UBE2T, the Fanconi Anemia Core Complex, and FANCD2 Are Recruited Independently to Chromatin: a Basis for the Regulation of FANCD2 Monoubiquitination (original) (raw)
Abstract
The Fanconi anemia (FA) nuclear core complex and the E2 ubiquitin-conjugating enzyme UBE2T are required for the S phase and DNA damage-restricted monoubiquitination of FANCD2. This constitutes a key step in the FA tumor suppressor pathway, and much attention has been focused on the regulation at this point. Here, we address the importance of the assembly of the FA core complex and the subcellular localization of UBE2T in the regulation of FANCD2 monoubiquitination. We establish three points. First, the stable assembly of the FA core complex can be dissociated of its ability to function as an E3 ubiquitin ligase. Second, the actual E3 ligase activity is not determined by the assembly of the FA core complex but rather by its DNA damage-induced localization to chromatin. Finally, UBE2T and FANCD2 access this subcellular fraction independently of the FA core complex. FANCD2 monoubiquitination is therefore not regulated by multiprotein complex assembly but by the formation of an active E2/E3 holoenzyme on chromatin.
Patients with the rare genetic disorder Fanconi anemia (FA) have a common defect in a DNA damage response pathway that contributes to the maintenance of genome stability (12). The FA pathway consists of a high-molecular-weight nuclear core complex which contains at least 10 subunits (FANCA, -B, -C, -E, -F, -G, -L, and -M, FAAP100, and FAAP24 proteins, known as the FA core complex) (20, 22, 27), as well as five additional proteins, FANCD2 (8, 38), FANCI (6, 35, 36), FANCD1 (BRCA2) (11), FANCN (PALB2) (33, 34), and FANCJ (BRIP1) (3, 16). DNA replication and DNA damage somehow activate the FA core complex to monoubiquitinate the FANCD2/FANCI heterodimer (9, 35, 36). The FA core complex and ubiquitinated FANCD2/FANCI are then thought to process DNA lesions. However, we still do not know how this FA core complex is activated and how it functions directly within such a DNA damage response pathway.
Most current studies that have determined interactions between FA core complex subunits suggest the existence of distinct subcomplexes in the cytosolic and nuclear compartments (5, 19, 32). Indeed, the stability of the FA core complex is severely compromised in most FA patients, and this is due to mutational inactivation of any of the FA core complex genes (8). In addition, the nuclear import of some of the FA core complex components is regulated and their localization is interdependent (15, 29). Cumulatively, all these observations have led to a compelling proposal that the FA core complex is sequentially assembled and that this process may regulate its activity. The FA core complex E3 ligase activity is tightly restricted to S and G2 phases of the cell cycle and can be further triggered by DNA damage (23, 25, 37). One way to ensure this restriction is to control the assembly of the FA core complex in response to such signals. In addition to complex assembly, some of the core complex components (such as FANCM, FANCE, and FANCG) are phosphorylated in response to DNA replication and damage (21, 24, 26, 31, 39). Such modifications may also be important for regulation.
The key function of the FA core complex is to stimulate the site-specific monoubiquitination of the FANCD2 and FANCI proteins (8, 36). Hence, the FA core complex likely functions as a multisubunit E3 ubiquitin ligase. Support for this comes from the fact that at least one intrinsic component of the FA core complex, FANCE, can bind and perhaps recruit FANCD2 for modification (29). More importantly, another essential component of the complex, FANCL, has a PHD/RING finger domain (20). This domain of FANCL binds to the E2-conjugating enzyme, UBE2T, and stimulates its autoubiquitination (18, 20). In addition, UBE2T has been shown to be necessary for the efficient DNA damage-induced monoubiquitination of FANCD2 (18). Thus, the FA core complex most likely assembles with UBE2T as an active E2/E3 holoenzyme. However, how and where such interactions occur and whether they contribute to regulation remain to be determined.
This study investigates the relevance of an assembly of the FA core complex as well as its subcellular localization with UBE2T to its E3 ubiquitin ligase activity. Using the model vertebrate genetic system DT40, we consolidate previous work and show that the E3 ligase activity of the FA core complex can be dissociated from its assembly. By creating novel DT40 strains, we also establish that the FA core complex is constitutively assembled and therefore stable throughout the cell cycle. Molecular size and abundance of the FA core complex are hardly affected, although the complex accumulates on chromatin in response to DNA replication and damage. Interestingly, UBE2T is constitutively present in the chromatin fraction, while the FA core complex and its substrate FANCD2 accumulate there independently in a restricted manner. Cumulatively, these observations allow us to put forward a model that may explain how the FA pathway is activated in response to DNA replication and DNA damage.
MATERIALS AND METHODS
Isolation of chicken FANCL cDNA and plasmid construction.
Full-length cDNA of chicken FANCL was derived from a screen of a chicken embryonic fibroblast cDNA library (gift from A. Neito and D. Wilkinson). FANCL cDNA with adapted XhoI/NotI restriction sites was amplified by PCR and ligated into pBluescript. N-terminal TAP-tag cDNA (encoding two protein A domains, a tobacco etch virus protease cleavage site, and a calmodulin binding domain) with flanking XhoI/SalI sites was cloned into the XhoI site of pBluescript-FANCL (SalI site destroyed). Subsequently, XhoI/NotI sites of the TAP-FANCL fragment were filled in with Klenow enzyme and ligated into the SmaI site of pExpress (gift from Jean-Marie Buerstedde). The expression cassette of TAP-FANCL (with a beta-actin promoter) from pExpress was then ligated into pLoxBsr (gift from Jean-Marie Buerstedde). Stable transfections of DT40 strains with TAP-FANCL were performed as described previously (28).
Cysteine point mutations, Cys305 to Ala and Cys357 to Ala, in TAP-FANCL were generated by site-directed mutagenesis (QuikChange II XL site-directed mutagenesis kit; Stratagene) using the following primer sets: AGTGACTTTA CTAAGGACGC TGGAATCTGC TATGCCGCCT AC and GTAGGCGGCA TAGCAGATTC CAGCGTCCTT AGTAAAGTC ACT for Cys305Ala; ATCTTTGGTG AATGTCCATA TGCCAACAAG CCACTGACAG TG and CACTGTCAGT GGCTTGTTGC AATATGGACA TTCACCAAAG AT for Cys357Ala.
