>C | ND6 | Hetero- or homoplasmic | Maternal | 166 | ||
| Myopathy and diabetes | Myopathy, weakness, diabetes | 14709T>C | TRNE | Hetero- or homoplasmic | Maternal | 167,168 |
| Sensorineural hearing loss | Deafness | 1555A>G | RNR1 | Homoplasmic | Maternal | 55 |
| Individual mutations | TRNS1 | Hetero- or homoplasmic | Maternal | 169,170 | ||
| Exercise intolerance | Fatigue, muscle weakness | Individual mutations | CYB | Heteroplasmic | Sporadic | 68 |
| Fatal, infantile encephalopathy; Leigh/Leigh-like syndrome | Encephalopathy, lactic acidosis | 10158T>C; 10191T>C | ND3 | Heteroplasmic | Sporadic | 66 |
Clinical syndromes that have a high probability of mitochondrial DNA involvement
Over the past decade, we have become aware of several clinical syndromes that might be associated with mtDNA mutations. The increasing recognition of mtDNA involvement in disease is partially due to the relative ease of sequencing the mitochondrial genome, although defining pathogenicity of specific base substitutions can be difficult60. Examples of such diseases include progressive external OPHTHALMOPLEGIA64, PEARSON SYNDROME65, LEIGH SYNDROME66,67 (see also Online links box), exercise-induced muscle pain, fatigue and RHABDOMYOLYSIS68 (see also Online links box), and amino-glycoside-induced hearing loss55. For some of these conditions, such as progressive external ophthalmoplegia, mtDNA disease is the most likely cause, whereas in others, such as Leigh syndrome, there is a long list of potential genetic causes.
Involvement in common disease phenotypes
The principal difficulty for clinicians is that patients with mtDNA disease rarely have a classic phenotype and mtDNA enters into the differential diagnosis of many common clinical syndromes. A good example of a common condition in which mtDNA is a potential cause is diabetes, the most common metabolic disease to affect humans69. mtDNA disease certainly is associated with diabetes, but these patients represent a tiny proportion of those affected with diabetes70–72. Because several mtDNA mutations cause diabetes63 it is clearly impractical and uneconomical to screen all patients with diabetes for causative mutations. From the list of common clinical features seen in patients with mtDNA disease (BOX 1), it is obvious that it is difficult to decide which patients to investigate for possible mtDNA disease. Further difficulties in this area have been highlighted by a recent report that describes a clear association between members of a large family that harbour a homoplasmic mitochondrial tRNA mutation and a metabolic syndrome that is characterized by hypertension, hypomagnesaemia and hypercholesterolaemia9. Because conditions such as hypertension and hyperc-holesterolaemia affect many individuals73, the number of patients with mtDNA disease is potentially very high. However, it must be kept in the context that most reports describe isolated families. In epidemiological surveys of populations74, the incidence of mtDNA mutations within a specific disease phenotype75 is small. Other factors that make the diagnosis of mtDNA difficult include the specific combination of clinical symptoms (BOX 2); for example, mitochondrial diabetes is often accompanied by deafness70, and stroke-like episodes are usually associated with migraine-like symptoms. There is no doubt that many patients with mtDNA mutations go undiagnosed for years and many are probably never recognized. Increased awareness among physicians and paediatricians is important, as is the investigation of patients in specialist centres that are able to provide a complete range of diagnostic tests.
Box 2 | Guidelines for the recognition of patients with mitochondrial DNA disease
A list of the clinical guidelines that have been generated from our experience of invesbtigating and managing patients with mtDNA mutations.
• Maternal inheritance
• Recognition of classic syndromes; for example, mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) or chronic progressive external ophthalmoplegia (CPEO)
• Recognition of characteristic clinical features; for example, biventricular cardiac hypertrophy
• Involvement of many organ systems; for example, diabetes and deafness
• Specific combinations of symptoms; for example, strokes and migraine and seizures and ataxia
• Abnormal clinical or laboratory investigations; for example, lactic acidaemia in children, characteristic magnetic-resonance-imaging findings, abnormal muscle biopsy that is associated with RAGGED-RED FIBRES or cytochrome c oxidase-deficient fibres
LEIGH SYNDROME
A disease that affects the brainstem and basal ganglia and is characterised by defects in mitochondrial oxidative phosphorylation.
RHABDOMYOLYSIS
The breakdown of muscle fibres owing to injury, toxins or metabolic disease, which leads to high concentrations of myoglobin in both plasma and urine
RAGGED3RED FIBRES
The pathological hallmark of mtDNA disease that is characterized by the subsarcolemmal accumulation of abnormal mitochondria in the muscle fibre, which stains red with a Gomori trichrome stain.
Mitochondrial DNA variants as a predisposition for common disease
There has been considerable interest in the possibility that mtDNA variants might predispose to common diseases; for example, diabetes, Alzheimer disease (AD) and Parkinson disease (PD)76. The known association of diabetes with mtDNA mutations and the involvement of reactive oxygen species in the aetiology of AD and PD has encouraged a large number of studies. The first approach involves identifying candidate pathogenic mutations by comparing mtDNA sequences between patients and control groups. Unfortunately, interpreting the results is limited by the relatively small number of individual mtDNA sequences in each group and by the high mutation rate of mtDNA, which results in a high frequency of rare variants or private polymorphisms77. Some variants are more common than others. For example, the 16189 variant, a variable length polycytosine tract in the non-coding control region, has been associated with several late-onset, multifactorial disorders, including susceptibility to cardiomyopathy78 and type 2 diabetes79, and has been proposed to affect mtDNA replication79. However, because this variant occurs frequently within the population, concerns arise (that are similar to the ‘haplogroup’ studies) over the control cohorts that are chosen for such population-based surveys. Human populations can be divided into several mtDNA haplogroups that are based on specific SNPs, reflecting mutations accumulated by a discrete maternal lineage80. Therefore, haplogroup association studies have been used to define the role of mtDNA mutations in complex diseases. In this approach, disease and control mtDNA sequence sets are assigned to haplo-groups, and the distribution of haplogroups is compared. A significant difference is interpreted to indicate that the mtDNA ‘background’ has an effect on expression or penetrance of the clinical abnormalities. Various results have been reported for both AD and PD81–84. Given the inconsistency among different studies, it is difficult to conclude unequivocally that the risk of either PD or AD is influenced by mitochondrial haplogroups. Some of the inconsistency could be due to population-substructure differences between disease and control groups. It is difficult to ‘match’ groups, even when one tries to control nationality and ethnicity85.
