A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis (original) (raw)
Abstract
Here, we demonstrate a role for the mitochondrial NAD-dependent deacetylase Sirt3 in the maintenance of basal ATP levels and as a regulator of mitochondrial electron transport. We note that Sirt3−/− mouse embryonic fibroblasts have a reduction in basal ATP levels. Reconstitution with wild-type but not a deacetylase-deficient form of Sirt3 restored ATP levels in these cells. Furthermore in wild-type mice, the resting level of ATP correlates with organ-specific Sirt3 protein expression. Remarkably, in mice lacking Sirt3, basal levels of ATP in the heart, kidney, and liver were reduced >50%. We further demonstrate that mitochondrial protein acetylation is markedly elevated in Sirt3−/− tissues. In addition, in the absence of Sirt3, multiple components of Complex I of the electron transport chain demonstrate increased acetylation. Sirt3 can also physically interact with at least one of the known subunits of Complex I, the 39-kDa protein NDUFA9. Functional studies demonstrate that mitochondria from Sirt3−/− animals display a selective inhibition of Complex I activity. Furthermore, incubation of exogenous Sirt3 with mitochondria can augment Complex I activity. These results implicate protein acetylation as an important regulator of Complex I activity and demonstrate that Sirt3 functions in vivo to regulate and maintain basal ATP levels.
Keywords: acetylation, sirtuins, complex I, electron transport
The sirtuins are a conserved family of proteins possessing NAD-dependent deacetylase activity. In yeast, Sir2 is involved in transcriptional silencing and acts as a regulator of life span (1, 2). Remarkably, increased dosage of Sir2 extends the life span of yeast as well as several other simple organisms (3, 4). Mammals have at least seven different Sir2 homologs, with mammalian Sirt1 being the closest structural relative of yeast Sir2. A number of Sirt1 protein deacetylase substrates have been previously identified including p53 and the Foxo family of transcription factors (5). Considerably less is known about the other sirtuin family members, three of which, Sirt3,Sirt4, and Sirt5 appear to localize primarily to the mitochondria (6–9). Evidence suggests that Sirt4 appears to function predominantly as an ADP-ribosyltransferase. One target for Sirt4 enzymatic activity appears to be glutamate dehydrogenase (GDH) (10). Consistent with a complex interaction between mitochondrial sirtuins, GDH also appears to be a deacetylase target for Sirt3 (11). In addition, Sirt3-dependent deacetylation regulates the activity of acetyl-CoA synthetase 2 (AceCS2) an important mitochondrial enzyme involved in generating acetyl-CoA for the tricarboxylic acid (TCA) cycle (12, 13).
A recent study demonstrated that mice deficient in Sirt3 are viable and metabolically unremarkable. In particular, these animals exhibit normal body weight as well as normal body fat composition, oxygen consumption, respiratory exchange ratios, and activity levels (11). Similarly, Sirt3−/− mice appeared outwardly healthy under both normal conditions and after mild stresses such as a 24-h period of starvation. In addition, although a previous report in a cell-culture model had implicated Sirt3 in adaptive thermogenesis (9), this physiological function was not apparently altered in Sirt3−/− mice (11).
A proteomic survey of intracellular proteins that had internal acetylation residues demonstrated that a disproportionate fraction of identified proteins were in the mitochondria and/or associated with energy metabolism (14). This has led to the notion that protein acetylation may represent an important mechanism to regulate overall mitochondrial function. Indeed, given the mechanism of action of the sirtuin family, levels of mitochondrial protein acetylation would seemingly be influenced and regulated by NAD+, a key mitochondrial energetic intermediate. Nonetheless, the reported unremarkable phenotype of Sirt3-deficient mice challenges the physiological importance of NAD-dependent deacetylation in regulating energy homeostasis. Here, using an independently generated model of Sirt3−/− mice, we provide evidence for Sirt3-dependent regulation of global mitochondrial function. In particular, we demonstrate that the absence of Sirt3 results in a marked alteration of basal in vivo ATP levels. Furthermore, we demonstrate that Sirt3 can reversibly bind to and regulate the acetylation and activity of Complex I of the electron-transport chain.
Results
The Sirt3 locus was inactivated by homologous recombination in ES cells leading to the deletion of exons 2–4 [supporting information (SI) Fig. S1]. Analysis of the tissues of homozygously deleted mice revealed undetectable levels of Sirt3 protein expression (Fig. S1). Consistent with a recent report (11), our own extensive survey of multiple organs by both gross and microscopic pathology revealed that Sirt3−/− mice appeared indistinguishable from wild-type littermates. In addition, we observed no increased or decreased mortality of these mice during the first year of life (H.-S.K. and C.-X.D., unpublished observations).