DT40 cell culture and transfections.
DT40 cells were cultured at 37°C in RPMI 1640 supplemented with 7% fetal calf serum (Gibco-BRL), 3% chicken serum (Gibco-BRL), 50 μM 2-mercaptoethanol, and penicillin-streptomycin. Transfections were carried out as described previously (28).
Generation of ΔFANCL, Δ_UBE2T_, in situ-tagged FANCB-HA, and ΔFANCL with in situ-tagged FANCC-TAP cell lines.
The scheme of the generation of gene disruptions is outlined in Fig. 1A. The targeting construct for FANCL disruption was made by cloning the 5′ arm, a KpnI/XhoI fragment amplified by PCR, with the primer pair 5′-CACAGTCAAG CTAAAAGCTG AAGACTC and 5′-AGTACCCACG TCTTCCCATC AATTTCATC, and the 3′ arm, a BamHI/NotI fragment, was amplified by PCR with the primer pair 5′-AACTTGAAAG ACCTCTTAG AAATTGA and 5′-GGTGCTTTTC TCAAGGTTTC TTCATG into a pBluescript vector. Drug resistance cassettes for neomycin (for the first allele knockout) and puromycin (for the second allele knockout) were cloned in as BamHI fragments. Targeted integrations were detected by Southern blotting of StuI/KpnI-digested genomic DNA. The probe was hybridized as illustrated in Fig. 1A.
FIG. 1.
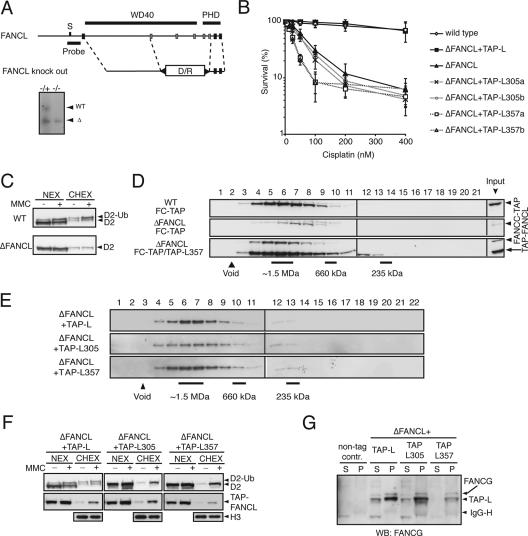
Functional separation of the FA core complex assembly and its E3 ubiquitin ligase activity. (A) Schematic representation of the genomic locus of FANCL with the exon configuration in correlation with the domain structure and the gene disruption construct with exons 2 to 6 removed. S, StuI. Southern blot analysis was performed on StuI/KpnI-digested genomic DNA from heterozygous and homozygous genotypes. WT, wild-type locus; Δ, gene knockout locus. The position of the probe used is indicated by the bar. (B) Sensitivity curves of the indicated cell lines. Cells were treated with various concentrations of cisplatin for 72 h and analyzed for survival in an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] assay. Standard errors of the means are given from three independent experiments. (C) Immunoblot analysis of FANCD2 in subfractionated Δ_FANCL_ cells either untreated (−) or treated with 150 ng/ml MMC (+). D2, FANCD2; D2-Ub, monoubiquitinated FANCD2; NEX, high-salt nuclear extract; CHEX, solubilized chromatin extract. (D) Nuclear extracts of the indicated cell lines (WT/C-TAP, ΔFANCL/C-TAP, and ΔFANCL/C-TAP + L357) were separated by size exclusion chromatography, and fractions were analyzed for FANCC-TAP and TAP-FANCL357 by immunoblot analysis. (E) Size exclusion chromatography of nuclear extracts of the indicated cell lines and immunoblot analyses for TAP-FANCC. (F) Subfractionation of either MMC-treated (+) or untreated (-) cells into high-salt NEX and CHEX and immunoblot analysis for FANCD2 and TAP-FANCL. D2, FANCD2; D2-Ub, monoubiquitinated FANCD2. Histone H3 was used as a CHEX loading control. (G) Coimmunoprecipitation of wild-type and mutant TAP-FANCL and immunoblot analysis for FANCG. Nontagged wild-type cells were used as a negative immunoprecipitation control. FANCG antibody cross-reacts with the protein A domain of TAP-tag. WB, Western blot; S/N, supernatant of nonprecipitated proteins; P, fraction of precipitated proteins; Ig-H, immunoglobulin heavy chain.
Figure 5A, below, gives a schematic overview of the UBE2T gene disruption. The 5′ and 3′ arms for the targeting construct were amplified by PCR with the primer pairs GGTGGAACGT CACATCCATC CATGCGTTT/GAGTCGATGT TAGGATGATA AATAGGGG and AATATTAGGT CAAATCGTTG TTGTTATG/GACCCGGCTG GTTTCCAGGA CATCGTGG, respectively. PCR fragments were subcloned into the pCR2.1 TOPO vector (Invitrogen). The 5′ arm as a NotI/BamHI fragment and the 3′ arm as a BamHI/EcoRI fragment were subsequently cloned into a pBluescript vector. Drug resistance cassettes for puromycin (first allele knockout) and blasticidin (second allele knockout) were cloned in as BamHI fragments. Targeted integrations were detected by Southern blot analysis of BamHI-digested genomic DNA. The probe used for hybridization is indicated below in Fig. 5A.
FIG. 5.