CYTOPLAST
An enucleated donor cell that contains patient mitochondria and that is used to generate cybrid fusions.
Models of mitochondrial DNA disease
The inability to genetically engineer or transform the mitochondrial genome in vitro or in vivo has severely hampered efforts to study the relationship between mtDNA genotype and clinical phenotype in animal models that bear heteroplasmic, pathogenic mtDNA mutations. Despite this, much has been learned of the biochemical and cellular consequences of specific mtDNA mutations through the generation of _trans_mitochondrial cytoplasmic cybrid cells. In this system, immortalized human cell lines that have been completely depleted of their endogenous mtDNA (ρ° cells) are repopulated with exogenous, patient mitochondria. As ρ° cells have no functional respiratory chain and are dependent on pyruvate and uridine for growth, the loss of either of these two metabolic requirements can be used to select for transformants that harbour complementing (exogenous) mtDNA86. This elegant system allows the functional and physiological consequences of different levels of heteroplasmy of certain mtDNA mutations to be tested87,88. In addition, it has also proved useful in determining the genetic origin of certain mitochondrial disorders89,90, the effect of nuclear background on the segregation of pathogenic mtDNA mutations91 and in identifying the first tRNA suppressor mutation in human mitochondria92. It has also been used to test rational, genetic therapies and more recently, to demonstrate the presence of heterologous mtDNA recombination in human cells44.
The generation of heteroplasmic mice that harbour pathogenic mtDNA mutations was frustratingly difficult. Heteroplasmic mice were first generated by fusing zygotes that carried one mtDNA haplotype with enucleated embryos that carried a different haplotype93,94, although these mice (BALB and NZB) harboured only neutral mtDNA variants. Further advances were made with the generation of chimeric, heteroplasmic chloramphenicol resistant (CAPR) mice, which could transmit mtDNA mutations to subsequent generations and showed signs of mitochondrial dysfunction95,96. By fusing CYTOPLASTS that harbour high levels of a somatic mtDNA rearrangement to one-cell embryos, Hayashi and colleagues generated a model that has a pathogenic deletion or duplication97 of the mitochondrial genome, which was transmitted through the germ line. These mice showed muscle and cardiac defects, but also, more prominently, renal failure together with lactic acidosis, anaemia and a mosaic distribution of COX deficiency. Human mtDNA-rearrangement mutations rarely result in renal disease, so this experiment highlighted the potential for species-specific symptoms and signs. Although subsequent studies that use this mouse model have improved our understanding of mitochondrial segregation and disease pathogenesis98, no reported studies specifically address the development of potential therapies.
The generation of mouse models that target nuclear genes involved in mtDNA maintenance or replication, such as the Tfam knockout mouse, has proved to be a more successful approach99–103. Using the Cre-loxP recombination system, Larsson and colleagues disrupted the Tfam gene in selected mouse tissues, yielding models that provide clues to potentially important factors in the development of specific clinical features associated with mtDNA disease. For example, the mice generated by the postnatal disruption of Tfam in neurons of the hippocampus and neocortex develop a late-onset neurodegeneration, but there is minimal cell loss until the mice developed seizures101. This finding indicates that stress or neuronal discharge resulted in an acute energy crisis, which highlights the importance of management issues such as the effective control of seizures in patients with mtDNA disease.
Management of mtDNA diseases
Although defining the clinical features and establishing the diagnosis of mtDNA disease remain considerable challenges, the development of effective curative treatment is also a challenging hurdle for experimental scientists. However, there are important issues associated with the clinical management of patients and many of these issues have recently been discussed104. Early detection of treatable symptoms is crucial.
SATELLITE CELLS
Quiescent cells that are located between the basal lamina and the plasmalemma of the muscle fibre, and are a main contributor to postnatal muscle growth.
ALLOTOPIC EXPRESSION
The expression of a gene in a different cellular compartment to its target location. In the context described here, it is the recoding of a mitochondrial gene to allow it to be expressed in the nucleus. The subsequent conjugation of a mitochondrial-targeting sequence promotes import and localization of the gene product to the organelle.
AMNIOCENTESIS
An aspiration of cells from the amniotic sac, using a needle, for biochemical and genetic analysis.
CHORIONIC VILLUS BIOPSY
A placental biopsy that is carried out in early pregnancy to collect fetal tissue for genetic and biochemical analysis.
POLAR BODY
A small haploid cell that is produced during oogenesis and that does not develop into a functional ovum.
Mitochondrial DNA mutation levels can be manipulated through exercise
In healthy individuals, lack of exercise (deconditioning) leads to an overall reduction in mitochondrial enzyme activity, whereas endurance training improves enzyme activity. Resistance training or muscle necrosis stimulates the incorporation of SATELLITE CELLS into existing muscle fibres105,106. It is postulated that for sporadic mutations, resistance training might lead to an overall reduction in the proportion of mutated mtDNA versus wild-type, as satellite cells contain a low or negligible amount of mutated mtDNA106,107. Endurance training might therefore improve function by increasing wild-type mtDNA levels. However, there are concerns that mutated mtDNA might be preferentially amplified, and that this increase might become clinically relevant after deconditioning105,108. Studies are currently underway to address both the improvements and the concerns arising from these earlier reports.
New avenues for treatment of mitochondrial diseases
The polyploid nature of the mitochondrial genome, the phenomenon of heteroplasmy, the inability to deliver potentially therapeutic nucleic acids to the organelle through mitochondrial transfection, and the slow realization of appropriate animal models of mtDNA disease are the main problems for the development of gene-based treatments. Despite these, several strategies that seek to rescue mitochondrial function through complementation of the genetic defect or direct manipulation of the levels of mutated mtDNA have been considered.
One way of complementing a dysfunctional mitochondrial protein is through ALLOTOPIC EXPRESSION. Using this approach, it was possible to express the wild-type ATPase 6 protein allotopically from nucleus-transfected constructs in _trans_mitochondrial cybrid cells that were homoplasmic for the 8993T>G MTATP6 (subunit 6 of mitochondrial ATP synthase) mutation (which causes neurogenic weakness, ataxia and retinitis pigmentosa (NARP) syndrome) and demonstrate a partial rescue of the biochemical defect109. A similar strategy has been used to express a modified NADH dehydrogenase subunit 4 (ND4) gene to complement the 11778G>A mutation that causes LHON110. To complement mitochondrial tRNA defects, an alternative approach has been proposed. Yeast cytosolic tRNALysCUU (tK1) and similar derivatives can be imported into isolated human mitochondria if the tRNA is amino-acylated and supplied with soluble factors, including lysl-tRNA synthetases111. Using _trans_mitochondrial cybrid cells and primary human fibroblasts that carry the 8344A>G TRNK (a tRNA gene) mutation, which causes myoclonic epilepsy and ragged-red fibres (MERRF) syndrome, it has recently been shown that the imported tRNALys is correctly amino-acylated and able to participate in mitochondrial translation, partially rescuing mitochondrial function112.