In an effort to more fully understand the role of Sirt3 in mitochondrial biology, Sirt3+/− mice were bred to generate multiple independent cultures of wild-type or Sirt3−/− mouse embryonic fibroblasts (MEFs). Given the central role of mitochondria in generating ATP via the electron-transport chain (ETC), we first asked whether deletion of Sirt3 altered basal ATP levels. We measured ATP levels in five independent lines of wild-type MEFs and a similar number of independent Sirt3−/− MEFs (Fig. 1A). These analysis demonstrated that, on average, Sirt3−/− MEFs had an ≈30% reduction in resting ATP levels (P < 0.01). We next asked whether transient reconstitution of Sirt3 could restore this ATP deficit. Sirt3−/− MEFs were transiently transfected with an empty vector, wild-type Sirt3, or a deacetylase-inactive form of Sirt3, Sirt3(HY). All transfections were performed with a cotransfected GFP expression vector to allow for subsequent purification of transfected cells using FACS sorting. As noted in Fig. 1B, expression of wild-type Sirt3 in the background of Sirt3−/− MEFs markedly increased basal ATP, whereas the deacetylase inactive mutant was ineffective in raising ATP levels. Similar results were obtained with transient expression of wild-type and deacetylase-inactive Sirt3 in HeLa cells that contain endogenous Sirt3 (Fig. 1C).
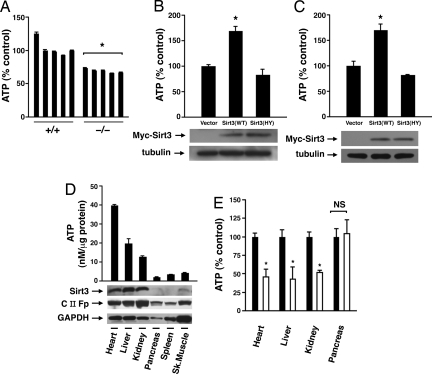
Fig. 1.
Sirt3 regulates basal ATP levels. (A) Levels of basal ATP in five independent isolates of primary wild-type MEFs and a similar number of independent Sirt3−/− MEF cell isolates. (B) Sirt3−/− MEFs were transfected with an expression vector encoding GFP along with an empty vector, epitope-tagged wild type, or a deacetylase inactive (HY) Sirt3. Thirty-six hours after transfection, GFP-positive cells were sorted by FACS and ATP determined. ATP levels are expressed relative to vector-transfected Sirt3−/− cells. Shown is the average of three independent experiments each performed in triplicate. (C) HeLa cells were transfected with an empty vector, epitope-tagged wild type, or Sirt3(HY) and levels of ATP determined 48 h after transfection. Shown is the mean ± SD of four independent experiments. (D) Absolute level of ATP in various tissues and organs of wild-type mice (n = 3; mean ± SD). Shown is the corresponding level of expressed Sirt3 within each tissue as well as the 70-kDa complex II-associated protein Fp (CII-Fp) as a general measure of mitochondrial number and GAPDH for protein loading. (E) Normalized levels of ATP in wild-type (black bars) and Sirt3−/− mice (white bars) in various organs with known high Sirt3 expression (heart, liver, and kidney) as well as an organ (pancreas) with low or absent endogenous Sirt3 expression (n = 4 animals per group). *, P < 0.01.
Based on these observations, we asked whether Sirt3 might also regulate ATP in vivo. We first took advantage of the known variations in resting ATP levels in various tissue and organ beds. In particular, we asked whether endogenous Sirt3 expression correlates with resting ATP levels in wild-type mice. As noted in Fig. 1D, resting ATP varied 10-fold in various distinct organ beds with lower levels in pancreas, spleen, and skeletal muscle and higher levels in heart, liver, and kidney. These resting levels of ATP appeared to roughly, albeit not perfectly, correlate with the degree of Sirt3 expression in these organs. We then asked whether the absence of Sirt3 could regulate organ-specific ATP levels. As noted in Fig. 1E, analysis of matched pairs of wild-type and Sirt3−/− mice demonstrated that in organs that normally express high amounts of Sirt3, the absence of this mitochondrial deacetylase results in a >50% reduction in resting ATP. These differences in observed ATP between wild-type and Sirt3−/− tissues could not be ascribed to obvious differences in the level of expression for various components of the mitochondrial electron transport (Fig. S2). In contrast to what we observed in tissues expressing high levels of Sirt3, in organs such as the pancreas, where we noted that normally there is little to no Sirt3 expression, levels of ATP were not appreciably different between wild-type and Sirt3−/− mice (Fig. 1E). We therefore conclude that, consistent with the overall appearance of the Sirt3−/− mice, the absence of Sirt3 does not produce a generalized energy deficiency but rather that Sirt3 is important in helping set and maintain tissue-specific levels of ATP.