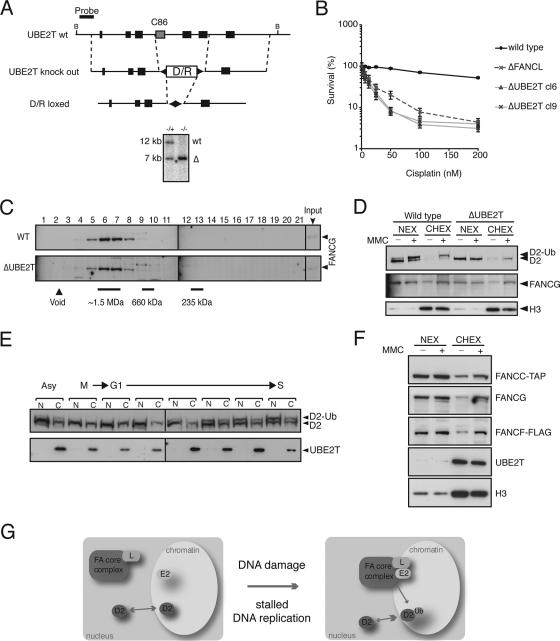
UBE2T is constitutively localized to chromatin. (A) Schematic presentation of the genomic locus of UBE2T with the exon configuration in correlation with the domain structure and the gene disruption construct, with exons 4 to 6 removed. B, BamHI. Southern blot analysis of BamHI-digested genomic DNA from heterozygous (+/−) and homozygous (−/−) genotypes. wt, wild type locus; Δ, gene knockout locus. The position of the probe used is indicated by the bar. (B) Sensitivity curves of two independent Δ_UBE2T_ cell line clones (cl6 and cl9). Cells were treated with increasing concentrations of cisplatin for 72 h and analyzed for survival. Standard errors of the means are given for three independent experiments. (C) Nuclear extracts of wild-type (WT) and Δ_UBE2T_ cells were separated by size exclusion chromatography, and fractions were immunoblotted for FANCG. (D) Cellular subfractionation of wild-type and Δ_UBE2T_ cells either untreated (−) or MMC treated (+). NEX, nuclear extract; CHEX, chromatin extract. Immunoblot detection of FANCG, histone H3 (H3), and FANCD2 was performed. (E) UBE2T distribution into nuclear extracts (N) and chromatin extracts (C) in asynchronous cells (Asy), mitosis-arrested cells (M), and synchronized cells from the G1-to-S-phase transition. Results show immunoblot detection of UBE2T and FANCD2. D2, FANCD2; D2-Ub, monoubiquitinated FANCD2. (F) Cellular subfractionation of the multitagged FA core complex cell line either untreated (−) or treated with MMC (+). Immunoblot detection of FANCC-TAP, FANCG, FANCF-FLAG, UBE2T, and histone H3 (H3) is shown. (G) Model for the S phase and DNA damage-induced activation of the FA core complex (see Discussion for details).
In situ tagging of FANCC with cDNA encoding a TAP-tag in the Δ_FANCL_ strain was done as described previously (26).
In situ tagging of FANCB with cDNA encoding 3× hemagglutinin (HA) directly after the last exon was generated by PCR of a 5′ arm using the following primer pairs: CCGTCGACGG GTTATCTTGG TAGCTTCAGT ACCAGTCC and TCTGATATGA TTGTGTGGAG ACTGAGCAAG TCCGCTAGCG G. The PCR product was cloned into pBluescript as a SalI/NheI fragment and ligated to NheI/NotI-adapted 3× HA cDNA. The NotI site was replaced with a BamHI linker. The 5′ arm was recovered as a SalI/BamHI fragment and ligated to the 3′ arm of the knockout construct described earlier. A drug resistance cassette for blasticidin was ligated into the BamHI site. Homologous targeting was detected by Southern blot analysis as for the Δ_FANCB_ knockout, and expression of FANCB-HA was determined by immunodetection with antibody specific for HA (Roche).
Cellular subfractionation, FANCD2 immunoblot analyses, and cisplatin sensitivity assays.
The cellular subfractionation, FANCD2 immunoblot analyses, and cisplatin sensitivity assays were done as described previously (26).
Cell synchronization, FANCC-TAP precipitation, and size exclusion chromatography.
To synchronize 1.5 × 109 cells in M phase, log-phase DT40 cells were cultured in medium containing 25 ng/ml nocodazole (Sigma) for 10 h at 37°C. More than 90% of the cells accumulated with a 4N DNA content as determined by fluorescence-activated cell sorter (FACS) analysis. The synchronized cells were washed three times with prewarmed RPMI medium and released into 1.5 liters of fresh RPMI medium (supplemented with 5 mM hydroxyurea [HU] where indicated). FACS analysis revealed that 85% of the cells were released as a synchronized population. A 250-ml aliquot of cells was harvested 0, 1, 4.5, 7, 9, and 12 h after release and washed once with ice-cold 1× phosphate-buffered saline. FANCC-TAP precipitations from whole-cell lysates, high-salt nuclear extracts, and soluble chromatin extracts (6 mg total protein) were done as described previously (26). Precipitates were separated by 8% Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis for immunoblot detection of FANCM and FANCD2 or on a 4 to 12% bis-Tris-PAGE gel (Invitrogen) for immunoblot detection with antibodies to Flag-tag (Sigma), HA-tag (Roche), TAP-tag (peroxidase-antiperoxidase soluble complex; Sigma), and FANCG protein. For size exclusion chromatography, 3 to 10 mg (total protein) of the indicated protein extract was separated on a Superose S6 (16/60) column (Amersham) using 150 ml running buffer (20 mM Tris-HCl pH 8, 400 mM NaCl, 10% glycine, 1 mM dithiothreitol). Four-milliliter fractions were collected, concentrated (Vivaspin), and immunoblotted for analysis. Size calibration of the Superose S6 column was performed for each experiment with a high-molecular-weight marker kit (Amersham).
FANCG and UBE2T antibodies.
The cDNA encoding the C-terminal portion of chicken FANCG (150 amino acids) was amplified with adapted oligonucleotides. The PCR product was cloned into the bacterial expression vector pET (Novagen), resulting in an N-terminal maltose binding protein-His6 tag fusion protein. Maltose binding protein purifications were done as described previously (26). Affinity-purified antibodies recognize a predominant band of 70 kDa in DT40 wild-type extracts but not in ΔFANCG knockout cells. Anti-human UBE2T antibody has recently been described (18). The antibody cross-reacts with endogenous chicken UBE2T when analyzed by immunoblotting. A dominant band corresponding to a protein of ∼25 kDa and with the same mobility as recombinant human UBE2T was detected. Competition tests with recombinant protein confirmed the specificity of the antibody (data not shown).
RESULTS
FANCL is the catalytic subunit of the FA core complex.
