Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 1.
Published in final edited form as: Nat Struct Mol Biol. 2009 Jan 18;16(2):114–123. doi: 10.1038/nsmb.1548
Abstract
Multisubunit tethering complexes are essential for intracellular trafficking and have been proposed to mediate the initial interaction between vesicles and the membranes with which they fuse. Here, we report initial structural characterization of the Dsl1p complex, whose three subunits are essential for trafficking from the Golgi apparatus to the ER. Crystal structures reveal that two of the three subunits, Tip20p and Dsl1p, resemble known subunits of the exocyst complex, establishing a structural connection among several multisubunit tethering complexes and implying that many of their subunits are derived from a common progenitor. We show, moreover, that Tip20p and Dsl1p interact directly via N-terminal α-helices. Finally, we establish that different Dsl1p complex subunits bind independently to different ER SNARE proteins. Our results map out two alternative protein interaction networks capable of tethering COPI-coated vesicles, via the Dsl1p complex, to ER membranes.
Introduction
Intracellular trafficking of proteins and lipids is accomplished in eukaryotes by means of vesicles that ferry cargo from one compartment to another, or to and from the plasma membrane. Cargo selection, vesicle formation, and vesicle docking and fusion require a large ensemble of cellular proteins and protein complexes1. Some of these, such as vesicle coats and SNAREs, play reasonably well-defined functional roles: the assembly of coat subunits helps drive vesicle formation, while the assembly of complexes between cognate SNARE proteins catalyzes the fusion of vesicles with appropriate target membranes. Precise functional roles have not, however, been assigned to other proteins that play essential roles in vesicle trafficking. A majority of these additional proteins are either small G proteins of the Rab family2, or members of a seemingly heterogeneous set of proteins and protein complexes collectively termed ‘tethering factors’3.
Tethering factors have been proposed to mediate an initial, reversible attachment between a transport vesicle and its proper intracellular target membrane3,4. Nonetheless, fundamental questions about tethering factors remain unanswered. First, how many different types of tethering factors are there? A strong distinction can be drawn between homo-dimeric tethering factors, which are highly elongated coiled coil proteins, and multisubunit tethering factors, which are composed of as many as ten different polypeptides5,6. Among the multisubunit tethering factors, there is clear evidence for structural diversity, and therefore mechanistic diversity, but the extent of this diversity is not understood. A second question concerns the extent to which multisubunit tethering factors actually mediate vesicle tethering. Considerable uncertainty remains on this central point, in part because, unlike budding and fusion, tethering has not been reconstituted using defined protein and lipid components. Moreover, structural information that could serve as a foundation for probing the function and mechanism of tethering factors has been, in many cases, unavailable. A third question is whether tethering factors fulfill additional functions beyond (or instead of) vesicle tethering. The multisubunit tethering factors, in particular, appear to be architecturally complex and might well possess functionality extending beyond simple membrane attachment. This seems especially plausible in light of the demonstrated genetic and/or physical interactions between multisubunit tethering factors and Rabs, vesicle coat proteins, SNAREs, and other components of the cellular trafficking machinery4,7. In several cases, multisubunit tethering factors appear to influence the assembly and/or stability of SNARE complexes8-11, but the mechanism by which this is accomplished is unknown.
To establish a basis for addressing some of these questions, we and others have initiated efforts to determine the structures of multisubunit tethering complexes, or their subunits or subassemblies. To date, eight conserved multisubunit complexes, containing 3-10 subunits each and functioning largely in discrete trafficking pathways, have been identified4. The most complete structural information is available for the 300-kDa TRAPP I (transport protein particle I) complex, which functions in ER-to-Golgi trafficking12,13. EM combined with x-ray crystallography established that TRAPP I is made up of seven subunits that assemble to form a flattened, two-lobed array14. More fragmentary structural information is available for the exocyst15 and COG16 (conserved oligomeric Golgi) complexes, which operate at the plasma membrane and Golgi, respectively. Both exocyst and COG complexes are hetero-octamers with molecular weights exceeding 500 kDa. Structures of five individual subunits - four exocyst subunits17-21 and one COG subunit22 - have been reported. Strikingly, while these structures all resemble one another, none of them resemble TRAPP I subunits. This observation divides the structurally characterized multisubunit tethering complexes into at least two different families. Whether the remaining complexes fit into either of these families is largely unknown, although sequence homology suggests that the GARP (Golgi-associated retrograde protein) complex probably belongs to the exocyst/COG family23,24. Also unknown, except for TRAPP I, is how the subunits within each complex interact with one another.
Here, we report initial biochemical and crystallographic analysis of the Dsl1p multisubunit tethering complex10. The Dsl1p complex has only three known subunits: in the yeast, Saccharomyces cerevisiae, they are Dsl1p, Tip20p, and Sec39p (also called Dsl3p). All three are essential for viability. Despite lacking predicted transmembrane domains, Dsl1p, Tip20p, and Sec39p all localize to ER membranes; temperature-sensitive mutations in any one of them blocks Golgi-to-ER retrograde trafficking10,25-29. We have determined x-ray structures for two subunits of the Dsl1p complex, Tip20p (full-length) and Dsl1p (residues 37-355). Both structures reveal unanticipated but significant similarity to subunits of the exocyst complex, providing direct structural evidence that the Dsl1p, exocyst, and COG complexes are derived from a common evolutionary precursor. Our results delineate a series of protein-protein interactions capable of tethering COPI vesicles to the ER via the t-SNAREs Sec20p and Use1p. They furthermore establish that the Dsl1p complex has two independent binding sites for two different SNARE proteins, suggesting a potential role in controlling SNARE assembly.