Because the recessive nature of many pathogenic mutations dictates that a small percentage of the wild-type genotype is protective, a minor adjustment to the balance of mutated to wild-type genomes in favour of the wild type could improve both biochemical function and resolve the clinical problem. The selective inhibition of mutated mtDNA replication by antigenomic agents has been applied to manipulate mtDNA heteroplasmy113. Another strategy involves targeting the organelle with restriction endonucleases that, by differentiating between mtDNA genotypes, can cause the preferential elimination of the mutated genotype and propagation of the wild-type genotype114,115. Pharmacological approaches to shift mtDNA heteroplasmy are also being investigated. Oligomycin, an irreversible inhibitor of mitochondrial ATP synthase, has been used to increase the fraction of wild-type molecules in cells that harbour the 8933T>G MTATP6 mutation under culture conditions (using galactose as the sole carbon source) that specifically select for the wild-type molecule116. This culture medium could also selectively kill cells that are homoplasmic for a common (4,977-bp deletion) mtDNA rearrangement — the molecular abnormality in many patients with KSS. In this case, a ketogenic medium has been used to shift the heteroplasmy of cells that contain a mixture of wild-type and partially deleted mtDNAs117. Finally, the revelation that overexpressing mitochondrial Tu translation elongation factor, TUFM, in yeast mutant counterparts of TRNL1 (a tRNA gene) MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) mutations facilitates the rescue of the respiratory defect might have implications for human disease-related mutations118.
Prevention of transmission of mtDNA disease
Improving genetic advice
The question often asked by both families and clinicians is whether there are any prospects for providing either better guidance for families or even preventing the transmission of mtDNA disease. AMNIOCENTESIS and CHORIONIC VILLUS BIOPSY are widely used to diagnose autosomal and chromosomal abnormalities, and might be of value in heteroplasmic mtDNA disorders. Chorionic villus biopsy has been used in some families that have heteroplasmic mtDNA disorders, where the results have influenced the clinical management119–121. In many heteroplasmic mtDNA64,122 disorders there are marked tissue-specific differences in the level of heteroplasmy, and therefore a reasonable concern is whether a prenatal sample will reflect the likely outcome for the fetus. The present evidence indicates that this should not be a problem94,123. Preimplantation genetic diagnosis (PGD) allows either analysis of mtDNA from the POLAR BODY of unfertilized oocytes, or from one to two single cells that are taken from 8-cell embryos, followed by the implantation of healthy embryos. The potential value of this technique is obvious, but there is little reported information of its application in patients with mtDNA mutations. The high number of mitochondrial genomes within oocytes124 indicates that PGD should be feasible for mtDNA disease. Experiments in heteroplasmic mice have shown the levels of heteroplasmy to be virtually identical between the ooplasm and polar body of the mature oocyte and also between blastomeres of each of the 2-, 4-, and 6–8-cell embryo125.
GERMINAL VESICLE
A stage during oocyte maturation in which the oocyte nucleus is located close to the surface of the egg cell and is clearly visible.
Alternative strategies associated with oocyte manipulation
Development of new strategies to prevent the transmission of mtDNA is a very important aspect of disease management. An obvious method to prevent the transmission of an mtDNA mutation to the offspring is oocyte donation. However, although the oocyte would be fertilized using the father’s sperm, the child would have the nuclear genotype of the donor female, a less-than-ideal option for many mothers. Cytoplasmic transfer involves the transfer of normal mitochondria into the abnormal oocyte to dilute the effect of any mtDNA defect126. Cytoplasmic transfer between human oocytes has been carried out to improve the outcome of assisted reproduction methods127. The effectiveness of this procedure is uncertain128, but some of the children born were heteroplasmic with low levels of mtDNA from the donor oocyte129. However, despite the observation that changes in heteroplasmy can occur, it is likely that this technique will have limited value for patients with mtDNA disease, as mouse experiments indicate that the amount of mtDNA that can be transferred is relatively small130.
Alternatively, nuclear chromosomes from an oocyte that contains mutated mtDNA could be transferred to an enucleated oocyte from a normal female131. This method has obvious advantages: the offspring will be genetically related to both parents and the chances of achieving very low levels of mutated mtDNA in the oocyte are high. However, important technical and ethical or legal issues make progress in this area difficult, principally because the technique involves the genetic manipulation of fertilized oocytes. To overcome these issues, the transfer could be carried out at different stages of oocyte maturation, although transfer from mature unfertilized oocytes is difficult because of the serious risk of aneuploidy that is due to chromosome loss during transfer or due to missegregation following the disruption of the meiotic spindle. Karyoplast transfer of the GERMINAL VESICLE between oocytes might be possible before in vitro maturation and subsequent fertilization (FIG. 4). However, although maturation and fertilization of normal germinal-vesicle-stage oocytes has been achieved with both mouse132–134 and human oocytes135,136, these methods are very inefficient. Pronuclear-stage transfer is possibly more feasible because human pronuclear oocytes can be manipulated to generate normal offspring137 and because this technique is feasible in the mouse138,139.
Figure 4. Nuclear transfer techniques.