In the chicken DT40 lymphoblastoid cell line, loss of the FA core complex protein FANCM results in the formation of unstable residual subcomplexes. Such subcomplexes have impaired E3 ligase activity and are defective in their accumulation in chromatin in response to DNA damage (26). To get further insight into the assembly and function of the FA core complex, we focused on FANCL, which probably provides the catalytic basis for the E3 ubiquitin ligase activity of the FA core complex. To study the function of FANCL in vivo, we first generated a DT40 strain where exons 2 to 6 (encoding the active cysteines Cys305 and Cys309 in the PHD/RING domain) were deleted (Fig. 1A). These cells are hypersensitive to the cross-linking agent cisplatin and are completely defective in their ability to monoubiquitinate FANCD2 (Fig. 1B and C). To study FA core complex assembly, we genetically engineered in situ a TAP (t_andem a_ffinity purification tag) cDNA into the FANCC locus in both wild-type and Δ_FANCL strains (see Materials and Methods for details). To test the assembly of FANCC-TAP into the FA core complex, we monitored complex formation by size exclusion chromatography. In wild-type cells, FANCC-TAP eluted in fractions corresponding to a molecular mass of ∼1.5 MDa. In contrast, the FANCC-TAP complex was much less abundant and shifted to fractions of a smaller size in Δ_FANCL cells (Fig. 1D, top and middle elution profiles).
In many ways, the FANCL disruption is indistinguishable from disruptions of any of the other FA core complex genes in human or chicken cells. In all these instances the FA core complex fails to assemble. It is therefore impossible to establish whether FANCL enzymatic activity or its role in stabilizing the FA core complex results in the cellular FA defects. We created a mutant FANCL cell line that could support complex assembly but was deficient in the monoubiquitination of FANCD2. The FANCL protein domain structure consists of three N-terminal WD40 repeats and a C-terminal RING-like PHD domain (20). Although the isolated PHD/RING domain is capable of autoubiquitination in vitro, these domains can also function in chromatin interactions and phosphoinositide binding. A recent study indicated that the WD40 repeats of FANCL may anchor this protein into the FA core complex (9). We mutated two highly conserved cysteine residues into alanines in the PHD/RING domain (C305A and C357A). cDNAs encoding TAP-tagged wild-type FANCL as well as TAP-tagged mutants FANCL-C305A and FANCL-C357A were stably expressed in the Δ_FANCL_ strain. TAP-FANCL point mutants failed to complement the cisplatin sensitivity and loss of FANCD2 monoubiquitination in the Δ_FANCL_ strain (Fig. 1B and F). Next, we studied the migration pattern of TAP-FANCL in the Δ_FANCL_-complemented strains. Indeed, wild-type TAP-FANCL, TAP-FANCL-C305A, and TAP-FANCL-C357A coeluted at 1.5-MDa fractions, indicating that they form a stable FA core complex (Fig. 1E). In addition, we introduced TAP-FANCL-C357A into the Δ_FANCL_/_FANCC_-TAP strain. Size exclusion chromatography analysis clearly showed that FANCC-TAP coelutes with TAP-FANCL-C357A (Fig. 1D, bottom elution profile). Coimmunoprecipitation of FANCG with wild-type and point-mutated TAP-FANCL gave supporting evidence for the presence of an intact complex (Fig. 1G).
The FA core complex accumulates on chromatin following DNA damage, a response that requires the FANCM protein in chicken cells (26). We therefore tested whether the TAP-FANCL mutant FA core complexes were also able to respond to DNA damage in this manner. Following exposure to the DNA cross-linking agent mitomycin C (MMC), wild-type and mutant TAP-FANCL accumulated in the chromatin fraction (Fig. 1F). We also noted that unmodified FANCD2 accumulated on chromatin in a DNA damage-dependent manner. Furthermore, this accumulation is independent of functional FANCL. Taken together, our observations provide strong supportive evidence that the PHD/RING finger domain of FANCL is the catalytic subunit of the FA core complex.
Establishing a multitagged FA core complex in DT40 cells.
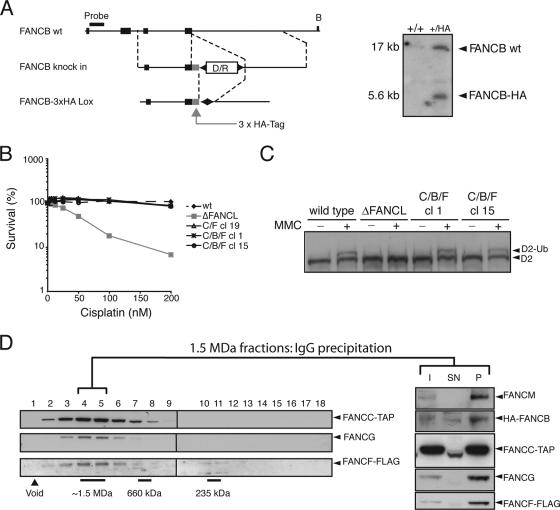
The experiments described above indicate that it is possible to dissociate FA core complex assembly from its enzymatic activity. As mentioned earlier, studies from patient-derived cell lines carrying mutations in FA core complex genes have shown the presence of subcomplexes. These studies further predict that a regulated sequential assembly process may underpin temporal regulation of the FA pathway. We therefore asked if sequential complex assembly determines where and how the pathway is activated. A key factor that limits the study of the core complex is the lack of antibodies of sufficient sensitivity and specificity to FA proteins. To circumvent this, we created a DT40 strain that only expresses a multiple-tagged FA core complex. This strain contains in situ-tagged FANCC (FANCC-TAP), FANCF (FANCF-2xFLAG) (described recently in reference 26), and FANCB (FANCB-3xHA) genes (Fig. 2A). These tags were engineered directly into the genomic locus so that the tagged FA gene is under normal physiological regulation. Gene targetings were confirmed by Southern blot analyses (Fig. 2A). The triple-tagged FA core complex cell line is indistinguishable from wild-type cells in terms of cisplatin sensitivity (Fig. 2B) and its ability to monoubiquitinate FANCD2 (Fig. 2C). Next, we determined whether the tagged FA core complex proteins are assembled in a large complex. Size exclusion chromatography of nuclear extracts clearly showed that FANCC-TAP, FANCF-FLAG, and untagged FANCG coelute in fractions that correspond to a 1.5-MDa complex (Fig. 2D). To emphatically establish that these fractions contain the FA core complex, a coprecipitation with FANCC-TAP from 1.5-MDa fractions was performed. The analyses for the presence of FANCC-TAP, FANCM, FANCB-HA, FANCG, and FANCF-FLAG confirmed a stable complex in the 1.5-MDa size exclusion chromatography fractions (Fig. 2D).
FIG. 2.