Results
Tip20p structure
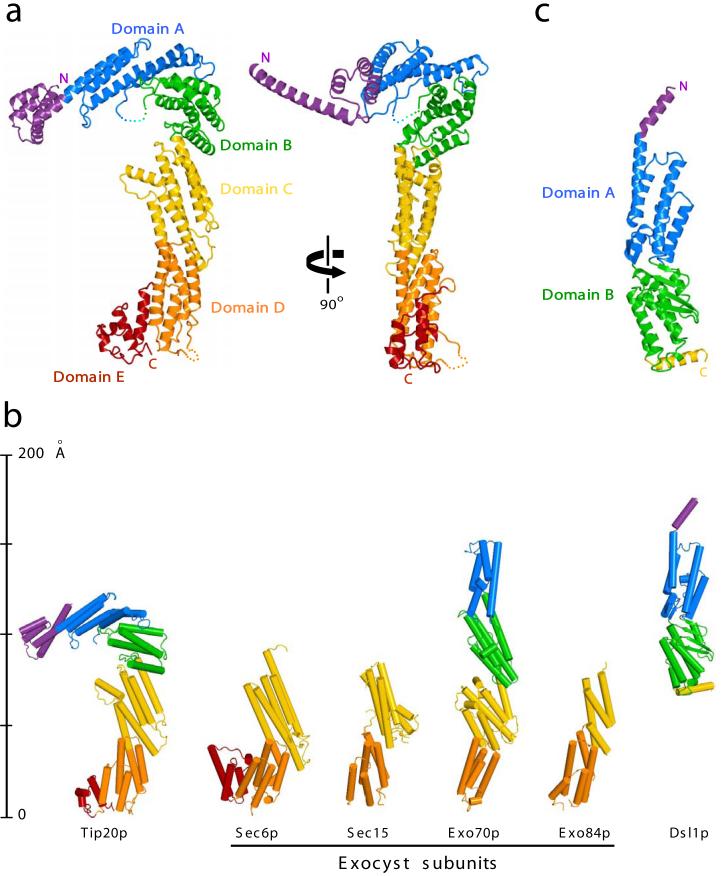
We began by determining the crystal structure of full-length yeast Tip20p (residues 1-701) at 3.0 Å resolution using MAD phasing (Fig. 1a). The structure consists entirely of α-helices and intervening loops of variable length, organized into a series of helix bundle domains. Despite the absence of any detectable sequence similarity23, there is a strong resemblance between Tip20p and each of the four exocyst subunits that have been structurally characterized (Fig. 1b). This resemblance establishes a structural link to the COG complex as well, since several COG subunits resemble exocyst subunits (ref. 22 and B. C. Richardson and F.M.H., unpublished results). Thus, the available structural data support the sorting of multisubunit tethering complexes into at least two unrelated families, one that includes TRAPP I and TRAPP II, and another that includes the exocyst, COG, and Dsl1p complexes.
Figure 1.
X-ray crystal structures of S. cerevisiae Dsl1p complex subunits. (a) Full-length Tip20p (residues 1-701), color-coded by domain. Two views are shown; it can be seen most clearly in the right panel that the N-terminal helix projects away from the remainder of the protein in a manner stabilized by crystal contacts. (b) Structural alignment of Tip20p and Dsl1ΔC to known exocyst subunits. Shown are S. cerevisiae Sec6p (PDB ID 2FJI, residues 411-805 out of 805)20, Drosophila melanogaster Sec15 (2A2F, residues 382-699 out of 766)21, S. cerevisiae Exo70p (2PFV, residues 67-623 out of 623)17-19, and S. cerevisiae Exo84p (2D2S, residues 525-753 out of 753)17. Pairwise alignment was performed with the program DaliLite to match each of the exocyst structures to domains C through E of Tip20p; Dsl1ΔC was then aligned to domains A and B of Exo70p. The DaliLite Z scores for the alignments shown were 11.5 (Tip20p-Exo70p), 9.0 (Tip20p-Exo84p), 16.0 (Tip20p-Sec6p), 12.4 (Tip20p-Sec15), and 7.1 (Dsl1ΔC-Exo70p). (c) Dsl1ΔC (residues 37-355 out of 754), color-coded by domain.
Tip20p is the first subunit of an exocyst/COG/Dsl1p tethering complex to be crystallized intact; previously reported structures17-22 were based on crystals (or NMR characterization) of N-terminally truncated subunits. The most nearly complete of the previously reported structures, lacking just 66 out of 623 residues, is the exocyst subunit Exo70p17-19. Comparing Exo70p and Tip20p reveals that they share a core structure consisting of helix-bundle domains (domains A-D; Fig. 1a-b). Tip20p has, in addition, a set of N-terminal helices, as well as an extra C-terminal domain (domain E). An analogous C-terminal domain is present in one other exocyst subunit (Sec6p20), but is lacking from others (Exo70p and Exo84p17). For Sec15, the potential presence of an extra C-terminal domain is ambiguous because the published structure21 lacks C-terminal regions in addition to N-terminal regions. Further details are provided in the Fig. 1 legend.
Comparing the intact Tip20p structure to the nearly intact Exo70p structure reveals a striking difference (Fig. 1b). Exo70p’s four domains are arranged in a linear array, giving rise to a rod-like shape. The corresponding domains of Tip20p, on the other hand, are arranged in a curving array, giving rise to a sharply bent, hook-like shape. The difference in global conformation between Exo70p and Tip20p is largely attributable to differences in the A-B and B-C hinge angles. The specific bent conformation observed for Tip20p is likely a thermodynamically favorable one, since it is adopted by all four independent Tip20p monomers in the crystallographic asymmetric unit (pairwise rmsd 1.3-2.9 Å). It is possible that the straight and bent conformations simply reflect static structural differences between Exo70p and Tip20p. An intriguing alternative is that Exo70p and Tip20p, and perhaps other exocyst/COG/Dsl1p subunits, are structurally dynamic molecules that adopt both straight and bent conformations during a functional cycle. Crystallographic evidence for modest flexibility at the B-C hinge of Exo70p was reported previously18.
The Tip20p structure offers a first opportunity to examine the conformation of the N-terminal region of an exocyst/COG/Dsl1p complex family subunit. The entire N-terminal region, except for residues 1-4, displays clear electron density. Strikingly, residues 5-38 form a long α-helix that projects away from the main body of the protein (Fig. 1a, right). This helix is stabilized, in the crystals, by forming an antiparallel coiled coil with the corresponding helix of a second monomer. This interaction is not, however, maintained in solution, as judged by sedimentation velocity analytical ultracentrifugation experiments (data not shown). Instead, as discussed below, the N-terminal helix is required for the interaction between Tip20p and another subunit of the Dsl1p complex, Dsl1p itself.
Dsl1p structure
We were able to produce soluble full-length yeast Dsl1p (residues 1-754), but could not generate diffraction-quality crystals, perhaps because the full-length protein contains a central region (residues 388-467) with an unusual concentration of charged residues28 and an absence of predicted regular secondary structure. We therefore tested truncated versions of Dsl1p, obtaining the highest quality crystals using an N-terminal fragment (residues 1-361) we named Dsl1ΔC. The x-ray structure of Dsl1ΔC, determined using MAD phasing and refined to 2.4 Å resolution, revealed a molecule with a significant resemblance to other exocyst/COG/Dsl1p complex family subunits (Fig. 1b-c). Like these structures, Dsl1ΔC consists primarily of α-helical bundles.
No electron density was discernable for the first 36 residues of Dsl1ΔC, suggesting that the extreme N-terminus, while present, is not crystallographically well ordered. Residues 38-73 form a long α-helix with a pronounced bend centered around residue 51 (Fig. 1c). The C-terminal portion of the helix, residues 57-73, forms the first helix of domain A. The N-terminal portion of the helix, by contrast, projects away from the rest of the protein and interacts in the crystals with the corresponding region of a second monomer via an antiparallel helix-helix interaction. Thus, both Tip20p and Dsl1ΔC crystallize in such a way that protruding N-terminal helices are paired and mutually stabilized. It seems likely that, in Tip20p or Dsl1p monomers, these N-terminal regions would be flexible. Such flexibility is consistent with the absence of N-terminal regions from previously reported exocyst and COG structures, all of which were based on stable fragments identified by limited proteolysis17-22.