We next asked whether these differences in ATP might result, at least in part, to an alteration in ETC function when Sirt3 is absent. To begin to address this hypothesis, we purified mitochondria from the livers of wild-type or Sirt3−/− mice. As demonstrated in Fig. 2A, biochemical analysis of hepatic mitochondria revealed a marked change in the protein acetylation of Sirt3−/− mice that involved multiple proteins. Similar results have been recently reported (11). To more specifically pursue alterations in the ETC, we took advantage of previously described methods that allow for the direct immunopurification of intact multisubunit ETC complexes (15, 16). In mice, Complex I is composed of >40 separate proteins that together function as an NADH dehydrogenase. Equal amounts of liver protein lysate obtained from either wild-type or Sirt3−/− mice was then used to immunocapture intact Complex I. These proteins were subsequently resolved on an SDS/PAGE and acetylation detected by using an antibody that recognizes internal acetyl-lysine residues. As seen in Fig. 2B, the absence of Sirt3 results in an increase in the acetylation of numerous proteins associated with Complex I. In contrast, similar analysis with immunocaptured Complex II revealed no substantial difference in protein acetylation between mitochondria isolated from the livers of wild-type or Sirt3−/− mice (Fig. 2C).
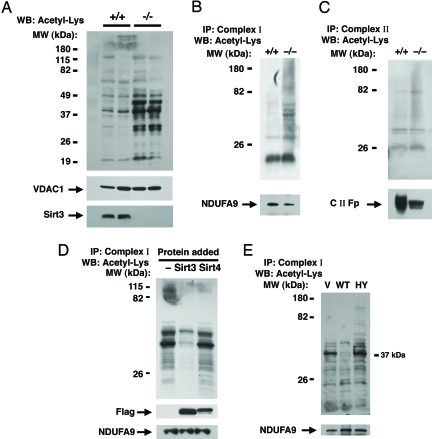
Fig. 2.
Sirt3 regulates mitochondrial protein acetylation. (A) Western blot (WB) analysis for internal acetyl-lysine residues using total liver mitochondrial protein extracts from two age-matched wild-type (+/+) or Sirt3−/− mice. The mitochondrial protein VDAC1 is used as a loading control, and Sirt3 expression is also shown. (B) Levels of acetylation from immunocaptured hepatic Complex I in a wild-type versus Sirt3−/− mouse. Subunit 9 (NDUFA9) of Complex I was used as a loading control for the immunocapture. (C) Similar acetylation analysis for immunocaptured Complex II. The 70-kDa Fp subunit of Complex II (C II-Fp) was used as a loading control. (D) Sirt3 deacetylates Complex I in vitro. Complex I was isolated from nicotinamide-treated HeLa cells and incubated for 2 hours in vitro with deacteylase reaction buffer only (−) or with reaction buffer containing purified Sirt3 or Sirt4. The level of the Complex I protein NDUFA9 is shown as a loading control as is the level of exogenously added Flag-tagged Sirt3 and Sirt4. (E) Level of in vivo Complex I acetylation in HeLa cells transfected with empty vector (V), wild-type Sirt3 (WT), or a deacetylase inactive form of Sirt3 (HY). IP, immunoprecipitation; MW, molecular mass.
Consistent with a role for mitochondrial sirtuins in regulating the ETC, treatment of HeLa cells with the broad-spectrum sirtuin inhibitor, nicotinamide, led to markedly enhanced Complex I acetylation (Fig. S3). In general, under basal conditions, the level of Complex I acetylation was higher in cultured human HeLa cells compared with isolated mouse liver mitochondria. We next asked whether Sirt3 could directly deacetylate Complex I. Nicotinamide-treated HeLa cells were used as a source to immunocapture Complex I that was then exposed for 2 h to either recombinant Sirt3 or Sirt4. Compared with samples exposed to Sirt4 or maintained in deacetylase buffer alone, Complex I incubated in the presence of exogenous Sirt3 protein demonstrated marked deacetylation of numerous proteins (Fig. 2D). This result is consistent with both a direct effect of Sirt3 on Complex I and with a previous result demonstrating that Sirt4−/− mitochondria do not have significant alterations in mitochondrial protein acetylation (11). Similar to what we observed in vitro, transient overexpression in HeLa cells of wild-type Sirt3 resulted in decreased acetylation of multiple proteins comprising Complex I (Fig. 2E). In contrast, expression of a deacetylase inactive form of Sirt3 [Sirt3(HY)] resulted in a modest increase in acetylation. This is consistent with other reports demonstrating a dominant-negative effect of such inactivating mutations (9, 17, 18).