Generation of a cell line expressing a multitagged FA core complex. (A) Schematic of the genomic locus of FANCB with the configuration of the last four exons. In situ tagging of the 3′ with 3× HA tag cDNA. wt, wild type; B, BamHI. Homologous targeting was detected by Southern blot analysis of BamHI-digested genomic DNA. The position of the probe used is indicated by the bar. (B) Sensitivity curves of the indicated cell lines. Cells were analyzed after 72 h of exposure to various concentrations of cisplatin. Standard errors of the means are given from three independent experiments. (C) MMC-induced FANCD2 monoubiquitination of the multitagged FA core complex strains (C/B/F) (clone 1 and clone 15). D2, FANCD2; D2-Ub, monoubiquitinated FANCD2. (D) Nuclear extracts of the multitagged FA core complex cell lines were separated by size exclusion chromatography, and the elution profiles for tagged complex proteins were analyzed by immunoblot detection. Peak fractions were subjected to TAP immunoprecipitation. Portions (10%) of the input material (I), nonprecipitated fraction (SN), and precipitated fraction were analyzed by immunoblotting for FANCM, FANCB-HA, FANCC-TAP, FANCG, and FANCF-FLAG.
The multitagged FA core complex is constitutively assembled.
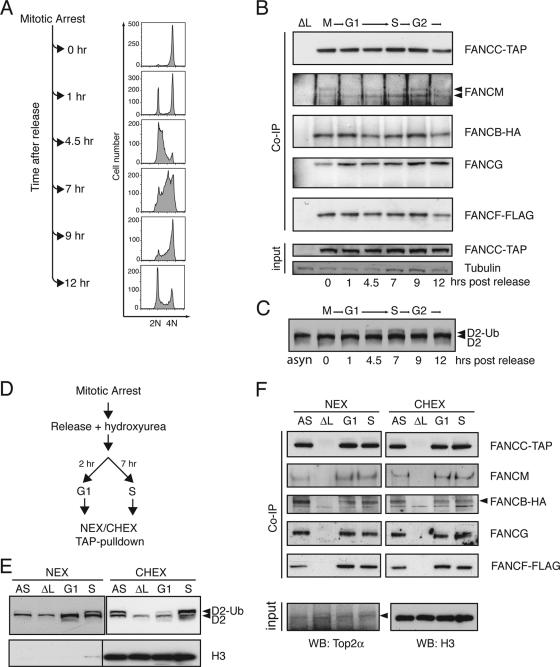
In order to study FA core complex formation during the cell cycle, the multitagged cell line was synchronized by nocodazole block in M phase and released into fresh medium. Synchronized cell populations were monitored and harvested at different time points as they transited through the G1, S, and G2/M phases of the cell cycle (Fig. 3A). Immunoprecipitations of FANCC-TAP from whole-cell lysates and immunoblot analyses of coprecipitated FA core complex components revealed that the FA core complex does not differ in its constituents in any of these phases, whereas FANCD2 monoubiquitination is at a maximum in late S/G2 (Fig. 3B and C).
FIG. 3.
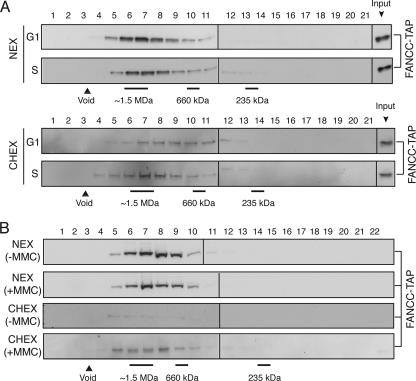
The FA core complex is constitutively assembled throughout the cell cycle. (A) Cells expressing the multitagged FA core complex were arrested at mitosis by nocodazole treatment and released into fresh medium. Synchronization was verified by FACS analysis. (B) At the indicated time points, whole-cell lysates of synchronized cell populations were subjected to TAP immunoprecipitation (IP), and the presence of FA core complex components was analyzed by immunoblotting. Δ_FANCL_ cell lysate was used as a negative IP control. Equal input was verified by immunoblotting for tubulin and FANCC-TAP. (C) FANCD2 immunoblot of total cell lysates from the indicated time points after nocodazole release. (D) Nocodazole-arrested cells were released into fresh medium supplemented with hydroxyurea. High-salt nuclear extracts (NEX) and soluble chromatin extracts (CHEX) of cells in G1 phase (2 h after release) and S phase (7 h after release) were used to perform TAP immunoprecipitations. (E) FANCD2 monoubiquitination was monitored in NEX and CHEX from the indicated cell cycle phases. (F) Coprecipitated proteins were identified by immunoblot analyses. Equal input was verified by immunoblotting for topoisomerase 2 (NEX) and histone H3 (CHEX). AS, asynchronous cells; G1, G1 phase synchronized; S, S phase synchronized; ΔL, Δ_FANCL_ cell extracts used as a negative IP control.
Another means by which the pathway can be activated is by exposure to DNA-damaging agents. Cells were released from nocodazole block in the presence of HU, an agent known to induce stalled replication forks during S phase (Fig. 3D). FANCC-TAP was immunoprecipitated from either high-salt nuclear and soluble chromatin extracts of G1-phase cells (when FANCD2-Ub is not detectable) or of S-phase cells (with maximal detectable FANCD2-Ub levels) (Fig. 3E). Immunoblot analysis showed no difference in the composition or abundance of FA core complex constituents (Fig. 3F). We did, however, notice that the FANCM protein resolves as two distinct bands as cells transit from late S to G2 and M phases (Fig. 3B) and that this protein in nuclear extracts is up-shifted to a slightly lower mobility band following HU treatment and S-phase entry (Fig. 3F). These studies show that FANCC-TAP is associated with most of the FA core complex components.
To determine whether FANCC-TAP complexes are indeed intact FA core complexes and not subcomplexes, we carried out size exclusion chromatography experiments. Indeed, a prominent portion of FANCC-TAP elutes at 1.5 MDa in samples prepared from nuclear and chromatin extracts obtained from both G1- and S-phase-transiting cells (Fig. 4A). We did, however, notice that the FANCC-TAP elution peak of G1 chromatin extracts showed a broader distribution, which may reflect a less stable FA core complex. In addition to the fractionation of synchronized cells, we also induced DNA damage in asynchronous cells by exposing them to MMC. Once again, we detected no obvious difference in the size of the FANCC-TAP complex in extracts prepared from both compartments following this treatment (Fig. 4B). However, the amount of FANCC-TAP on chromatin in the absence of MMC is very low and can therefore hardly be detected in 1.5-MDa fractions. In conclusion, a 1.5-MDa FA core complex is assembled regardless of DNA damage or cell cycle transitions.