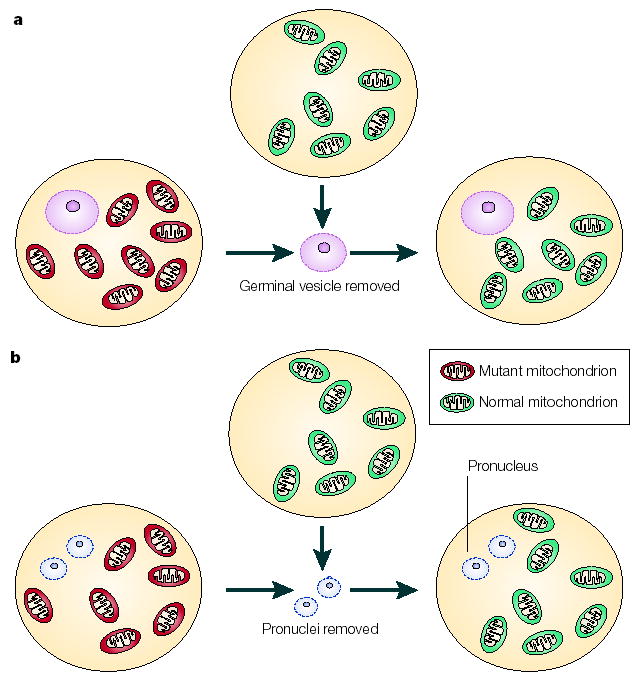
The prevention of mutated maternal mtDNA transmission could be achieved by nuclear transfer techniques. In theory, this could be carried out at the germinal vesicle stage (a), when clearly identifiable structures are visible. The germinal vesicle contains nuclear chromosomes and could be transferred to an oocyte that contains normal mtDNA (green). Alternatively, pronuclear transfer can be carried out at the fertilized oocyte stage (b); this involves removal of the male and female pronuclei from the oocyte that contains mutated mtDNA (red) and their subsequent transfer to an oocyte that has normal mtDNA (green).
Acquired mtDNA mutations in ageing and cancer
Mitochondrial theory of ageing
The mitochondrial theory of ageing proposes that progressive accumulation of somatic mutations in mtDNA during a lifetime leads to an inevitable decline in mitochondrial function140. The most important biochemical factors in this process are considered to be reactive oxygen species (ROS), which are generated at very low levels during the normal function of the mitochondrial respiratory chain. It has been proposed that ROS can cause somatic mutations in mtDNA140. These mutations then result in impaired function of the respiratory chain, leading to increased ROS production and the subsequent accumulation of more mutations. There is evidence that some pathogenic mutations, such as at 8993T>G, lead to increased ROS production141; interestingly, for 8993T>G, free radical-mediated inhibition of OXPHOS can be restored by antioxidants142. The ROS vicious cycle is believed to account for an exponential increase in oxidative damage during ageing, which results in the eventual loss of cellular and tissue functions through a combination of energy insufficiency, signalling defects, apoptosis and replicative senescence.
Studies in human and mouse models
Although the mitochondrial theory of ageing was first suggested in the 1950s, the first convincing evidence of a biochemical defect in humans was the detection of COX-deficient cells in ageing post-mitotic tissues143 (FIG. 2). These cells had identical characteristics to those found in patients with inherited mitochondrial disease37. Surprisingly, these cells show the clonal expansion of an individual mutation, which is often an mtDNA rearrangement, rather than many individual mtDNA mutations144,145. In addition, studies from patients showed that, in the presence of many mtDNA genomes within a cell, the mutation is functionally recessive and a threshold effect is observed37. More recently, mtDNA mutations were found to accumulate to high levels in dividing cells and presumably stem cells39,146, and cause an observable biochemical defect39.
GENETIC DRIFT
Changes in the frequency of a genetic variant in a population owing to chance alone.
How a mutation in an individual genome can clonally expand to high levels in individual cells is a crucial question. So far, several theories have been proposed, including the selective replication of mutated mtDNA, the longevity of mitochondria that contain mutated mtDNA and the ‘sick mitochondrion’ hypothesis (with preferential replication of mitochondria that have impaired OXPHOS activities and higher mutant load)147–149. However, mathematical-modelling studies have also indicated that this process might occur by GENETIC DRIFT150.
Studies from ageing humans and animal models showed good correlation between ageing and the presence of mtDNA mutations. To test the mitochondrial ageing hypothesis, knock-in mice expressing a proofreading-deficient version of PolgA were developed10. These mice acquire mtDNA mutations at a higher rate than is normal because the introduction of a point mutation into the exonuclease region of the PolgA gene drastically reduces its proof-reading exonuclease activity. The phenotype of the PolgA mutator mouse has many hallmarks of premature ageing, including a characteristic hunched appearance that is indicative of osteoporosis, reduced activity, severe weight loss with virtually no remaining adipose tissue and a marked reduction in muscle mass. Although this study provides strong evidence that somatic mtDNA mutations contribute to ageing, further experiments are required to confirm that somatic mtDNA mutations are causal in the ageing process.
Mitochondrial DNA mutations and cancer
In a landmark paper in 1998, Vogelstein and colleagues reported that mtDNA mutations were present in 7 out of 10 colorectal cancer cell lines that were studied151. Although these mtDNA mutations were somatic and absent in other patient tissues, many were homoplasmic in the tumour tissue. Since then, numerous other studies have documented the presence of somatic mtDNA mutations in both solid tumours and leukaemias152–156. How these mutations accumulate to high levels in individual tumours is still unclear, although both modelling and molecular studies indicate that they possibly accumulate without selection38,39. In addition, there is no evidence of whether mtDNA mutations themselves contribute to the development of the tumour. Original work on colorectal tumours showed that many of the mutations detected were not associated with a mitochondrial defect. Moreover, some of the mutations seen in other tumours are recognized polymorphic variants151. Therefore, although a direct link between the presence of the mtDNA mutation and the development of the tumour has not been made, the presence of a mtDNA mutation might prove significant in the detection of tumour recurrence152 and possibly in the detection of genotoxic damage157.
Conclusions and future prospects
It is nearly 25 years since Sanger and colleagues sequenced the human mitochondrial genome, and significant progress in the mitochondrial field continues to be made. Our understanding of the pathogenesis of mtDNA disease will greatly improve by studying the basic processes involved in replication, transcription and translation of the mitochondrial genome.
Unequivocally, mtDNA mutations are an important cause of genetic disease. The clinical variability of these disorders makes the recognition of patients with mtDNA disease a real challenge. Clinicians must be aware of its impact; accurate diagnosis requires a combination of different studies and should be carried out in specialist centres. Although the most disappointing area has been the lack of treatment for patients with mtDNA disease, several new experimental approaches are currently under investigation. It is crucial that further work and ideas are forthcoming to realistically treat or prevent the transmission of mtDNA disease to future generations. The involvement of mtDNA mutations in neurodegenerative disease, ageing and cancer is under debate. Although the development of an animal model should clarify a possible role of mtDNA mutations in ageing, further work in both human tissues and animal models is required to explain the role of mtDNA in these areas of medicine.