Given that Sirt3 appears to preferentially regulate Complex I acetylation, we asked whether there might be a physical association between the deacetylase and the ETC. Using equal amounts of HeLa cell protein lysate, we immunocaptured Complex I or Complex II. These immunocaptured ETC complexes were then probed for associated Sirt3. As noted in Fig. 3A, endogenous Sirt3 protein clearly associated with the immunoprecipitated Complex I, whereas a similar association was not detected with Complex II. It should be noted that, in contrast to mouse Sirt3, human Sirt3 exists in two forms, a short form (equivalent to mouse Sirt3) and a longer form containing a 142-aa human-specific N-terminal extension (7). Our data would suggest that in HeLa cells, the association between Complex I and Sirt3 involved both the long and short form of human Sirt3. To begin to understand the dynamics of this association, we purified Complex I from HeLa cells exposed to various stresses. As noted in Fig. 3B, Sirt3 was still associated with Complex I after either a 2- or 6-h period of nutrient withdrawal (no serum or glucose). In contrast, brief treatment with hydrogen peroxide or with the Complex I inhibitor rotenone led to the apparent dissociation of the Sirt3–Complex I interaction. These observations suggest that Sirt3−/− cells might have different sensitivities to either rotenone or hydrogen peroxide. Consistent with this notion, treatment with the Complex I inhibitor rotenone, as expected, produced a significant fall in ATP levels in wild-type MEFs (Fig. 3C). In contrast, as previously noted, although basal levels of ATP were reduced ≈30% in Sirt3−/− MEFs, the addition of rotenone had relatively little effect on these Sirt3-deficient cells. A similar differential sensitivity in the reduction of ATP levels was also observed after treatment with hydrogen peroxide. In contrast, treatment with the mitochondrial poison cyanide, a cytochrome c oxidase (Complex IV) inhibitor, produced a similar magnitude reduction in ATP levels in wild-type and Sirt3−/− MEFs.
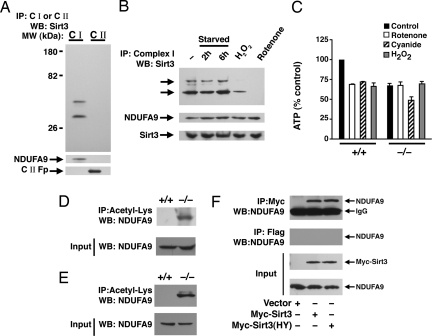
Fig. 3.
Sirt3 associates with Complex I of the ETC. (A) Sirt3 associates with Complex I. Equal amounts of HeLa cell lysate was used to immunocapture either Complex I or Complex II. These ETC components were then resolved on SDS/PAGE and probed for association with Sirt3. Both the short and long form of human Sirt3 associates with Complex I. The purity of the immunocapture complexes are demonstrated by probing the stripped blot for the Complex I component NDUFA9 and the 70-kDa Fp subunit of Complex II. (B) Reversible association of endogenous Sirt3 with Complex I. Immunocaptured Complex I was probed for associated Sirt3 under fed conditions (−), after 2 or 6 h of starvation, after hydrogen peroxide (0.5 mM, 30 min) treatment, or after rotenone (10 μM, 30 min) treatment. The arrows indicate the short and long form of human Sirt3. Below, total levels of Sirt3 or the Complex I component NDUFA9 were assessed for each condition by using 30 μg of mitochondrial protein lysate. (C) ATP levels in wild-type (+/+) or Sirt3−/− MEFs under basal conditions (black bars), or after a 30-min exposure to rotenone (50 μM; white bars), cyanide (20 μM; hatched bars), or hydrogen peroxide (0.5 mM; gray bars). Basal levels of ATP are reduced in the Sirt3−/− cells and are relatively resistant to rotenone or hydrogen peroxide challenge but exhibit normal ATP sensitivity to cyanide. (D and E) Levels of acetylation of NDUFA9 in MEFs (D) or liver protein lysates from wild type (+/+) or Sirt3−/− mice (E). (F) HeLa cells were transfected with a myc-tagged empty vector, wild-type Sirt3, or a deacetylase-inactive Sirt3(HY) and equal amounts of lysate immunoprecipitated with a myc-epitope antibody or an irrelevant Flag-epitope antibody. Sirt3 immunoprecipitation reveals the presence of coprecipitated endogenous NDUFA9. IP, immunoprecipitation; WB, Western blotting.
To further assess the association between Sirt3 and Complex I, we noted that whereas the acetylation of multiple proteins in Complex I were effected by the absence or overexpression of Sirt3, in HeLa cells, a protein of ≈39 kDa appeared to be affected in the most dramatic fashion. To pursue the identity of this protein, we relied on a recent general proteomic survey of acetylated proteins that identified 10 of the ≈40 subunits of Complex I as having one or more acetylation sites (14). One previously identified acetylated component of Complex I with an estimated molecular mass of 39 kDa is subunit 9 (NDUFA9), in which lysine 370 is acetylated (14). To ascertain whether acetylation of NDUFA9 was indeed regulated by Sirt3, we immunoprecipitated all acetylated protein from either MEFs or liver protein lysates and subsequently analyzed these immunoprecipitated proteins for NDUFA9. As evident in Fig. 3 D and E, both in vitro and in vivo, the absence of Sirt3 resulted in a marked increase in NDUFA9 acetylation. Similarly, treatment of cells with nicotinamide resulted in increased NDUFA9 acetylation, whereas in HeLa cells, overexpression of wild type but not Sirt3(HY) reduced NDUFA9 acetylation (Fig. S4). These data support the notion that NDUFA9 is one of several components of Complex I, whose acetylation is regulated by Sirt3. Although, unfortunately, the available antibodies for NDUFA9 and Sirt3 were not suitable for immunoprecipitation, after transfection of an epitope-tagged Sirt3, we could also demonstrate a direct interaction between Sirt3 and endogenous NDUFA9 (Fig. 3F).