FIG. 4.
The overall size of the FA core complex is not markedly affected during the cell cycle or after DNA damage. (A) Size exclusion chromatography of high-salt nuclear extracts (NEX) and soluble chromatin extracts (CHEX) of G1- and S-phase-synchronized cells expressing the multitagged FA core complex (Fig. 3) and immunoblot detection for FANCC-TAP. (B) Size exclusion chromatography of NEX and CHEX, either MMC treated (+MMC) or untreated (-MMC), and immunoblot detection for FANCC-TAP.
The FA core complex is stable and accumulates on chromatin in the absence of UBE2T.
Most gene products that are essential for the E3 ligase activity of the FA core complex are also integral components of this complex. One exception is the recently identified FANCI gene, which is not essential for the stability of the core complex but is required for FANCD2 monoubiquitination. However, it is unclear whether UBE2T, the E2-conjugating enzyme for FANCD2 monoubiquitination, is also required for the stability of the FA core complex and its chromatin accumulation after DNA damage. To address this, we disrupted the UBE2T gene in DT40 cells. The gene disruption construct is outlined in Fig. 5A and removes exons 4 to 6, including the active cysteine Cys86. Two independently derived Δ_UBE2T_ cell lines show hypersensitivity to cisplatin (Fig. 5B) and are defective in the monoubiquitination of FANCD2 (Fig. 5D). To determine whether the FA core complex is compromised in the absence of UBE2T, we fractionated cell extracts from wild-type and Δ_UBE2T_ cells and analyzed these fractions for the elution profile for FANCG. FANCG can be detected in the 1.5-MDa fractions in the Δ_UBE2T_ cell line, indicating that the FA core complex is intact and stable (Fig. 5C), like in wild-type cells. We then tested to see if UBE2T is required for the FA core complex or FANCD2 to accumulate on chromatin. Following exposure to MMC, wild-type cells demonstrate robust FANCD2 monoubiquitination and both FANCD2 and FANCG accumulate on chromatin (Fig. 5D). FANCD2 is not monoubiquitinated in Δ_UBE2T_ strains; however, FANCD2 and the FA core complex subunit FANCG still accumulate on chromatin following damage (Fig. 5D). In conclusion, UBE2T is essential for FANCD2 monoubiquitination but is not required for either FA core complex stability or accumulation on chromatin.
UBE2T accumulates constitutively on chromatin.
The FA core complex and UBE2T must form a transient holoenzyme complex on chromatin for FANCD2 monoubiquitination to occur. We therefore determined how UBE2T gains access to this compartment. When we tested nuclear and soluble chromatin extracts from cells exposed to MMC, we noted that UBE2T is constitutively present on chromatin (Fig. 5F). Moreover, we compared extracts from G1 to S phase for the presence of UBE2T and FANCD2. UBE2T always fractionated with chromatin, whereas monoubiquitinated FANCD2 accumulated on chromatin during the S phase only (Fig. 5E). Cumulatively, these studies indicate that UBE2T is predominantly localized on chromatin in DT40 cells independently of DNA damage or cell cycle transitions.
DISCUSSION
The main conclusion of this study is that a constitutively assembled FA core complex with an associated E3 ubiquitin ligase activity responds to DNA damage by migrating to chromatin. Here, the FA core complex likely interacts with UBE2T and FANCD2. These two proteins access chromatin independently of the FA core complex. An important implication of this work is that in chicken B cells the FA core complex E3 ubiquitin ligase activity is not regulated by an assembly/disassembly cycle of the FA core subunits. This is an unexpected conclusion, as studies on FA patient-derived cell lines led to the hypothesis that the sequential assembly of this complex underpins the regulation of FANCD2 monoubiquitination (17, 19). In addition, early studies indicated that some of the FA proteins were regulated in a cell cycle-dependent manner (10). However, we found that the assembled FA core complex is very stable. It would seem to be wasteful, from the point of view of a cell's energy, to disassemble such a complex when it is not needed, only to reassemble it later. Furthermore, many of the FA core complex proteins are of very low abundance in cells, suggesting that assembly of the complete FA core complex may be limited by the concentration of its constituents. An important caveat is that our studies in chicken DT40 cells may not apply to mammalian cells. Nevertheless, having concluded that the FA core complex is not regulated by an assembly process checkpoint, there are certain points of discussion raised by the presence of such a constitutive FA core complex. How does it respond to DNA damage and how does it get recruited to chromatin? What enables such a large complex to be retained or gain access to the nucleus after mitosis is completed? Finally, how stable is this complex, and does it turn over rapidly once activated or is it recycled?
The first point we discuss here is the model we have put forward explaining how the activity of the FA core complex might be restricted (Fig. 5G). Central to the model is that the E3 ubiquitin ligase, the FA core complex, and the substrates FANCD2/FANCI independently accumulate on chromatin during DNA replication or damage. The E2-conjugating enzyme UBE2T is constitutively present in this compartment. Hence, it can then form a transiently active E2/E3 holoenzyme for the monoubiquitination of FANCD2/FANCI. How the FA core complex and its substrate FANCD2/FANCI translocate to chromatin needs to be established. We have recently shown that the FANCM component of the FA core complex is required not only for the FA core complex to assemble but also for it to accumulate on chromatin (26). In addition, this protein has two domains that provide it with an ability to recognize DNA structures that occur at stalled DNA replication forks (13, 14). It has recently been shown that this interaction of FANCM with DNA structures is mediated by the FA core complex component FAAP24 that binds to the C-terminal part of FANCM (4). How the interaction is regulated is still an open question, but FANCM is also hyperphosphorylated upon DNA damage, and it is plausible that this modification alters its affinity or activity towards DNA (21). The manner in which FANCD2 gains access to the chromatin compartment deserves attention. In this study, we did find a requirement for a stable FA core complex for nonubiquitinated FANCD2 to accumulate on chromatin. However, it was clear that monoubiquitinated FANCD2 did strongly associate with chromatin. Biochemical studies suggest that FANCD2 is able to bind directly to DNA (30). In addition, this protein is phosphorylated by both ATR and ATM DNA checkpoint kinases, which are induced by different kinds of DNA lesions (1, 37). Since disruption of some of the phosphorylation sites weakens the ability of FANCD2 to be ubiquitinated and to accumulate in DNA damage-induced foci, it is therefore possible that these modifications are critical signals that allow the unmodified protein to transiently accumulate on chromatin. In addition, a recent study provides evidence that γ-H2AX at sites of DNA damage is essential for the accumulation of FANCD2 into foci (2). Future studies using single point mutants of key FA core complex proteins and FANCD2 should enable a dissection of how both components accumulate onto chromatin.