Tip20p-Dsl1p interaction
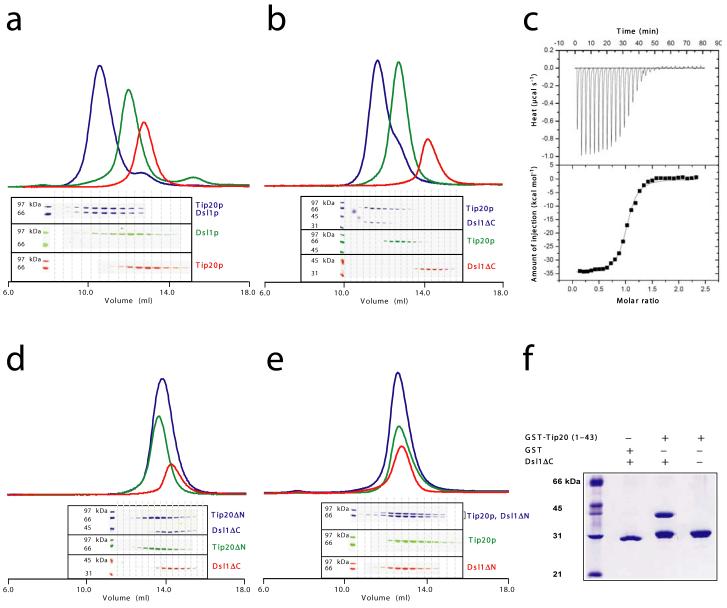
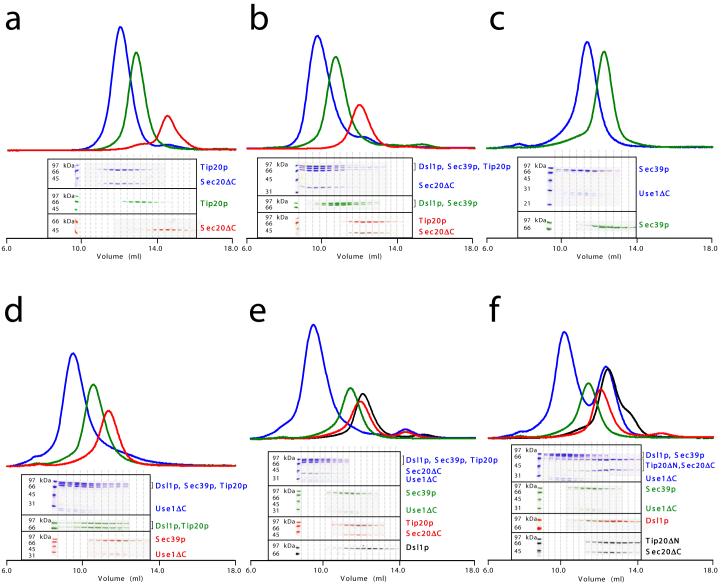
Pure recombinant Tip20p bound in vitro to both full-length Dsl1p (Fig. 2a) and Dsl1ΔC (Fig. 2b), as judged by comparing gel filtration chromatography profiles of the individual proteins to their equimolar mixture. Although this finding is consistent with a wealth of previous data10,25,26,30, it represents the first demonstration of a direct physical interaction between Tip20p and Dsl1p. Sedimentation velocity analytical ultracentrifugation demonstrated that Tip20p and Dsl1ΔC form 1:1 complexes (data not shown). Isothermal titration calorimetry yielded the same 1:1 stoichiometry, together with an equilibrium dissociation constant (Kd) of 100 nM (Fig. 2c). Control ultracentrifugation experiments, using Tip20p or Dsl1ΔC alone, revealed little or no homodimerization. Therefore, the pairing of N-terminal helices observed in both crystal structures is not sufficient to stabilize either homodimer in solution. It remained possible that an analogous interaction between antiparallel N-terminal helices might mediate the formation of Tip20p-Dsl1p heterodimers. We therefore tested whether N-terminal truncations affected the ability of Tip20p and Dsl1p to bind one another. Indeed, deleting the N-terminal region of either Tip20p (residues 1-81) or Dsl1p (residues 1-56) eliminated heterodimer formation (Fig. 2d-e). On the other hand, removing just those N-terminal Dsl1p residues (1-36) that were poorly ordered in the crystal structure had no effect on complex formation (data not shown). A GST fusion protein containing only the first 43 residues of Tip20p, corresponding to the N-terminal helix, was sufficient to bind Dsl1ΔC (Fig. 2f). Together, these results provided a strong indication that Tip20p-Dsl1p heterodimerization entails the pairing of one N-terminal helix from each protein.
Figure 2.
The Tip20p and Dsl1p subunits of the Dsl1p complex form stoichiometric heterodimers. (a) Tip20p binds full-length Dsl1p (residues 1-754). Tip20p alone, Dsl1p alone, or an equimolar mixture were sized on a Superdex 200 gel filtration column. Protein-containing fractions were analyzed using SDS-PAGE gels stained with Coomassie Blue, false-colored to match the corresponding gel filtration profiles. (b) Tip20p binds Dsl1ΔC (residues 1-361). A slight molar excess of Tip20p was present in the mixture (blue gel filtration profile) and accounts for the apparent trailing of the peak. (c) As judged by isothermal titration calorimetry, Tip20p binds Dsl1ΔC with a dissociation constant of 100 nM to form 1:1 complexes. (d) Tip20ΔN (residues 82-701) does not bind Dsl1ΔC, demonstrating that the N-terminal region of Tip20p is essential for heterodimer formation. (e) Tip20p does not bind Dsl1ΔN (residues 57-754), demonstrating that the N-terminal region of Dsl1p is essential for heterodimer formation. (f) The N-terminus of Tip20p (residues 1-43), fused to GST, is sufficient to bind Dsl1ΔC (residues 1-361).