Because Sirt3 appeared to selectively regulate the acetylation of Complex I with little or no effect on Complex II, we next sought to functionally test whether the absence of Sirt3 could lead to selective alterations in ETC function. Mitochondria were isolated from age-matched wild-type and Sirt3−/− mice, and respiration was determined by using Complex I- and Complex II-dependent substrates. As noted in Fig. 4 A and B, the use of the Complex I-dependent substrates glutamate and malate and the absence of Sirt3 results in a noticeable decrease in State 3-dependent respiration. Analysis of multiple wild-type and Sirt3−/− littermates revealed an ≈20% reduction in measured Complex I activity, with no corresponding change in Complex II activity (Fig. 4 C and D). Respiratory control ratios and uncoupled respirations were similar between wild-type and Sirt3−/− mitochondria (Table S1). Next, in a fashion analogous to our previous experiments where we had demonstrated efficient in vitro deacetylation (see Fig. 2D), we sought to assess the direct effects of Sirt3 on Complex I activity by incubating permeabilized HeLa cell mitochondria with purified sirtuin proteins. In this case, after mitochondria were exposed to purified sirtuin proteins, we assessed the level of rotenone-sensitive NADH oxidation, a direct measurement of Complex I activity. Although Complex I activity was roughly similar in vector-treated samples and Sirt4-treated mitochondria, incubation of HeLa cell mitochondria with exogenous Sirt3 led to increased Complex I activity (Fig. 4E and Fig. S5).
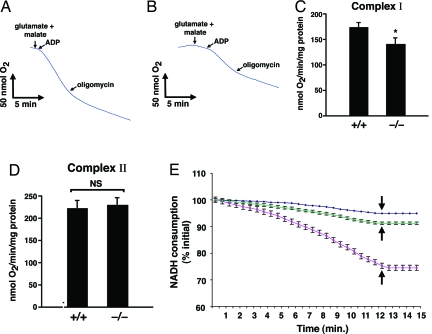
Fig. 4.
The absence of Sirt3 selectively affects Complex I activity. (A and B) Representative rate of oxygen consumption using Complex I-dependent substrates for liver mitochondria obtained from a wild-type (A) or Sirt3−/− (B) mouse. (C) Calculated State 3 respiration rate for Complex I from intact liver mitochondria of four wild-type and four Sirt3−/− mice (mean ± SD; *, P < 0.02). (D) Calculated State 3 respiration rate for Complex II-dependent substrate (succinate + rotenone) in wild-type and knockout mice. NS, not significant. (E) Rates of NADH consumption for purified HeLa cell mitochondria after in vitro incubation with deacetylase buffer containing purified Flag-Sirt4 (green), Flag-Sirt3 (red), or Flag-vector (purple). Normalized NADH absorbance was monitored at 340 nm. The arrow indicates the time of addition of rotenone (4 μM final concentration). Shown is the mean rate (±SD) of NADH consumption of triplicate determinations from one of two similar experiments.
Discussion
In summary, we demonstrate that Sirt3 is an important regulator of basal ATP and, hence, overall energy homeostasis. In tissues such as the liver, heart, and kidney that normally express high levels of Sirt3, the absence of the deacetylase leads to marked reduction in ATP. Surprisingly, we observed no outwardly observable untoward effects of this marked energy reduction. A recent study also concluded that Sirt3−/− mice appear to have normal weight, body fat, oxygen consumption, activity levels, and food consumption (11). These normal general metabolic parameters stand in contrast to the significant reduction in ATP we observed within certain Sirt3−/− tissues. It should be noted, however, that in organs such as the heart, concentrations of ATP are normally in the millimolar range (19). As such, even with a 50% reduction in ATP concentration, levels of ATP would be substantially above the k m of almost all intracellular enzymes. It will, however, be interesting to test in future experiments whether the function of Sirt3−/− mice are impaired under certain stress conditions, especially those conditions that might result in increased energetic demand or reduced nutrient or energy supply.
Based on reconstitution with either wild type or the deacetylase-inactive mutant of Sirt3, our data would suggest that the deacetylase function of Sirt3 is necessary for maintaining ATP levels. Given previous observations demonstrating that Sirt3 deacetylates GDH and AceCS2 (11–13) and our observations here regarding Complex I, the effects of Sirt3 on ATP levels are undoubtedly complex and presumably represent the coordinated regulation of both substrate flow to the mitochondria and the ETC itself. In this regard, the data presented here (Fig. 2A) as well as elsewhere (11, 14) suggest that the list of Sirt3-dependent mitochondrial targets undoubtedly will expand. Nonetheless, our results are supportive of the notion that Complex I activity can be regulated by acetylation and deacetylation, a posttranslational modification that heretofore had no known role in ETC function.