At mitosis, the nuclear envelope breaks down and essentially the strict demarcation between nucleus and cytoplasm ceases to exist. The FA core complex is a large complex and as such is present in low abundance in cells. A key problem raised by the current work is how a constitutively assembled complex gains access to a newly formed nucleus. It is very unlikely that nuclear import of FA core complexes that are dispersed in the cytoplasm occurs after mitosis. Because of the molecular size, ∼1.5 MDa, of this complex, it would seem beyond the capacity of the nuclear pore size to allow free diffusion of the FA core complex. One could imagine that the FA core complex is tethered to a structural component (such as nuclear lamin, lipid membrane, or indeed, condensed chromosomes) at mitosis; thus, when the nucleus reforms it is carried into the structure directly. This model is supported by recent structural insights into FANCE. This FA protein has a putative membrane-adsorbing amphipathic α-helix which is shared among some membrane-associated proteins and which is thought to recognize curved lipid membranes (7). Hence, FANCE might be the anchor for the FA core complex to remain associated with nuclear membrane subunits during mitosis. Another possibility is that natural loss of the FA core complex occurs, and only by some virtue of even distribution does it get captured into a new nucleus. If this were the case, a high turnover rate of core complex formation might be expected.
The ability to monitor a physiologically tagged FA core complex should enable us to establish its natural turnover. While we could not detect any major differences in the steady-state levels of the FA core complex during DNA replication or DNA damage, this does not give us much idea regarding the rate of turnover. It is important to note that the work presented here does not deny the existence of subcomplexes. Such subcomplexes have been detected for certain pairs of FA proteins and could be intermediates of a subsequent assembly (17, 19). Another possibility is that these subcomplexes might confer unique and distinct biochemical functions. Since ablating any one of the many FA core complex proteins severely compromises the complex stability, as such, as well that of individual FA proteins, it remains possible that activated FA core complexes may be less stable. Moreover, as more components of the FA core complex are identified, it will be possible to test their contribution to stability by gene ablation in the multiple-tagged FA core complex cell line. Future studies to determine the precise half-lives and turnover rates of the FA core complex and complex components may lead to important insights into the function of the FA core complex.
Acknowledgments
A.A. was supported by the Leukemia Research Fund and F.L. by the Children with Leukemia Fund.
We are grateful to Paul Pace and other members of K. J. Patel's laboratory for critical comments and technical advice.
Footnotes
▿
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Andreassen, P. R., A. D. D'Andrea, and T. Taniguchi. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18**:**1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogliolo, M., A. Lyakhovich, E. Callen, M. Castella, E. Cappelli, M. J. Ramirez, A. Creus, R. Marcos, R. Kalb, K. Neveling, D. Schindler, and J. Surralles. 2007. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 26**:**1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridge, W. L., C. J. Vandenberg, R. J. Franklin, and K. Hiom. 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37**:**953-957. [DOI] [PubMed] [Google Scholar]
- 4.Ciccia, A., C. Ling, R. Coulthard, Z. Yan, Y. Xue, A. R. Meetei, H. el Laghmani, H. Joenje, N. McDonald, J. P. de Winter, W. Wang, and S. C. West. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25**:**331-343. [DOI] [PubMed] [Google Scholar]
- 5.de Winter, J. P., L. van der Weel, J. de Groot, S. Stone, Q. Waisfisz, F. Arwert, R. J. Scheper, F. A. Kruyt, M. E. Hoatlin, and H. Joenje. 2000. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum. Mol. Genet. 9**:**2665-2674. [DOI] [PubMed] [Google Scholar]
- 6.Dorsman, J. C., M. Levitus, D. Rockx, M. A. Rooimans, A. B. Oostra, A. Haitjema, S. T. Bakker, J. Steltenpool, D. Schuler, S. Mohan, D. Schindler, F. Arwert, G. Pals, C. G. Mathew, Q. Waisfisz, J. P. de Winter, and H. Joenje. 2007. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 29**:**211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drin, G., J. F. Casella, R. Gautier, T. Boehmer, T. U. Schwartz, and B. Antonny. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14**:**138-146. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7**:**249-262. [DOI] [PubMed] [Google Scholar]
- 9.Gurtan, A. M., P. Stuckert, and A. D. D'Andrea. 2006. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J. Biol. Chem. 281**:**10896-10905. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich, M. C., K. V. Silvey, S. Stone, A. J. Zigler, D. J. Griffith, M. Montalto, L. Chai, Y. Zhi, and M. E. Hoatlin. 2000. Posttranscriptional cell cycle-dependent regulation of human FANCC expression. Blood 95**:**3970-3977. [PubMed] [Google Scholar]
- 11.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. A. Fox, and A. D. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297**:**606-609. [DOI] [PubMed] [Google Scholar]
- 12.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2**:**446-457. [DOI] [PubMed] [Google Scholar]
- 13.Komori, K., R. Fujikane, H. Shinagawa, and Y. Ishino. 2002. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 77**:**227-241. [DOI] [PubMed] [Google Scholar]
- 14.Komori, K., M. Hidaka, T. Horiuchi, R. Fujikane, H. Shinagawa, and Y. Ishino. 2004. Cooperation of the N-terminal helicase and C-terminal endonuclease activities of archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 279**:**53175-53185. [DOI] [PubMed] [Google Scholar]