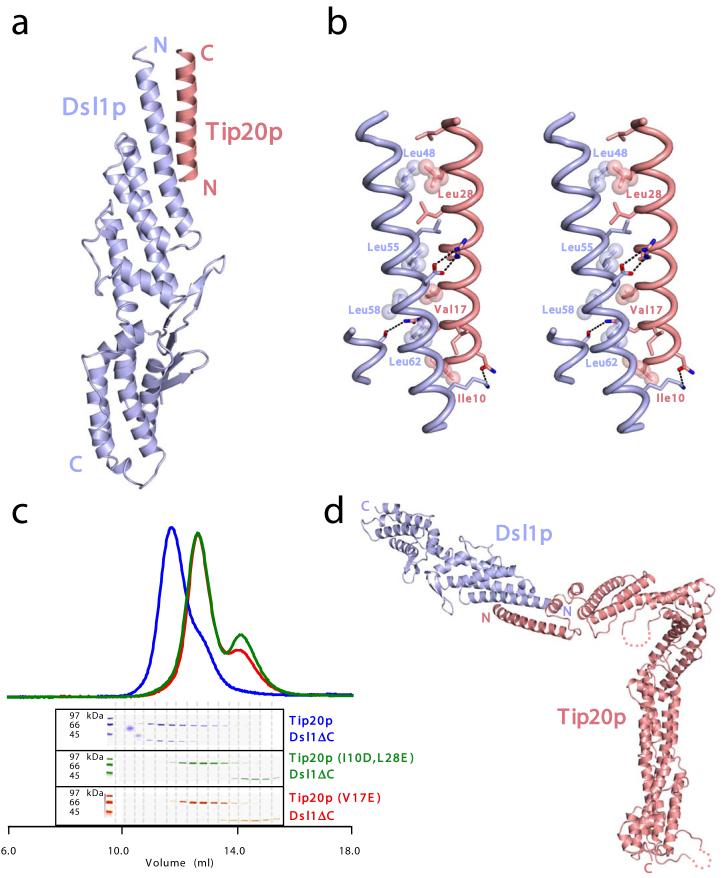
To further analyze the Tip20p-Dsl1p interaction, we sought to determine the crystal structure of a Tip20p-Dsl1p complex. Because all of the crystals we obtained from mixtures of Tip20p and Dsl1ΔC contained only one of the two proteins, we took an alternative approach, fusing the N-terminal helix of Tip20p to the N-terminus of Dsl1ΔC. High-quality crystals were obtained from a fusion protein that linked residues 1-40 of Tip20p to the well-ordered region of Dsl1ΔC (residues 37-339) via an eight-residue Gly/Ser linker. The resulting structure, determined by molecular replacement and refined to a resolution of 1.9 Å, includes residues 9-32 of Tip20p and residues 42-338 of Dsl1p (Fig. 3a-b). These are connected by a tether, not visible in electron density maps, consisting of residues 33-40 of Tip20p, the eight-residue Gly/Ser linker, and residues 37-41 of Dsl1p; this 21-residue tether is capable of reaching 60 Å or more. In the crystal structure, residues 9-32 of Tip20p form an α-helix about 35 Å in length that packs, in an antiparallel orientation, against the N-terminal helix of Dsl1p. The presence of the Tip20p helix eliminates the bend in the Dsl1p helix (compare Figs. 1c and 3a). Also noticeable, upon comparing the Dsl1ΔC structure to the fusion protein structure, is a small reorientation of domain B relative to domain A (not shown). Otherwise, neither the presence of the Tip20p helix nor the change in crystal packing environment causes significant perturbation in the Dsl1ΔC structure.
Figure 3.
Structural and biochemical characterization of the Tip20p-Dsl1p interaction. (a) X-ray crystal crystal structure of the Tip20p-Dsl1ΔC fusion protein (see text for details). (b) The antiparallel interaction between N-terminal helices of Tip20p and Dsl1p. Side chains are shown for residues in the Tip20p-Dsl1p interface. The side chains of the residues selected for site-directed mutagenesis are labeled and shown as spheres. ‘Intermolecular’ polar interactions are highlighted with black dashed lines. (c) Representative results of Tip20p-Dsl1ΔC binding experiments. Dsl1ΔC binds wild-type Tip20p (top panel; see also Fig. 2b) but not the mutant proteins Tip20p (I10D, L28E) or Tip20p (V17E). A slight molar excess of Dsl1ΔC accounts for the trailing of the blue gel filtration profile. (d) Model for Tip20p-Dsl1ΔC complex generated by replacing Tip20p residues 9-32 in the Tip20p-Dsl1ΔC fusion protein with full-length Tip20p. The model contains a single steric clash, just to the left of the blue “N”, involving a presumably flexible region of Tip20p (see text for details).
To test whether the antiparallel helix-helix interaction observed in the crystal structure of the fusion protein was required for Tip20p-Dsl1ΔC heterodimer formation, site-directed mutagenesis was used to change interfacial hydrophobic residues to either glutamate or aspartate (see Fig. 3b). In excellent agreement with the structure, each of the following mutations abolished heterodimer formation: Tip20p (I10D, L28E), Tip20p (V17E), Dsl1ΔC (L48E), Dsl1ΔC (L55E), and Dsl1ΔC (L58D); representative results are shown in Fig. 3c. Dsl1ΔC (L41E), on the other hand, modifies a residue that is not ordered in the crystal structure (and therefore not present in Fig. 3b); as expected, this modification had no effect on binding. The only unexpected result was that the buried interfacial residue Leu 62 could be replaced by Glu without eliminating binding. However, closer inspection revealed that the mutant Glu 62 side chain could, with minor structural readjustment, salt bridge with Arg 13 of Tip20p. Thus, structure-based mutagenesis appears to be fully consistent with the x-ray structural analysis of the Tip20p-DslΔC interaction.
Full-length Tip20p was docked onto the structure of the Tip20p-Dsl1ΔC fusion protein by overlaying residues 9-32 of Tip20p (Fig. 3d). This docking exercise revealed a steric clash between the N-terminus of Dsl1ΔC (residues 42-46) and a short helix in the N-terminal region of Tip20p (residues 46-55). This clash is readily resolved, however, by allowing flexibility in the loop connecting the Tip20p N-terminal helix (residues 5-38) to the clashing helix (residues 46-55). Allowing this flexibility is justified by the high likelihood that the specific positioning of this region is dictated by lattice contacts in the Tip20p crystals. Thus, we propose that Tip20p and Dsl1p interact via sequences at (Tip20p) or near (Dsl1p) their N-termini. Furthermore, we suggest that this interaction mode likely results in a pliable connection, because of flexibility between the N-terminal helix of Tip20p and the bulk of the Tip20p molecule.
In light of the importance of the N-terminal regions in mediating the interaction of Tip20p and Dsl1p, it was surprising that replacing the full-length Tip20p or Dsl1p subunits with N-terminally truncated versions had been reported to cause only relatively mild growth and trafficking defects in yeast10,31. Nonetheless, we were able to obtain additional evidence for these earlier conclusions by using plasmid shuffling to replace wild-type subunits with mutant subunits incapable of forming stable heterodimers. We tested Tip20p (I10D, L28E), Tip20p (V17E), Tip20ΔN, and Dsl1p (L55E, L58D); in no case did we observe a growth defect (data not shown). To attempt to resolve this apparent conundrum, we carried out additional experiments to investigate the network of protein interactions centered around Tip20p and Dsl1p.