Our results also suggest that the association of Sirt3 with Complex I is reversible and that either the Complex I inhibitor rotenone or hydrogen peroxide exposure can lead to the apparent release of Sirt3 from the ETC. Interestingly, strengthening the connection between Sirt3 binding to the ETC and ATP homeostasis, treatment with rotenone or hydrogen peroxide significantly reduces ATP in wild-type, but not Sirt3−/−, MEFs. In contrast, treatment with the Complex IV inhibitor cyanide resulted in nearly equivalent reduction in ATP levels in wild-type and Sirt3−/− MEFs. These observations are consistent with our other data demonstrating that the association of Sirt3 with Complex I participates in setting and maintaining optimal ATP levels. These results also raise the hypothesis that Sirt3 might protect the respiratory chain from oxidative stress and that ETC function after hydrogen peroxide might be different in wild-type and Sirt3−/− MEFs. Surprisingly, however, we observed that total whole-cell oxygen consumption fell to a similar degree in both wild-type and Sirt3−/− MEFs after hydrogen peroxide exposure (Fig. S6). Similarly, basal levels of total glutathione before and after hydrogen peroxide were similar in wild-type and Sirt3−/− MEFs (Fig. S7). Thus, we conclude that, under the conditions used, the absence of Sirt3 does not appear to alter the inhibition of mitochondrial oxygen consumption that occurs after hydrogen peroxide exposure (20–22). Additional experiments with extended ranges of concentrations of hydrogen peroxide and detailed kinetics of recovery to more formally test the role of Sirt3 in the cellular response to oxidative stress await completion.
Finally, it is of interest to note that Complex I is an NADH dehydrogenase and that Sirt3 is, in turn, an NAD-dependent and -regulated deacetylase. The direct physical association of Sirt3 with Complex I suggests that Sirt3 might act as a rheostat for the ETC, using NAD generated from Complex I activity to, in turn, modulate overall energy homeostasis (Fig. S8). This is also supported by our observation that the interaction of Sirt3 with Complex I is sensitive to the inhibitor rotenone. Given that Sirt3 can also regulate AceCS2 function and, hence, regulate the synthesis of acetyl CoA, Sirt3 is uniquely poised to sense and/or respond to two separate intermediates, NAD and acetyl CoA, that are, in turn, critical outputs of mitochondrial metabolism. Given that the absence of Sirt3 leads to a dramatic increase in the acetylation of numerous mitochondrial proteins and that this is accompanied by significant alterations in basal ATP levels, our results suggest that reversible protein acetylation might represents an underappreciated but fundamental mechanism to regulate overall mitochondrial activity.
Materials and Methods
Generation of Sirt3 Knockout Animals.
To construct the targeting vector, we isolated recombinant phages containing overlapping regions of genomic DNA comprising the Sirt3 locus from a 129SVEV-mouse library (23). A 4.5-kb NotI-XhoI fragment originating 5′ to exon 2 of Sirt3 was subcloned into the NotI and XhoI sites of ploxPneo1 (24). The resulting construct was cleaved with EcoR1, allowing for the insertion of a 4.4-kb EcoR1 fragment that originates 3′ to exon 4. A single loxP site was inserted into the NotI-XhoI fragment at its Nsi1 site located in intron 1 before formation of the final targeting construct designated ploxPneoSirt3. TC1 ES cells were then transfected with NotI-linearized pLoxPneoSirt3 and selected with G418 and FIAU (25). Of the 113 clones analyzed, 6 were subsequently confirmed positive by Southern blot analysis. In brief, Southern blot analysis involved digestion of genomic DNA with EcoRV and subsequent hybridization with a 3′ flanking probe. Wild-type allele generated an ≈20-kb band, whereas the recombined allele generated a 5-kb band.
Chimeric mice were mated with NIH Black Swiss females (Taconic) to screen for germ-line transmission. Male mice bearing germ-line transmission were subsequently mated with female FVB EII-Cre mice (26) to generate complete deletion of exons 2–4 of Sirt3. The animals were genotyped by either Southern blot analysis or by PCR analysis using the following primers: F1; 5′-gagatccatcagcttctgtg, R1; 3′-ccctcaatcacaaatgtcgg, F2; 5′-gggagcactctcatactcta, R2; 3′-ttactgctgcctaacgttcc. Primers F1 and R1 are located within intron 4 and amplify the wild-type allele (450 bp). Primers F2 and R2 are located within intron 1, and the combination of primers F2 and R1 amplify the deleted allele (486 bp).
Cell Culture and Transfection.
HeLa cells (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% FCS. MEFs were generated by standard methods and subsequently maintained in DMEM containing 15% FCS. To transfect HeLa cells or MEFs, we used either Fugene HD (Roche) or Effectene (Qiagen) according to the manufacture's recommendations. For determining the effects of Sirt3 expression on HeLa cell ATP levels, we transfected a single 10-cm plate with 10 μg of the indicated plasmid DNA. Using a GFP expression vector, we determined that this strategy resulted in an ≈70% transfection efficiency. For isolation of Complex I from transfected HeLa cells, we, in general, transfected four 15-cm plates per indicated condition and, when indicated, used 20 μg of plasmid per plate. Forty-eight hours after transfection, cells were lysed for ETC immunocapture as described below. For assessment of ATP levels in transfected MEF cells, two 10-cm plates per condition were transfected with 1 μg of GFP expression plasmid along with 5 μg of an empty vector, wild-type Sirt3, or the deacetylase-inactive Sirt3. Thirty-six hours later, GFP-positive cells (≈5–10%) were purified by FACS sorting and then immediately lysed for ATP analysis.