- 15.Leveille, F., M. Ferrer, A. L. Medhurst, H. el Laghmani, M. A. Rooimans, P. Bier, J. Steltenpool, T. A. Titus, J. H. Postlethwait, M. E. Hoatlin, H. Joenje, and J. P. de Winter. 2006. The nuclear accumulation of the Fanconi anemia protein FANCE depends on FANCC. DNA Repair (Amsterdam) 5**:**556-565. [DOI] [PubMed] [Google Scholar]
- 16.Levitus, M., Q. Waisfisz, B. C. Godthelp, Y. de Vries, S. Hussain, W. W. Wiegant, E. Elghalbzouri-Maghrani, J. Steltenpool, M. A. Rooimans, G. Pals, F. Arwert, C. G. Mathew, M. Z. Zdzienicka, K. Hiom, J. P. De Winter, and H. Joenje. 2005. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37**:**934-935. [DOI] [PubMed] [Google Scholar]
- 17.Ling, C., M. Ishiai, A. M. Ali, A. L. Medhurst, K. Neveling, R. Kalb, Z. Yan, Y. Xue, A. B. Oostra, A. D. Auerbach, M. E. Hoatlin, D. Schindler, H. Joenje, J. P. de Winter, M. Takata, A. R. Meetei, and W. Wang. 2007. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 26**:**2104-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machida, Y. J., Y. Machida, Y. Chen, A. M. Gurtan, G. M. Kupfer, A. D. D'Andrea, and A. Dutta. 2006. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23**:**589-596. [DOI] [PubMed] [Google Scholar]
- 19.Medhurst, A. L., H. el Laghmani, J. Steltenpool, M. Ferrer, C. Fontaine, J. de Groot, M. A. Rooimans, R. J. Scheper, A. R. Meetei, W. Wang, H. Joenje, and J. P. de Winter. 2006. Evidence for subcomplexes in the Fanconi anemia pathway. Blood 108**:**2072-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. van de Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35**:**165-170. [DOI] [PubMed] [Google Scholar]
- 21.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37**:**958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23**:**3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi, J., and G. M. Kupfer. 2005. The Fanconi anemia core complex associates with chromatin during S phase. Blood 105**:**759-766. [DOI] [PubMed] [Google Scholar]
- 24.Mi, J., F. Qiao, J. B. Wilson, A. A. High, M. J. Schroeder, P. T. Stukenberg, A. Moss, J. Shabanowitz, D. F. Hunt, N. J. Jones, and G. M. Kupfer. 2004. FANCG is phosphorylated at serines 383 and 387 during mitosis. Mol. Cell. Biol. 24**:**8576-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montes de Oca, R., P. R. Andreassen, S. P. Margossian, R. C. Gregory, T. Taniguchi, X. Wang, S. Houghtaling, M. Grompe, and A. D. D'Andrea. 2005. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105**:**1003-1009. [DOI] [PubMed] [Google Scholar]
- 26.Mosedale, G., W. Niedzwiedz, A. Alpi, F. Perrina, J. B. Pereira-Leal, M. Johnson, F. Langevin, P. Pace, and K. J. Patel. 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 12**:**763-771. [DOI] [PubMed] [Google Scholar]
- 27.Niedernhofer, L. J., A. S. Lalai, and J. H. Hoeijmakers. 2005. Fanconi anemia (cross)linked to DNA repair. Cell 123**:**1191-1198. [DOI] [PubMed] [Google Scholar]
- 28.Niedzwiedz, W., G. Mosedale, M. Johnson, C. Y. Ong, P. Pace, and K. J. Patel. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15**:**607-620. [DOI] [PubMed] [Google Scholar]
- 29.Pace, P., M. Johnson, W. M. Tan, G. Mosedale, C. Sng, M. Hoatlin, J. de Winter, H. Joenje, F. Gergely, and K. J. Patel. 2002. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21**:**3414-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, W. H., S. Margossian, A. A. Horwitz, A. M. Simons, A. D. D'Andrea, and J. D. Parvin. 2005. Direct DNA binding activity of the Fanconi anemia D2 protein. J. Biol. Chem. 280**:**23593-23598. [DOI] [PubMed] [Google Scholar]
- 31.Qiao, F., J. Mi, J. B. Wilson, G. Zhi, N. R. Bucheimer, N. J. Jones, and G. M. Kupfer. 2004. Phosphorylation of fanconi anemia (FA) complementation group G protein, FANCG, at serine 7 is important for function of the FA pathway. J. Biol. Chem. 279**:**46035-46045. [DOI] [PubMed] [Google Scholar]
- 32.Qiao, F., A. Moss, and G. M. Kupfer. 2001. Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 276**:**23391-23396. [DOI] [PubMed] [Google Scholar]
- 33.Rahman, N., S. Seal, D. Thompson, P. Kelly, A. Renwick, A. Elliott, S. Reid, K. Spanova, R. Barfoot, T. Chagtai, H. Jayatilake, L. McGuffog, S. Hanks, D. G. Evans, D. Eccles, D. F. Easton, and M. R. Stratton. 2007. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39**:**165-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid, S., D. Schindler, H. Hanenberg, K. Barker, S. Hanks, R. Kalb, K. Neveling, P. Kelly, S. Seal, M. Freund, M. Wurm, S. D. Batish, F. P. Lach, S. Yetgin, H. Neitzel, H. Ariffin, M. Tischkowitz, C. G. Mathew, A. D. Auerbach, and N. Rahman. 2007. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 39**:**162-164. [DOI] [PubMed] [Google Scholar]
- 35.Sims, A. E., E. Spiteri, R. J. Sims III, A. G. Arita, F. P. Lach, T. Landers, M. Wurm, M. Freund, K. Neveling, H. Hanenberg, A. D. Auerbach, and T. T. Huang. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14**:**564-567. [DOI] [PubMed] [Google Scholar]
- 36.Smogorzewska, A., S. Matsuoka, P. Vinciguerra, E. R. McDonald III, K. E. Hurov, J. Luo, B. A. Ballif, S. P. Gygi, K. Hofmann, A. D. D'Andrea, and S. J. Elledge. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129**:**289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109**:**459-472. [DOI] [PubMed] [Google Scholar]
- 38.Timmers, C., T. Taniguchi, J. Hejna, C. Reifsteck, L. Lucas, D. Bruun, M. Thayer, B. Cox, S. Olson, A. D. D'Andrea, R. Moses, and M. Grompe. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7**:**241-248. [DOI] [PubMed] [Google Scholar]
- 39.Wang, X., R. D. Kennedy, K. Ray, P. Stuckert, T. Ellenberger, and A. D. D'Andrea. 2007. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 27**:**3098-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]