Dsl1p ternary complex
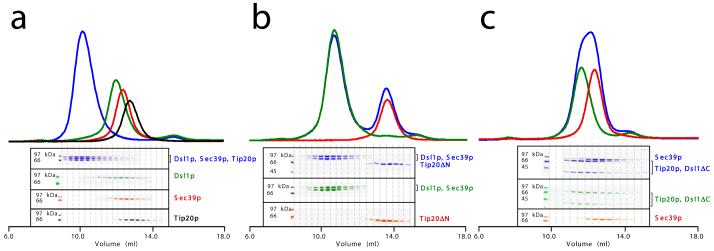
The only other known component of the Dsl1p complex, in addition to Tip20p and Dsl1p itself, is Sec39p10,32. We were able to reconstitute stoichiometric Tip20p-Dsl1p-Sec39p complexes by combining the three full-length recombinant proteins in an equimolar ratio (Fig. 4a). When only two of the three proteins were combined, we found that Sec39p bound directly to Dsl1p but not to Tip20p (Supplementary Fig. 1a and data not shown). These results are consistent with a model in which the Dsl1p subunit lies at the center of a ternary Sec39p-Dsl1p-Tip20p complex, where it serves to link Sec39p to Tip20p. Further experiments indicated that Tip20p and Sec39p bind to non-overlapping regions of Dsl1p. As discussed above, Tip20p binds a helix near the N-terminus of Dsl1p. Moreover, Tip20ΔN did not bind to Dsl1p-Sec39p complexes (Fig. 4b), demonstrating that the inclusion of Tip20p into Sec39p-Dsl1p-Tip20p complexes requires its N-terminal helix. Sec39p, on the other hand, binds to a C-terminal region of Dsl1p. This conclusion is based on the observation that a C-terminal fragment of Dsl1p, Dsl1ΔN1 (residues 340-754) bound efficiently to Sec39p (Supplementary Fig. 1b). Furthermore, neither Dsl1ΔC (Supplementary Fig. 1c) nor Tip20p-Dsl1ΔC complexes (Fig. 4c) were able to bind Sec39p.
Figure 4.
Reconstitution of the heterotrimeric Dsl1p complex. (a) Full-length Tip20p, Dsl1p, and Sec39p (residues 1-709) form stoichiometric heterotrimers. (b) Tip20ΔN (residues 82-701) does not bind to Dsl1p-Sec39p heterodimers, demonstrating that the N-terminal region of Tip20p is essential for its incorporation into the Dsl1p complex. (c) Tip20p-Dsl1ΔC heterodimers do not bind Sec39p, demonstrating that a C-terminal region of Dsl1p is essential for incorporation of Sec39p into the Dsl1p complex.
Interaction of the Dsl1p complex with ER-localized SNAREs
The subunits of the Dsl1p complex, although they lack potential transmembrane domains, localize to ER membranes10,25-27. This localization may be mediated, at least in part, by an interaction between Tip20p and the ER SNARE protein Sec20p10,27,33,34. Indeed, Tip20p (originally named Tip1p) was first discovered in a screen for “SEC twenty interacting protein” genes27. We attempted to recapitulate this interaction by testing whether Tip20p and the cytoplasmic domain of the SNARE protein (residues 1-275, denoted Sec20ΔC) bind to one another directly. As predicted, they indeed formed Sec20ΔC-Tip20p complexes (Fig. 5a). Sec20ΔC also bound efficiently to Tip20ΔN (Supplementary Fig. 1d), demonstrating that the N-terminal region of Tip20p is not required for the interaction. Most importantly, Sec20ΔC bound the intact Dsl1p complex, forming a stoichiometric complex containing all four polypeptides (Fig. 5b).
Figure 5.
ER SNAREs Sec20p and Use1p bind Dsl1p complex via different subunits. (a) Sec20ΔC (cytoplasmic domain, residues 1-275) binds directly to Tip20p. (b) Sec20ΔC binds the intact Dsl1p complex to form stoichiometric heterotetramers. (c) Use1ΔC (cytoplasmic domain; see text for details) binds directly to Sec39p. (d) Use1ΔC, Sec39p, Dsl1p, and Tip20p form a heterotetrameric complex. (e) Use1ΔC, Sec39p, Dsl1p, Tip20p, and Sec20ΔC form a heteropentameric complex. (f) Use1ΔC-Sec39p-Dsl1p does not bind Tip20ΔN-Sec20ΔC.
Our findings imply that a chain of binary protein-protein interactions give rise to a heterotetrameric Sec39p-Dsl1p-Tip20p-Sec20ΔC assembly. A strong prediction of the model is that disrupting the interaction between Dsl1p and Tip20p would cause the heterotetrameric complex to dissociate into two binary complexes, Sec39p-Dsl1p and Tip20p-Sec20ΔC. We tested this prediction in two different ways: by replacing full-length Tip20p with Tip20ΔN and by replacing full-length Dsl1p with Dsl1ΔN. In both cases (Supplementary Fig. 1e-f), only the two binary complexes were observed. These results provide strong support for the proposed arrangement of the Dsl1p complex subunits. Importantly, they also establish that the Dsl1p complex interacts with the t-SNARE Sec20p primarily, if not exclusively, through the Tip20p subunit.
Previous immunoprecipitation experiments using TAP-tagged proteins10 suggested that the Dsl1p complex associates stoichiometrically with a second ER SNARE protein, Use1p. Like Sec20p, Use1p is required for Golgi to ER trafficking35,36. Although we were unable to overexpress the cytoplasmic domain (residues 1-217) of Use1p in soluble form in E. coli, we found that co-expressing it with Sec39p yielded heterodimers that could be purified to near homogeneity. Unfortunately, despite the addition of protease inhibitors, Use1p was invariably cleaved - presumably by a cellular protease - during purification. Nonetheless, both of the fragments (comprising residues 1-167 and 1-175) bind Sec39p (Fig. 5c). It is worth noting that both of these Use1p fragments lack a substantial portion of the membrane proximal SNARE motif and are therefore unlikely to form stable SNARE complexes. Gel filtration suggests that the purified Sec39p-Use1ΔC complex, which elutes at a different volume than Sec39p alone, contains little if any unbound Sec39p (Fig. 5c). Thus, the relatively faint Coomassie Blue staining of Use1ΔC is not an indication of sub-stoichiometric binding, but is rather a consequence of its small size relative to Sec39p and the fact that it migrates as two distinct bands. We were able to reconstitute complexes containing the full Dsl1p complex plus either Use1ΔC alone (Fig. 5d) or both Use1ΔC and Sec20ΔC (Fig. 5e). These results support the model shown in Fig. 6. As expected, based on this model, deletion of the Tip20p N-terminus severed the heteropentameric complex into two parts, Use1ΔC-Sec39p-Dsl1p and Tip20p-Sec20ΔC (Fig. 5f).
Figure 6.