Acknowledgments
The authors work is supported by the Wellcome Trust, Medical Research Centre, Alzheimer’s Research Trust, Muscular Dystrophy Campaign, Newcastle upon Tyne Hospitals NHS Trust and the European Union (FP6, EUmitocombat). We thank R.N. Lightowlers, N. Howell, R. McFarland and M. Herbert for valuable input, and L. Craven and M. Barron for help with the figures.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov PL, et al. Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nature Genet. 1996;12:417–420. doi: 10.1038/ng0496-417. [DOI] [PubMed] [Google Scholar]
- 3.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. This paper describes the sequence and organization of human mtDNA — the first complete mitochondrial genome to be sequenced. [DOI] [PubMed] [Google Scholar]
- 4.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 5.Anderson S, et al. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 6.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genet. 1999;23:147. doi: 10.1038/13779. A re-analysis of the original human placental DNA sample used by Fred Sanger and colleagues, using automated, fluorescent DNA sequencing, which allowed a consensus human mtDNA sequence (the ‘revised Cambridge Reference Sequence’) to be made available. [DOI] [PubMed] [Google Scholar]
- 7.Majamaa K, et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am J Hum Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders — past, present and future. Biochim Biophys Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Wilson FH, et al. A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science. 2004;306:1190–1194. doi: 10.1126/science.1102521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. This report describes a knock-in mouse model that expresses a proof-reading-deficient version of PolgA, the catalytic subunit of mtDNA polymerase, and provides the first in vivo data in mammals to establish a causative link between mtDNA mutations and ageing phenotypes. [DOI] [PubMed] [Google Scholar]
- 11.Muller-Hocker J. Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age-related alteration. J Neurol Sci. 1990;100:14–21. doi: 10.1016/0022-510x(90)90006-9. [DOI] [PubMed] [Google Scholar]
- 12.Mootha VK, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 13.Petersen KF, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 15.Shoubridge EA. Nuclear genetic defects of oxidative phosphorylation. Hum Mol Genet. 2001;10:2277–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- 16.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 17.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase-γ, Pol-γB, functions as a homodimer. Mol Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 19.Spelbrink JN, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 20.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. This paper outlines the strand-displacement model of mammalian mtDNA replication, in which the two strands of mtDNA are each replicated in a continuous fashion from widely separated origins, requiring extensive displacement of parental DNA strands during leading-strand synthesis. [DOI] [PubMed] [Google Scholar]
- 21.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. Using two-dimensional agarose gel electrophoresis, the demonstration of duplex replication intermediates led the authors to propose a further mechanism for mammalian mtDNA replication that involves strand-coupled synthesis. [DOI] [PubMed] [Google Scholar]
- 22.Yang MY, et al. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 23.Bowmaker M, et al. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 24.Bogenhagen DF, Clayton DA. The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:357–360. doi: 10.1016/S0968-0004(03)00132-4. [DOI] [PubMed] [Google Scholar]
- 25.Holt IJ, Jacobs HT. Response: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:355–356. doi: 10.1016/S0968-0004(03)00133-6. [DOI] [PubMed] [Google Scholar]
- 26.Bogenhagen DF, Clayton DA. Concluding remarks: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:404–405. doi: 10.1016/S0968-0004(03)00165-8. [DOI] [PubMed] [Google Scholar]
- 27.Fish J, Raule N, Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 28.Gaspari M, Larsson NG, Gustafsson CM. The transcription machinery in mammalian mitochondria. Biochim Biophys Acta. 2004;1659:148–152. doi: 10.1016/j.bbabio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 30.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 31.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 33.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs HT. Disorders of mitochondrial protein synthesis. Hum Mol Genet. 2003;12:R293–R301. doi: 10.1093/hmg/ddg285. [DOI] [PubMed] [Google Scholar]
- 35.Miller C, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 36.Coenen MJ, et al. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- 37.Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet. 1994;3:13–19. doi: 10.1093/hmg/3.1.13. A study of individual muscle fibres from patients with mitochondrial myopathy caused by large-scale mtDNA rearrangements revealed that, in addition to levels of deleted mtDNA that are above a critical threshold, respiratory-deficient fibres exhibit a marked reduction in the absolute amount of wild-type mtDNA. [DOI] [PubMed] [Google Scholar]
- 38.Coller HA, et al. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nature Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 39.Taylor RW, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gyllensten U, Wharton D, Josefsson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 42.Shitara H, Hayashi JI, Takahama S, Kaneda H, Yonekawa H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics. 1998;148:851–857. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awadalla P, Eyre-Walker A, Smith JM. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science. 1999;286:2524–2525. doi: 10.1126/science.286.5449.2524. [DOI] [PubMed] [Google Scholar]
- 44.D’Aurelio M, et al. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet. 2004;13:3171–3179. doi: 10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- 45.Kraytsberg Y, et al. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- 46.Elson JL, et al. Analysis of European mtDNAs for recombination. Am J Hum Genet. 2001;68:145–153. doi: 10.1086/316938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 48.Taylor RW, et al. Genotypes from patients indicate no paternal mitochondrial DNA contribution. Ann Neurol. 2003;54:521–524. doi: 10.1002/ana.10673. [DOI] [PubMed] [Google Scholar]
- 49.Filosto M, et al. Lack of paternal inheritance of muscle mitochondrial DNA in sporadic mitochondrial myopathies. Ann Neurol. 2003;54:524–526. doi: 10.1002/ana.10709. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz M, Vissing J. No evidence for paternal inheritance of mtDNA in patients with sporadic mtDNA mutations. J Neurol Sci. 2004;218:99–101. doi: 10.1016/j.jns.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Danan C, et al. Evaluation of parental mitochondrial inheritance in neonates born after intracytoplasmic sperm injection. Am J Hum Genet. 1999;65:463–473. doi: 10.1086/302484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchington DR, et al. No evidence for paternal mtDNA transmission to offspring or extra-embryonic tissues after ICSI. Mol Hum Reprod. 2002;8:1046–1049. doi: 10.1093/molehr/8.11.1046. [DOI] [PubMed] [Google Scholar]