Constructs and Antibodies.
The human wild-type Sirt3 expression and the catalytically inactive HY mutant in which a single amino acid residue has been modified (histidine-to-tyrosine at amino acid residue 248) were generated by standard PCR amplification and subsequent cloning into a Myc-tagged expression vector (Invitrogen). Constructs were confirmed by direct sequencing. For the in vitro deacetylase assay, we used Flag- rather than Myc-tagged Sirt3 or Sirt4 (Addgene). For immunodetection of human Sirt3, we used a commercial antibody (Cell Signaling Technology), whereas for mouse Sirt3, we used a previously described rabbit polyclonal antisera (11) that was a generous gift (B. Schwer and E. Verdin, University of California at San Francisco, CA). In addition, the following antibodies were used: acetylated-lysine polyclonal antibody, and tubulin (Cell Signaling Technology), c-Myc monoclonal antibody (Santa Cruz Biotechnology), the 39-kDa Complex I component NDUFA9 and COX IV antibody (Abcam), the Complex II 70-kDa Fp subunit antibody, the Complex I 30-kDa subunit NDUFS3, the Complex III 49-kDa Core1 subunit, and the Complex I and Complex II Immunocapture kit (Mitosciences), VDAC1/porin (Invitrogen), GAPDH (Ambion), and Flag antibodies (Sigma).
Mitochondrial Isolation, Acetylation Assays, and Immunoprecipitation.
Mitochondria were isolated from cultured cells by incubating cells in Isolation Buffer I [1% Triton X-100, 0.5% Nonidet P-40, 150 mM NaCl, 50 mM Tris (pH 7.4), 10% Glycerol] containing Complete EDTA-free protease inhibitor mixture (Roche). Mitochondria were then purified by using a mitochondrial rapid-isolation kit and procedures in accord with the manufacturer's recommendation (Pierce). For isolation of functional mitochondria from mouse tissues, we rapidly harvested, washed, and minced mouse livers in ice-cold Isolation Buffer II containing 210 mM mannitol, 70 mM sucrose, 5 mM Hepes (pH 7.4), 1 mM EGTA, and 0.5 mg/ml BSA. The livers were then homogenized in this buffer with a glass–Teflon motorized homogenizer. The mitochondrial fraction was isolated by differential centrifugation and subsequently washed twice before resuspension in Isolation Buffer II at a concentration of 0.5 mg/ml before functional assessment.
The isolation of either Complex I or Complex II was achieved by using isolated mitochondria with the relevant immunocapture beads (Mitoscience) according to the manufacturer's recommendations. In general, we began with 2 mg of mouse hepatic mitochondria or 1 mg of isolated HeLa cell mitochondria for ETC complex isolation. For measurement of basal tissue expression of Sirt3 by Western blotting, we used 50 μg of protein lysate from various mouse organs and probed with the rabbit polyclonal antibody described above. For detection of NDUFA9 acetylation, we immunoprecipitated overnight either 0.5 mg of total MEF cell lysate or 1 mg of solubilized mitochondrial liver lysate derived from either wild-type or Sirt3−/− mice with a polyclonal acetyl-lysine antibody. All immunoprecipitated acetyl-lysine proteins were washed five times in Isolation Buffer I and then resolved by SDS/PAGE and processed for Western blotting to assess for specific levels of NDUFA9 acetylation.
To assess the in vitro deacetylation of Complex I, we used immunocaptured Complex I purified from nicotinamide-treated HeLa cells (10 mM nicotinamide for 16 h before harvest). Each reaction contained equal amounts of purified Complex I and was derived initially from 2 mg of purified HeLa cell mitochondria. The immunocaptured ETC complexes were then resuspended in 50 μl of deacetylase reaction buffer [50 mM Tris, 150 mM NaCl (pH 8.0), 1 mM MgCl2 with 10 mM NAD) for 2 h at 37°C along with the addition of purified Sirt3 or Sirt4. These purified sirtuins were prepared by transfecting HeLa cells with expression constructs encoding either Flag vector, Flag-Sirt3, or Flag-Sirt4 (Addgene). We used 4 μg of each plasmid construct per 10-cm HeLa cell dish. Twenty-four hours after transfection, HeLa cells were harvested in Isolation Buffer I, and 2 mg of transfected protein lysates were subsequently incubated with an anti-FLAG M2 affinity gel (Sigma). Purified proteins were subsequently eluted competitively with five 1-column volumes of a solution containing 100 μg/ml FLAG peptide (Sigma) in TBS [50 mM Tris·HCl, 150 mM NaCl (pH 7.4)]. Isolated Complex I was then incubated with Sirt3 or Sirt 4 (300 ng) or with and equivalent volume of TBS from samples purified as above but initially derived from HeLa cells transfected with Flag-vector only. The reaction was stopped after 2 h by the addition of SDS-loading buffer. The eluted Complex I was analyzed for acetylation by Western blotting using the acetyl-lysine polyclonal antibody.