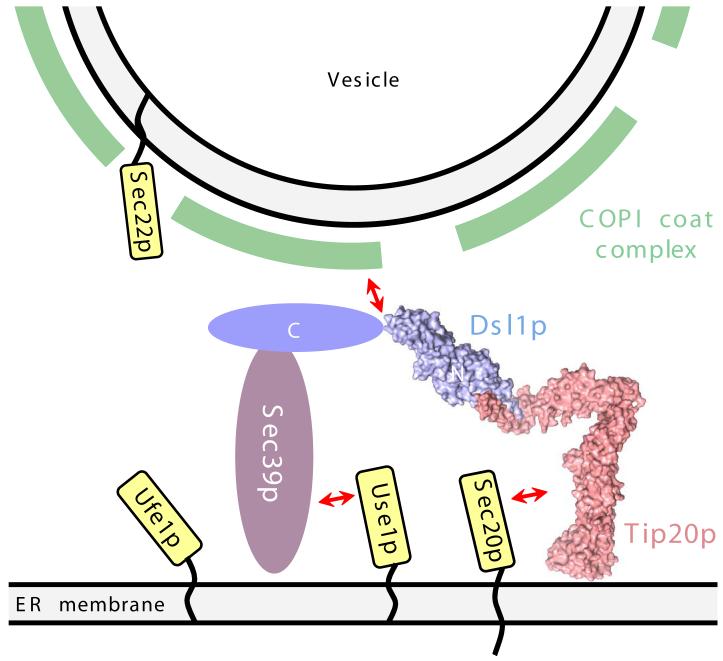
Schematic model for the tethering of Golgi-derived retrograde trafficking vesicles to the ER via bivalent attachment of the Dsl1p complex to the ER SNAREs Use1p and Sec20p. Also shown are two additional SNAREs, Ufe1p and Sec22p, that together with Use1p and Sec20p are thought to form the quaternary SNARE complex that mediates membrane fusion. A central, potentially disorderd region of Dsl1p (residues 388-467) contains binding sites for COPI coat proteins30.
Discussion
The Dsl1p complex is composed of only three subunits, fewer than any of the other known multisubunit tethering complexes4. Here we have shown that these subunits - Tip20p, Dsl1p, and Sec39p - combine to form stoichiometric binary and ternary complexes. The Dsl1p subunit itself lies at the center of the complex, interacting via its N-terminal region with the Tip20p subunit and via its C-terminal region with the Sec39p subunit. By determining the x-ray structures for approximately half of the Dsl1p complex, including the entire Tip20p subunit and domains A-B of the Dsl1p subunit, we have been able to place these Dsl1p complex subunits into the same structural family as the known exocyst and COG complex subunits, and distinguish them from the unrelated TRAPP I complex subunits. A fourth multisubunit tethering complex, GARP, probably belongs to the exocyst/COG/Dsl1p structural family, as judged by distant sequence homology among GARP, exocyst, and COG subunits23,24. Perhaps most strikingly, comparison of Tip20p and Exo70p reveals a structural homology extending over all four domains of the Exo70p structure. Owing to large differences in the relative orientations of domains A-C, the overall shapes of Tip20p and Exo70p are nonetheless very different, with Exo70p adopting a straight, rodlike conformation, while Tip20p displays a sharply bent conformation. Only one other protein bears a strong structural resemblance to the known exocyst and Dsl1p subunits: the cargo-binding domain of the yeast myosin V molecular motor Myo2p37. Remarkably, this domain too functions in tethering processes; these include, for example, the tethering of yeast secretory vesicles to actin filaments38.
Despite the emerging evidence for widespread structural homology among the exocyst/COG/Dsl1p family of multisubunit tethering complexes, it remains difficult to discern the extent to which the various complexes are homologous at the quaternary structural level or to which they operate using homologous mechanisms. At present, the most distinctive property shared by all of these tethering factors is a relatively large array of interacting partners. The yeast exocyst complex, for example, interacts with small GTP-binding proteins on both vesicles and the plasma membrane, and in addition binds the plasma membrane t-SNARE Sec9 and the plasma membrane lipid phosphatidylinositol 4,5-bisphosphate39-41. The COG complex interacts genetically and physically with the Rab protein Ypt1, Golgi SNAREs, and COPI coat subunits42. A similar density of interaction partners is emerging for the Dsl1p complex: although no Rab interaction has been reported, each of the three Dsl1p complex subunits binds directly to either a SNARE protein or, as discussed below, the COPI coat complex.
A potential role in catalyzing SNARE assembly is implied by the finding that the Dsl1p complex uses distinct sites to bind two different ER-localized SNARE proteins. For example, the Dsl1p complex could orient Use1p and Sec20p for facile assembly, or it could modify their conformations to release autoinhibitory interactions, or it could simply increase the local concentration of Use1p relative to Sec20p. Further developments in our ability to generate the relevant recombinant SNARE proteins will be necessary to enable in vitro tests of these possibilities; unfortunately, to date we have not been able to produce the full-length cytoplasmic domains of Use1p or the third t-SNARE, Ufe1p (Fig. 6). Nonetheless, published evidence is consistent with a role for the Dsl1p complex in SNARE assembly. Specifically, mutations or truncations in any of the Dsl1p complex subunits cause severe reductions in the amount of Use1p and Sec20p that can be co-immunoprecipitated from yeast lysates10. It is intriguing to speculate that the potential ability of Tip20p to adopt both bent and Exo70p-like extended conformations might be important for mediating SNARE assembly. By controlling SNARE assembly, tethering complexes might orchestrate the events leading to membrane fusion.
The Dsl1p subunit contains a central region with overlapping binding sites for two different subunits of the COPI vesicle coat protein complex25,30. This observation, in conjunction with our findings, immediately suggests a mechanism for Dsl1p-complex-mediated tethering of COPI vesicles to the ER, via bivalent recognition of the ER SNAREs Sec20p and Use1p (Fig. 6). Indeed, our biochemical analysis establishes that the Tip20p subunit binds directly to Sec20p, as predicted based on earlier studies10,27,33,34, while the Sec39p subunit binds directly to Use1p. Thus, the Dsl1p complex contains a COPI coated vesicle binding site at the center and one SNARE binding site at each ‘end’. This constellation of binding sites provides a mechanism for vesicular tethering through the simultaneous recognition of vesicles (via direct interactions with the COPI coat itself) and the ER (via direct interactions with Sec20p, Use1p, or both). Even in the absence of a Dsl1p-Tip20p interaction, this tethering function could, in principle, be mediated by Use1p-Sec39p-Dsl1p, potentially explaining the lack of an observed growth defect when the Dsl1p-Tip20p interaction is disrupted (refs. 10, 31, and data not shown). An alternative explanation, of course, is that the Dsl1p-Tip20p interaction is stabilized in vivo by additional factors not present in our reconstituted system. The Tip20p-Dsl1p interaction could also be sensitive to the assembly state of the SNARE proteins. In the future, additional structural information about the Dsl1p complex and its binding partners should allow these and other models to be tested directly.