- 53.Chinnery PF, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet. 2004;364:592–596. doi: 10.1016/S0140-6736(04)16851-7. [DOI] [PubMed] [Google Scholar]
- 54.Man PY, et al. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am J Hum Genet. 2003;72:333–339. doi: 10.1086/346066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prezant TR, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nature Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 56.Battersby BJ, Loredo-Osti JC, Shoubridge EA. Nuclear genetic control of mitochondrial DNA segregation. Nature Genet. 2003;33:183–186. doi: 10.1038/ng1073. [DOI] [PubMed] [Google Scholar]
- 57.Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am J Hum Genet. 2000;68:533–536. doi: 10.1086/318190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poulton J, Turnbull DM. 74th ENMC international workshop: mitochondrial diseases 19–20 November 1999, Naarden, the Netherlands. Neuromuscul Disord. 2000;10:460–462. doi: 10.1016/s0960-8966(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 59.Taylor RW, Schaefer AM, Barron MJ, McFarland R, Turnbull DM. The diagnosis of mitochondrial muscle disease. Neuromuscul Disord. 2004;14:237–245. doi: 10.1016/j.nmd.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 60.McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: when ‘definitely maybe’ is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. The first demonstration that heteroplasmic, large-scale rearrangements of the mitochondrial genome could cause human disease. [DOI] [PubMed] [Google Scholar]
- 62.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. This paper describes the first example of a single-nucleotide change in the mitochondrial genome (11778G>A) as the cause of a maternally inherited, neurological disorder in multiple families. [DOI] [PubMed] [Google Scholar]
- 63.Brandon MC, et al. MITOMAP: a human mitochondrial genome database — 2004 update. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moraes CT, et al. Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns–Sayre syndrome. N Engl J Med. 1989;320:1293–1299. doi: 10.1056/NEJM198905183202001. [DOI] [PubMed] [Google Scholar]
- 65.Rotig A, Cormier V, Blanche S, Bonnefont J-P, Ledeist F. Pearson’s marrow pancreas syndrome. A multisystem mitochondrial disorder of infancy. J Clin Invest. 1990;86:1601–1608. doi: 10.1172/JCI114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McFarland R, et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- 67.de Vries DD, van Engelen BG, Gabreels FJ, Ruitenbeek W, van Oost BA. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- 68.Andreu AL, et al. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N Engl J Med. 1999;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- 69.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 70.Maassen JA, et al. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes. 2004;53(Suppl 1):103–109. doi: 10.2337/diabetes.53.2007.s103. [DOI] [PubMed] [Google Scholar]
- 71.Saker PJ, et al. UKPDS 21: low prevalence of the mitochondrial transfer RNA gene (tRNALeu(UUR)) mutation at position 3243bp in UK Caucasian type 2 diabetic patients. Diabet Med. 1997;14:42–45. doi: 10.1002/(SICI)1096-9136(199701)14:1<42::AID-DIA295>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 72.Ohkubo K, et al. Mitochondrial gene mutations in the tRNALeu(UUR) region and diabetes: prevalence and clinical phenotypes in Japan. Clin Chem. 2001;47:1641–1648. [PubMed] [Google Scholar]
- 73.Kearney PM, et al. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 74.Chinnery PF, et al. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol. 2000;48:188–193. [PubMed] [Google Scholar]
- 75.Choo-Kang AT, et al. Defining the importance of mitochondrial gene defects in maternally inherited diabetes by sequencing the entire mitochondrial genome. Diabetes. 2002;51:2317–2320. doi: 10.2337/diabetes.51.7.2317. [DOI] [PubMed] [Google Scholar]
- 76.Wallace D. C. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 77.Herrnstadt C, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khogali SS, et al. A common mitochondrial DNA variant associated with susceptibility to dilated cardiomyopathy in two different populations. Lancet. 2001;357:1265–1267. doi: 10.1016/S0140-6736(00)04422-6. [DOI] [PubMed] [Google Scholar]
- 79.Poulton J, et al. Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case-control study. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- 80.Torroni A, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chagnon P, et al. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am J Med Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 82.van der Walt JM, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 83.Ross OA, et al. mt4216C variant in linkage with the mtDNA TJ cluster may confer a susceptibility to mitochondrial dysfunction resulting in an increased risk of Parkinson’s disease in the Irish. Exp Gerontol. 2003;38:397–405. doi: 10.1016/s0531-5565(02)00266-8. [DOI] [PubMed] [Google Scholar]
- 84.van der Walt JM, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helgason A, Yngvadóttir B, Hrafnkelsson B, Gulcher J, Stefánsson K. An Icelandic example of the impact of population structure on association studies. Nature Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- 86.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. This paper highlights the technology that has made transmitochondrial cybrids — which are generated by fusing human cell lines that lack mtDNA to enucleated cytoplasts from patients’ cells that harbour mtDNA mutations and then growing them under selection — such an elegant cell culture system to study the bioenergetic and cellular consequences of pathogenic mtDNA mutations. [DOI] [PubMed] [Google Scholar]
- 87.Chomyn A, et al. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci USA. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayashi J, et al. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tiranti V, et al. Maternally inherited hearing loss, ataxia and myoclonus associated with a novel point mutation in mitochondrial tRNASer(UCN) gene. Hum Mol Genet. 1995;4:1421–1427. doi: 10.1093/hmg/4.8.1421. [DOI] [PubMed] [Google Scholar]
- 90.Taanman JW, et al. Molecular mechanisms in mitochondrial DNA depletion syndrome. Hum Mol Genet. 1997;6:935–942. doi: 10.1093/hmg/6.6.935. [DOI] [PubMed] [Google Scholar]
- 91.Dunbar DR, Moonie PA, Jacobs HT, Holt IJ. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci USA. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.El Meziane A, et al. A tRNA supressor mutation in human mitochondria. Nature Genet. 1998;18:350–353. doi: 10.1038/ng0498-350. [DOI] [PubMed] [Google Scholar]