To assess the in vitro effects of Sirt3 on Complex I activity, we isolated mitochondria from HeLa cells in Isolation Buffer I as described above. Permeabilized mitochondria (200 μg of protein) were then incubated with an equal volume of 2× deacetylase reaction buffer in a total reaction buffer of 250 μl. Where indicated, this reaction mixture contained ≈700 ng of Flag-purified Sirt3 or Sirt4 or a similar volume obtained from Flag elution of Flag vector-transfected cells. The mitochondria were incubated with Flag-eluted proteins for 45 min at 37°C before being placed on ice. To assess Complex I activity, 80 μl of the reaction mix was transferred to Complex I assay buffer [25 mM KH2PO4 (pH 7.2), 5 mM MgCl2, 1.2% (wt/vol) BSA, 0.15 mM Coenzyme Q1 and 0.36 mM NADH] in a total reaction volume of 500 μl. Absorbance of NADH was measured at 340 nm for 12 min, whereupon rotenone (final concentration of 4 μM) was added to assess the level of NADH metabolism that was rotenone sensitive.
ATP Measurements and Mitochondrial Respiration.
ATP was measured by using the ATP determination kit (Molecular Probes). For in vivo measurements, mice tissues were rapidly harvested and immediately placed in ice-cold ATP buffer [20 mM Tris (pH 7.5), 0.5% Nonidet P-40, 25 mM NaCl, 2.5 mM EDTA) for 5 min. Tissue samples then underwent two 10-s rounds of sonication. Lysates were than centrifuged at 13,000 × g for 30 min and the supernatant measured for protein concentration. ATP concentration was measured from 1 μg of this protein lysate. We always measured ATP from freshly isolated, not frozen, tissues. A similar procedure was used for cultured cells, although no sonication was used, and only 0.5 μg of protein per determination was used. Each ATP sample was measured in triplicate. Glutathione measurements were performed by using the Glutathione Assay kit II (Calbiochem) according to the manufacturer's recommendation.
Mitochondrial respiration from intact mitochondria was measured by standard protocols using a respiration buffer containing 225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris·HCl, and 5 mM KH2PO4 at pH 7.2. Glutamate (5 mM), malate (5 mM), and ADP (1 mM) were used to assay respiration through complex I. Succinate (5 mM), rotenone (1 μM), and ADP (1 mM) were used to assay complex II respiration. Respiratory control ratios (state 3/state 4) were determined after inhibition of mitochondrial ATPase with oligomycin.
Supplementary Material
Supporting Information
Acknowledgments.
We are grateful to I. Rovira for help with manuscript preparation. We thank B. Schwer, M. Hirschey, and E. Verdin (University of California at San Francisco, CA) for the gift of Sirt3 antibody. This work was supported by National Institutes of Health Intramural funds (to C.D. and T.F.) and a grant from the Ellison Medical Foundation (to T.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae life span by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends life span in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 5.Haigis MC, Guarente LP. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 6.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 10.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Murray J, Yonally S, Aggeler R, Marusich MF, Capaldi RA. Focused proteomics: Towards a high throughput monoclonal antibody-based resolution of proteins for diagnosis of mitochondrial diseases. Biochim Biophys Acta. 2004;1659:206–211. doi: 10.1016/j.bbabio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Schilling B, et al. Proteomic analysis of succinate dehydrogenase and ubiquinol-cytochrome c reductase (Complex II and III) isolated by immunoprecipitation from bovine and mouse heart mitochondria. Biochim Biophys Acta. 2006;1762:213–222. doi: 10.1016/j.bbadis.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 19.Ingwall JS, Weiss RG. Is the failing heart energy starved? On using chemical energy to support cardiac function. Circ Res. 2004;95:135–145. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- 20.Romeo C, Eaton S, Quant PA, Spitz L, Pierro A. Neonatal oxidative liver metabolism: Effects of hydrogen peroxide, a putative mediator of septic damage. J Pediatr Surg. 1999;34:1107–1111. doi: 10.1016/s0022-3468(99)90577-8. [DOI] [PubMed] [Google Scholar]
- 21.Cardoso SM, Pereira C, Oliveira R. Mitochondrial function is differentially affected upon oxidative stress. Free Radic Biol Med. 1999;26:3–13. doi: 10.1016/s0891-5849(98)00205-6. [DOI] [PubMed] [Google Scholar]
- 22.Cortes-Rojo C, et al. Electron transport chain of Saccharomyces cerevisiae mitochondria is inhibited by H2O2 at succinate-cytochrome c oxidoreductase level without lipid peroxidation involvement. Free Radic Res. 2007;41:1212–1223. doi: 10.1080/10715760701635082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Li C, Xu X, Deng C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc Natl Acad Sci USA. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 26.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information