Methods
Protein production
We constructed expression plasmids derived from pQLink (Addgene plasmid43 13670, 13667; for Use1ΔC/Sec39p co-expression), pGEX-4T1 (GE Healthcare; for GST-Tip20(1-43)), or pProExHTb (Gibco; for all other proteins) using PCR. Mutations were introduced using QuickChange Mutagenesis (Stratagene). All expression constructs were confirmed by DNA sequencing. N-terminally His6-tagged proteins were overproduced in either Rosetta or BL21 E. coli (Novagen) grown in Luria-Bertani media at 37°C to an OD600 of 0.6-0.8 and induced by the addition of 0.3-0.5 mM IPTG. Cells were harvested after an additional five hours of growth at 23°C, and the tagged proteins were purified from cell lysates by Ni2+-affinity chromatography followed by removal of the His6 tag by digestion with rTEV protease. The cleaved proteins were then further purified by anion exchange (MonoQ; GE Healthcare) and, for crystallization, size exclusion (S200; GE Healthcare) chromatography. Purified proteins were stored at -80°C in 15 mM Tris, pH 8.0, 150 mM NaCl, 1-2 mM dithiothreitol. For the preparation of selenomethionine-labeled (Se-Met) Dsl1p (residues 1-361) and Tip20p (residues 1-701), methionine synthesis was suppressed by metabolic inhibition essentially as described44. Each protein was expressed in Rosetta E. coli cells grown in expression medium (M9 media supplemented with 5% (w/v) dextrose and 0.7% (w/v) yeast nitrogen base without amino acids (DIFCO)) to an OD600 of approximately 0.8. L-selenomethionine (Acros Organics) was added to a final concentration of 50 mg l-1, together with a mixture of amino acids intended to inhibit the methionine biosynthetic pathway (lysine, phenylalanine, threonine, arginine, isoleucine, leucine, valine; final concentrations, 50 mg l-1; Sigma). After 20 min, protein expression was induced by adding 1 mM IPTG and shaking overnight at 18°C (Tip20p) or 23°C (Dsl1p). Se-Met proteins were purified as above, with 6 mM β-mercaptoethanol present throughout.
Crystallization and data collection
We obtained crystals of full-length Se-Met substituted Tip20p by vapor diffusion at 23°C using a 3:1 ratio of protein (2 mg ml-1) and well buffer (0.1 M ADA, pH 6.0, 10% (w/v) PEG monomethyl ether 5K, 0.2 M LiSO4, 3% (v/v) isopropanol, 5 mM DTT). After three days, crystals of dimensions 200 × 100 × 75 μm were obtained and were subsequently cryoprotected using well buffer supplemented with sequentially increasing amounts of glycerol (up to 22.5% (v/v)) before flash freezing in liquid nitrogen. Crystals of native and Se-Met substituted Dsl1p (residues 1-361) were obtained by vapor diffusion at 23°C using a 2:3 ratio of protein (4 mg ml-1) and well buffer (0.1 M HEPES, pH 7.5, 0.45-0.50 M sodium citrate). Crystals were cryoprotected by a brief soak in well buffer supplemented with 30% (v/v) glycerol and flash frozen in liquid nitrogen. A Tip20p-Dsl1p fusion protein (residues 1-40 of Tip20p linked to residues 37-339 of Dsl1p by the linker GGGSGGGS) formed plate-like crystals by vapor diffusion at 23°C using a 5:5:1 ratio of protein (8 mg ml-1), well buffer (0.1 M sodium acetate, pH 5.0, 0.2 M ammonium acetate, 20% (w/v) PEG 4000), and additive (1.0 M lithium chloride). The crystals were flash frozen without additional cryoprotection. All data were collected at NSLS beamlines X25 or X29 and processed using the HKL suite45.
Structure determination and refinement
Tip20p crystallized in space group P1, with four molecules in the asymmetric unit (Table 1). The structure was determined using MAD phasing methods from selenomethionine-substituted protein to a maximum resolution of 3.0 Å. The SHELX46 suite of programs were used to find the SeMet sites and calculate the initial electron density maps. The program SHARP47 was then used to further improve the phases. Electron density maps calculated from solvent-flattened experimental phases showed clear density for a number of α-helices (Supplementary Fig. 2). Non-crystallographic symmetry (NCS) was determined from the SeMet sites located by SHELXD and from experimentally-phased anomalous difference maps. The four copies of the monomer did not share single NCS relationships over the entire length of each of the molecules; therefore, NCS restraints and averaging needed to be defined on a local basis rather than globally over each chain. Sequence assignment was made on the basis of both model- and experimentally-phased electron density maps, with reference to four-fold averaged maps where necessary. Building was done using the programs O48 and COOT49. The structure was refined using the program CNS50 against data in the range 30-3.0 Å from the peak SeMet dataset, which exhibited the least radiation damage. NCS restraints were applied between molecules on main-chain atoms (including Cβ). The final model (Supplementary Fig. 3) spans nearly the entire molecule, comprising residues 5-701, with residues 217-234 and 546-551 missing because of disordered loops that are not visible in any electron density map. Data collection and structure refinement statistics are summarized in Table 1. 92.0% of the residues are in the most favored regions of the Ramachandran plot, while 7.0% fall in additional allowed regions, as judged using MolProbity51.
Table 1. Data collection, phasing and refinement statistics.
| Tip20p SeMet | Dsl1ΔC | Dsl1ΔC SeMet | Tip20p (1-40) - Dsl1 (37-339) | |||||
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P1 | P3221 | P3221 | C2 | ||||
| Cell dimensions | ||||||||
| a, b, c (Å) | 85.5, 111.6, 149.8 | 110.6, 110.6, 77.2 | 110.9, 110.9, 78.3 | 168.4, 61.4, 37.3 | ||||
| α, β, γ (°) | 77.1, 88.1, 70.4 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 92.0, 90.0 | ||||
| Peak | Inflection | Remote | Peak | Inflection | Remote | |||
| Wavelength (Å) | 0.9792 | 0.9794 | 0.9640 | 1.1000 | 0.9795 | 0.9797 | 0.9641 | 1.0400 |
| Resolution (Å) | 50-3.00 (3.11-3.00) | 50-3.00 (3.11-3.00) | 50-3.20 (3.31-3.20) | 100-2.40 (2.49-2.40) | 100-3.00 (3.11-3.00) | 100-3.00 (3.11-3.00) | 100-3.00 (3.11-3.00) | 100-1.94 (1.99-1.94) |
| Rsym (%) | 7.7 (49.7) | 8.3 (67.0) | 8.1 (56.4) | 7.3 (43.3) | 9.1 (39.1) | 8.9 (48.4) | 9.0 (61.7) | 5.7 (46.7) |
| I/σ_I_ | 12.2 (2.6) | 10.8 (1.7) | 10.9 (2.4) | 28.7 (2.8) | 34.0 (5.3) | 30.5 (3.6) | 27.4 (2.7) | 18.0 (2.0) |
| Completeness (%) | 98.5 (98.2) | 97.5(98.4) | 98.5 (98.3) | 99.2 (96.8) | 99.9 (99.5) | 99.8 (99.2) | 99.8 (99.1) | 90.7 (97.0) |
| Redundancy | 3.7 (3.6) | 3.7(3.3) | 3.7 (3.5) | 7.1 (6.6) | 10.4 (9.9) | 10.1(9.1) | 9.5 (8.4) | 3.1 (2.9) |
| Refinement | ||||||||
| Resolution (Å) | 30-3.0 | 40.0-2.40 | 40.0-1.94 | |||||
| No. reflections | 99,867 | 20,321 | 24,166 | |||||
| _R_work/_R_free (%) | 22.0/26.4 | 22.9/26.6 | 22.5/27.7 | |||||
| No. atoms | ||||||||
| Protein | 22,048 a | 2,549 | 2,566 | |||||
| Water | 49 | 221 | ||||||
| B-factors (Å2) | ||||||||
| Protein | 79.4 | 69.2 | 28.3 | |||||
| Water | 62.5 | 31.3 | ||||||
| R.m.s deviations | ||||||||
| Bond lengths (Å) | 0.0084 | 0.009 | 0.009 | |||||
| Bond angles (°) | 1.31 | 1.126 | 1.091 |
We initially determined the structure of Dsl1ΔC by multiwavelength anomalous dispersion (MAD) at 3.0 Å resolution using a Se-Met substituted crystal (Table 1). Initial phases were calculated using SHELX and subsequently improved using SHARP (Supplementary Fig. 2). Refinement using REFMAC552 against native data to 2.4 Å resolution yielded a model containing residues 37-238 and 245-355; no interpretable electron density was observed for residues 1-36, 239-244, or 356-361 (Supplementary Fig. 3). 96.8% of the residues are in the most favored regions of the Ramachandran plot, while 2.9% fall in additional allowed regions.