- 93.Jenuth J, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germ line explains the rapid segregation of mammalian mitochondrial DNA. Nature Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. By generating heteroplasmic mice for two (neutral) mtDNA genotypes, the authors demonstrated that random genetic drift in early oogenesis was the reason for the observed rapid segregation of mtDNA sequence variants that occurs between generations. [DOI] [PubMed] [Google Scholar]
- 94.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nature Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 95.Marchington DR, Barlow D, Poulton J. Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nature Med. 1999;5:957–960. doi: 10.1038/11403. [DOI] [PubMed] [Google Scholar]
- 96.Sligh JE, et al. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci USA. 2000;97:14461–14466. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue K, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nature Genet. 2000;26:176–181. doi: 10.1038/82826. By isolating mouse cybrid clones with high levels of a somatic mtDNA rearrangement and fusing these with fertilized mouse eggs, these authors generated the first mouse model of a pathogenic mtDNA mutation (mtDNA deletion or duplication), which was transmitted from mother to offspring. [DOI] [PubMed] [Google Scholar]
- 98.Nakada K, et al. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nature Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- 99.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. The first Tfam knockout mouse model that demonstrates a role for the TFAM nuclear protein in maintaining mtDNA copy number. [DOI] [PubMed] [Google Scholar]
- 100.Silva JP, et al. Impaired insulin secretion and β-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nature Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 101.Sorensen L, et al. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wredenberg A, et al. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci USA. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 104.Chinnery PF, Bindoff LA. 116th ENMC international workshop: the treatment of mitochondrial disorders, 14th–16th March 2003, Naarden, The Netherlands. Neuromuscul Disord. 2003;13:757–764. doi: 10.1016/s0960-8966(03)00097-x. [DOI] [PubMed] [Google Scholar]
- 105.Taivassalo T, et al. Gene shifting: a novel therapy for mitochondrial myopathy. Hum Mol Genet. 1999;8:1047–1052. doi: 10.1093/hmg/8.6.1047. [DOI] [PubMed] [Google Scholar]
- 106.Clark KM, et al. Reversal of a mitochondrial DNA defect in human skeletal muscle. Nature Genet. 1997;16:222–224. doi: 10.1038/ng0797-222. [DOI] [PubMed] [Google Scholar]
- 107.Fu K, et al. A novel heteroplasmic tRNAleu(CUN) mtDNA point mutation in a sporadic patient with mitochondrial encephalomyopathy segregates rapidly in skeletal muscle and suggests an approach to therapy. Hum Mol Genet. 1996;5:1835–1840. doi: 10.1093/hmg/5.11.1835. [DOI] [PubMed] [Google Scholar]
- 108.Taivassalo T, et al. Aerobic conditioning in patients with mitochondrial myopathies: physiological, biochemical, and genetic effects. Ann Neurol. 2001;50:133–141. doi: 10.1002/ana.1050. [DOI] [PubMed] [Google Scholar]
- 109.Manfredi G, et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nature Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 110.Guy J, et al. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- 111.Kolesnikova OA, et al. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- 112.Kolesnikova OA, et al. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 113.Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nature Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 114.Tanaka M, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 115.Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum Mol Genet. 2001;10:3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 116.Manfredi G, et al. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J Biol Chem. 1999;274:9386–9381. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 117.Santra S, Gilkerson RW, Davidson M, Schon EA. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann Neurol. 2004;56:662–669. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- 118.Feuermann M, et al. The yeast counterparts of human ‘MELAS’ mutations cause mitochondrial dysfunction that can be rescued by overexpression of the mitochondrial translation factor EF-Tu. EMBO Rep. 2003;4:53–58. doi: 10.1038/sj.embor.embor713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harding AE, Holt IJ, Sweeney MG, Brockington M, Davis MB. Prenatal diagnosis of mitochondrial DNA 8993T>G disease. Am J Hum Genet. 1992;50:629–633. [PMC free article] [PubMed] [Google Scholar]
- 120.Leshinsky-Silver E, et al. Prenatal exclusion of Leigh syndrome due to T8993C mutation in the mitochondrial DNA. Prenat Diagn. 2003;23:31–33. doi: 10.1002/pd.516. [DOI] [PubMed] [Google Scholar]
- 121.Jacobs LJ, et al. Transmission and prenatal diagnosis of the T9176C mitochondrial DNA mutation. Mol Hum Reprod. 2005;11:223–228. doi: 10.1093/molehr/gah152. [DOI] [PubMed] [Google Scholar]
- 122.Weber K, et al. A new mtDNA mutation showing accumulation with time and restriction to skeletal muscle. Am J Hum Genet. 1997;60:373–380. [PMC free article] [PubMed] [Google Scholar]
- 123.White SL, et al. Two cases of prenatal analysis for the pathogenic T to G substitution at nucleotide 8993 in mitochondrial DNA. Prenat Diagn. 1999;19:1165–1168. [PubMed] [Google Scholar]
- 124.Lin DP, et al. Comparison of mitochondrial DNA contents in human embryos with good or poor morphology at the 8-cell stage. Fertil Steril. 2004;81:73–79. doi: 10.1016/j.fertnstert.2003.05.005. [DOI] [PubMed] [Google Scholar]
- 125.Dean NL, et al. Prospect of preimplantation genetic diagnosis for heritable mitochondrial DNA diseases. Mol Hum Reprod. 2003;9:631–638. doi: 10.1093/molehr/gag077. [DOI] [PubMed] [Google Scholar]
- 126.Kagawa Y, Hayashi JI. Gene therapy of mitochondrial diseases using human cytoplasts. Gene Ther. 1997;4:6–10. doi: 10.1038/sj.gt.3300347. [DOI] [PubMed] [Google Scholar]
- 127.Cohen J, Scott R, Schimmel T, Levron J, Willadsen S. Birth of infant after transfer of anucleate donor oocyte cytoplasm into recipient eggs. Lancet. 1997;350:186–187. doi: 10.1016/S0140-6736(05)62353-7. [DOI] [PubMed] [Google Scholar]
- 128.Hawes SM, Sapienza C, Latham KE. Ooplasmic donation in humans: the potential for epigenic modifications. Hum Reprod. 2002;17:850–852. doi: 10.1093/humrep/17.4.850. [DOI] [PubMed] [Google Scholar]
- 129.Brenner CA, Barritt JA, Willadsen S, Cohen J. Mitochondrial DNA heteroplasmy after human ooplasmic transplantation. Fertil Steril. 2000;74:573–578. doi: 10.1016/s0015-0282(00)00681-6. [DOI] [PubMed] [Google Scholar]
- 130.Thorburn DR, Dahl HHM. Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet. 2001;106:102–114. doi: 10.1002/ajmg.1380. [DOI] [PubMed] [Google Scholar]
- 131.Roberts RM. Prevention of human mitochondrial (mtDNA) disease by nucleus transplantation into an enucleated donor oocyte. Am J Med Genet. 1999;87:265–266. doi: 10.1002/(sici)1096-8628(19991126)87:3<265::aid-ajmg14>3.0.co;2-s. This paper describes the possibility of preventing transmission of mitochondrial DNA disease. [DOI] [PubMed] [Google Scholar]
- 132.Liu H, Wang CW, Grifo JA, Krey LC, Zhang J. Reconstruction of mouse oocytes by germinal vesicle transfer: maturity of host oocyte cytoplasm determines meiosis. Hum Reprod. 1999;14:2357–2361. doi: 10.1093/humrep/14.9.2357. [DOI] [PubMed] [Google Scholar]
- 133.Liu H, Zhang J, Krey LC, Grifo JA. In-vitro development of mouse zygotes following reconstruction by sequential transfer of germinal vesicles and haploid pronuclei. Hum Reprod. 2000;15:1997–2002. doi: 10.1093/humrep/15.9.1997. [DOI] [PubMed] [Google Scholar]
- 134.Takeuchi T, Ergun B, Huang TH, Rosenwaks Z, Palermo GD. A reliable technique of nuclear transplantation for immature mammalian oocytes. Hum Reprod. 1999;14:1312–1317. doi: 10.1093/humrep/14.5.1312. [DOI] [PubMed] [Google Scholar]
- 135.Barnes FL, et al. Blastocyst development and birth after in-vitro maturation of human primary oocytes, intracytoplasmic sperm injection and assisted hatching. Hum Reprod. 1995;10:3243–3247. doi: 10.1093/oxfordjournals.humrep.a135896. [DOI] [PubMed] [Google Scholar]
- 136.Goud PT, et al. In-vitro maturation of human germinal vesicle stage oocytes: role of cumulus cells and epidermal growth factor in the culture medium. Hum Reprod. 1998;13:1638–1644. doi: 10.1093/humrep/13.6.1638. [DOI] [PubMed] [Google Scholar]
- 137.Kattera S, Chen C. Normal birth after microsurgical enucleation of tripronuclear human zygotes: case report. Hum Reprod. 2003;18:1319–1322. doi: 10.1093/humrep/deg262. [DOI] [PubMed] [Google Scholar]
- 138.Meirelles F, Smith LC. Mitochondrial genotype segregation in a mouse heteroplasmic lineage produced by embyonic karyoplast transplantation. Genetics. 1997;145:445–451. doi: 10.1093/genetics/145.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meirelles F, Smith LC. Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics. 1998;148:877–883. doi: 10.1093/genetics/148.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harman D. Free radical theory of aging. Mutat Res. 1992;275:257–266. doi: 10.1016/0921-8734(92)90030-s. This is a description of the theory of ageing in which mitochondria have a key role. [DOI] [PubMed] [Google Scholar]
- 141.Geromel V, et al. Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum Mol Genet. 2001;10:1221–1228. doi: 10.1093/hmg/10.11.1221. [DOI] [PubMed] [Google Scholar]
- 142.Mattiazzi M, et al. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 143.Muller-Hocker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart —an age-related phenomenon. A histochemical ultracytochemical study. Am J Pathol. 1989;134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- 144.Muller-Hocker J, Seibel P, Schneiderbanger K, Kadenbach B. Different in situ hybridization patterns of mitochondrial DNA in cytochrome c oxidase-deficient extraocular muscle fibres in the elderly. Virchows Arch A Pathol Anat Histopathol. 1993;422:7–15. doi: 10.1007/BF01605127. [DOI] [PubMed] [Google Scholar]
- 145.Brierley EJ, Johnson MA, Lightowlers RN, James OF, Turnbull DM. Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann Neurol. 1998;43:217–223. doi: 10.1002/ana.410430212. [DOI] [PubMed] [Google Scholar]
- 146.Nekhaeva E, et al. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci USA. 2002;99:5521–5526. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 148.de Grey AD. A proposed refinement of the mitochondrial free radical theory of aging. Bioessays. 1997;19:161–166. doi: 10.1002/bies.950190211. [DOI] [PubMed] [Google Scholar]
- 149.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nature Genet. 1998;20:291–293. doi: 10.1038/3108. The first paper to describe the presence of somatic mtDNA mutations in solid human tumours, in this case colon cancer. In many cases, the mtDNA mutations had accumulated to homoplasmic levels and were not evident in the matched normal tissue from the same patient. A causal relationship between mtDNA mutations and tumorigenesis is yet to be established. [DOI] [PubMed] [Google Scholar]
- 152.Fliss MS, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 153.Jeronimo C, et al. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195–5198. doi: 10.1038/sj.onc.1204646. [DOI] [PubMed] [Google Scholar]
- 154.Jones JB, et al. Detection of mitochondrial DNA mutations in pancreatic cancer offers a ‘mass’-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 155.Kirches E, et al. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93:534–538. doi: 10.1002/ijc.1375. [DOI] [PubMed] [Google Scholar]
- 156.He L, et al. Somatic mitochondrial DNA mutations in adult-onset leukaemia. Leukemia. 2003;17:2487–2491. doi: 10.1038/sj.leu.2403146. [DOI] [PubMed] [Google Scholar]
- 157.Wardell TM, et al. Changes in the human mitochondrial genome after treatment of malignant disease. Mutat Res. 2003;525:19–27. doi: 10.1016/s0027-5107(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 158.Zeviani M, et al. Deletions of mitochondrial DNA in Kearns–Sayre syndrome. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 159.Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 160.Santorelli FM, et al. Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun. 1997;238:326–328. doi: 10.1006/bbrc.1997.7167. [DOI] [PubMed] [Google Scholar]
- 161.Kirby DM, et al. Mutations of the mitochondrial ND1 gene as a cause of MELAS. J Med Genet. 2004;41:784–789. doi: 10.1136/jmg.2004.020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shoffner JM, et al. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 163.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 164.van den Ouweland JWM, Lemkes HHPJ, Ruitenbeek K. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nature Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 165.Howell N, et al. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49:939–950. [PMC free article] [PubMed] [Google Scholar]
- 166.Johns DR, Neufeld MJ, Park RD. An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem Biophys Res Commun. 1992;187:1551–1557. doi: 10.1016/0006-291x(92)90479-5. [DOI] [PubMed] [Google Scholar]
- 167.Hao H, Bonilla E, Manfredi G, DiMauro S, Moraes CT. Segregation patterns of a novel mutation in the mitochondrial tRNA glutamic acid gene associated with myopathy and diabetes mellitus. Am J Hum Genet. 1995;56:1017–1025. [PMC free article] [PubMed] [Google Scholar]
- 168.McFarland R, et al. Familial myopathy: New insights into the T14709C mitochondrial tRNA mutation. Ann Neurol. 2004;55:478–484. doi: 10.1002/ana.20004. [DOI] [PubMed] [Google Scholar]
- 169.Reid FM, Vernham GA, Jacobs HT. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum Mutat. 1994;3:243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- 170.Sue CM, et al. Maternally inherited hearing loss in a large kindred with a novel T7511C mutation in the mitochondrial DNA tRNASer(UCN) gene. Neurology. 1999;52:1905–1908. doi: 10.1212/wnl.52.9.1905. [DOI] [PubMed] [Google Scholar]
- 171.Strachan T, Read AP. Human Molecular Genetics. 2nd edn. John Wiley and Sons; New York: 1999. [Google Scholar]