We determined the Tip20p-Dsl1p fusion protein structure by molecular replacement using PHASER53 (Table 1). Dsl1ΔC, broken into separate domains, was used as the search model. The final model, built using COOT and refined against native data to 1.94 Å resolution using REFMAC5, includes Tip20p residues 9-32 and Dsl1p residues 42-238 and 245-338. 99.0% of the residues are in the most favored regions of the Ramachandran plot, with the remaining 1.0% falling in additional allowed regions.
Structure-based sequence alignment was guided by DaliLite54. Molecular graphics were rendered using PyMOL55.
Binding experiments
For gel filtration binding experiments, we prepared binding reactions by mixing proteins at 8 μM final concentration in a total volume of 300 μl in 15 mM Tris, pH 8.0, 150 mM NaCl, 2 mM DTT. After incubating on ice for 30 min, samples were loaded onto a Superdex 200 10/30 column (GE Healthcare) equilibrated with the same buffer and run at 4°C using a flow rate of 0.6 ml min-1. Equal volumes from individual 0.3-ml fractions were analyzed using Coomassie Blue stained SDS-PAGE gels. For measuring binding by isothermal titration calorimetry, we used a VP-ITC titration microcalorimeter (Microcal). 100 μM Dsl1ΔC in 15 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM Tris[2-carboxyethyl] phosphine (TCEP) was injected into the sample cell containing 10 μM Tip20p in the same buffer. The resulting titration data were subjected to least squares fitting using Origin version 7.0 (Origin Laboratories). For measuring binding to immobilized GST fusion proteins, cell lysates containing GST or GST-Tip20p (residues 1-43) were loaded onto glutathione resin (Clontech Laboratories). After washing the beads with buffer (15 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT), purified Dsl1ΔC was added and binding was allowed to proceed for 1 h at 23°C. Beads were washed extensively with the same buffer, after which bound proteins were analyzed using Coomassie-stained SDS-PAGE gels.
Supplementary Material
1
Acknowledgements
We gratefully acknowledge Bashkim Kokona and Robert Fairman for sedimentation velocity analytical ultracentrifugation; the staff of the National Synchrotron Light Source X25 and X29 beamlines for assistance with x-ray data collection; Melanie Diefenbacher and Anne Spang for many fruitful discussions and for communicating results prior to publication; Konrad Büssow (Max Planck Institute for Molecular Genetics, Berlin) for reagents; and Sean Munro, Mary Munson, Anne Spang, and members of the Hughson lab for critical comments on the manuscript. This work was supported by NIH grant GM071574.
Footnotes
Accession codes. Protein Data Bank: Crystallographic coordinates for Tip20p, Dsl1ΔC, and the Tip20p-Dsl1p fusion protein have been deposited with assession codes 3FHN, 3ETU, and 3ETV, respectively.
References
- 1.Pfeffer SR. Unsolved mysteries in membrane traffic. Annu. Rev. Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- 2.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 4.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Gillingham AK, Munro S. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta. 2003;1641:71–85. doi: 10.1016/s0167-4889(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 6.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 7.Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am. J. Physiol. Cell Physiol. 2006;290:C11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 8.Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 9.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraynack BA, et al. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell. 2005;16:3963–3677. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J. Cell Biol. 2007;179:1179–1192. doi: 10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 13.Sacher M, et al. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 15.Terbush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 16.Ungar D, et al. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J. Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 18.Hamburger ZA, Hamburger AE, West AP, Jr., Weis WI. Crystal structure of the S. cerevisiae exocyst component Exo70p. J. Mol. Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 19.Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J. Mol. Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat. Struct. Mol. Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 22.Cavanaugh LF, et al. Structural analysis of conserved oligomeric Golgi complex subunit 2. J. Biol. Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 23.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 25.Andag U, Neumann T, Schmitt HD. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- 26.Reilly BA, Kraynack BA, VanRheenen SM, Waters MG. Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol. Biol. Cell. 2001;12:3783–3796. doi: 10.1091/mbc.12.12.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweet DJ, Pelham HR. The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 1993;12:2831–2840. doi: 10.1002/j.1460-2075.1993.tb05944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.VanRheenen SM, Reilly BA, Chamberlain SJ, Waters MG. Dsl1p, an essential protein required for membrane traffic at the endoplasmic reticulum/Golgi interface in yeast. Traffic. 2001;2:212–231. doi: 10.1034/j.1600-0854.2001.020307.x. [DOI] [PubMed] [Google Scholar]
- 29.Kamena F, Spang A. Tip20p prohibits back-fusion of COPII vesicles with the endoplasmic reticulum. Science. 2004;304:286–289. doi: 10.1126/science.1095049. [DOI] [PubMed] [Google Scholar]
- 30.Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- 31.Frigerio G. The Saccharomyces cerevisiae early secretion mutant tip20 is synthetic lethal with mutants in yeast coatomer and the SNARE proteins Sec22p and Ufe1p. Yeast. 1998;14:633–646. doi: 10.1002/(SICI)1097-0061(199805)14:7<633::AID-YEA267>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 32.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Sweet DJ, Pelham HR. The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 1992;11:423–432. doi: 10.1002/j.1460-2075.1992.tb05071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 35.Burri L, et al. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dilcher M, et al. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schott D, Ho J, Pruyne D, Bretscher A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaram MV, Saporita JA, Furgason ML, Boettcher AJ, Munson M. Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry. 2005;44:6302–6311. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- 40.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat. Struct. Mol. Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, et al. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1998;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 47.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 48.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Brunger AT, et al. Crystallography & NMR System (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Lovell SC, et al. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 54.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 55.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1