Frequency-modulated nuclear localization bursts coordinate gene regulation (original) (raw)
. Author manuscript; available in PMC: 2009 Jun 14.
Published in final edited form as: Nature. 2008 Sep 25;455(7212):485–490. doi: 10.1038/nature07292
Abstract
In yeast, the transcription factor Crz1 is dephosphorylated and translocates into the nucleus in response to extracellular calcium. Using time-lapse microscopy, we found that Crz1 exhibited short bursts of nuclear localization (∼2 minutes) that occurred stochastically in individual cells and propagated to the expression of downstream genes. Strikingly, calcium concentration controlled the frequency, but not duration, of localization bursts. Using an analytic model, we found that this frequency modulation (FM) of bursts ensures proportional expression of multiple target genes across a wide dynamic range of expression levels, independent of promoter characteristics. We experimentally confirmed this theory with natural and synthetic Crz1 target promoters. Another stress response transcription factor, Msn2, exhibits similar, but largely uncorrelated, localization bursts under calcium stress. These results suggest that FM regulation of localization bursts may be a general control strategy utilized by the cell to coordinate multi-gene responses to external signals.
Cells sense extracellular signals and respond by regulating the expression of target genes1,2,3. This process requires two stages of information processing: First, cells encode extracellular signals internally, in the states and localization of transcription factors. Second, transcription factors activate the expression of downstream genes that will implement cellular responses1,2,3. Although many signal transduction systems have been studied extensively, it often remains unclear how signals are encoded dynamically in transcription factor activities at the single-cell level. In addition, cellular responses often involve many proteins acting in concert, rather than individually. But it is not known in general how the expression levels of target genes are coordinated, allowing them to be regulated together, despite diverse promoter architectures3. Here we investigate how signal encoding and protein coordination are achieved in individual cells.
We examined the calcium stress response pathway in Saccharomyces cerevisiae, or budding yeast. Cellular response to extracellular calcium is mediated by Crz1, the calcineurin-responsive zinc finger transcription factor4. The activity of Crz1 is modulated by phosphorylation and dephosphorylation, resulting in changes in Crz1 nuclear localization4 (Fig. 1a), rather than changes in its protein abundance (Supplementary Fig. 1). To understand how Crz1 phosphorylation dynamics respond to calcium and regulate the more than 100 different targets necessary for calcium adaptation5, we acquired time-lapse movies of Crz1 localization dynamics, using a strain in which the Crz1 protein was tagged with GFP6. In each movie, we tracked the response of Crz1 localization in individual cells to step changes in extracellular calcium concentration. We find that Crz1 dynamics connect the encoding of signals and the coordination of target gene expression.
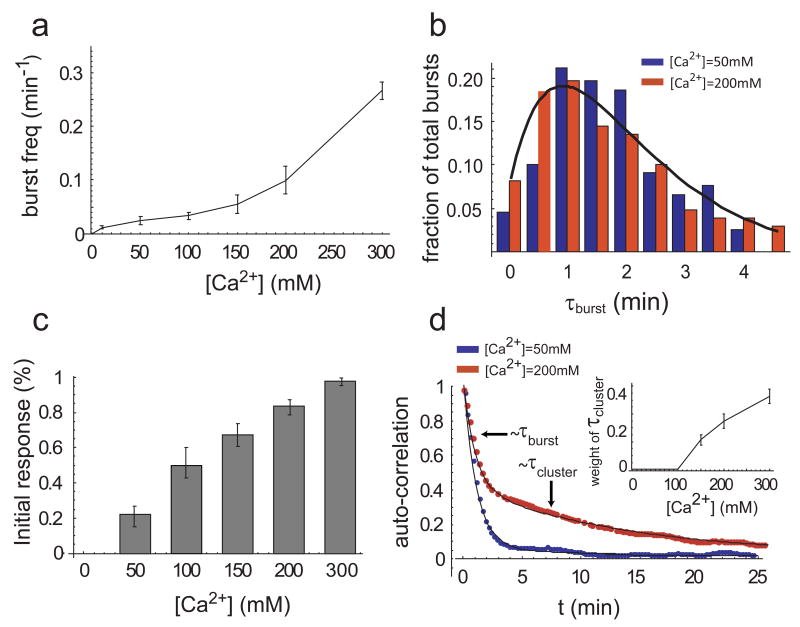
Figure 1.
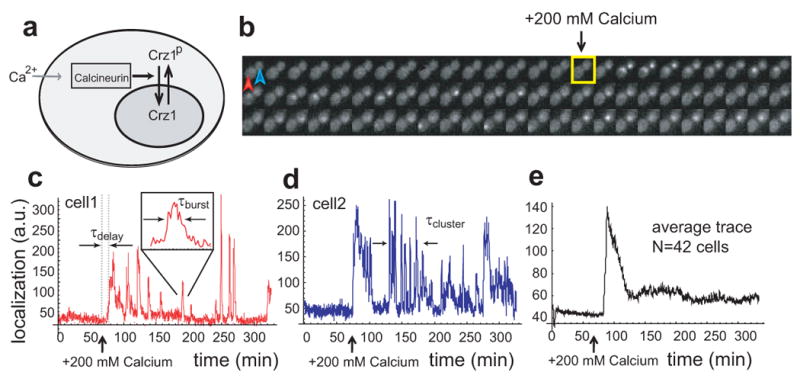
Crz1 undergoes bursts of nuclear localization in response to calcium. a, In the presence of extracellular Ca2+, Crz1 is dephosphorylated and translocates into the nucleus. b, Filmstrip showing yeast cells with Crz1-GFP before and after addition of 200mM extracellular calcium (yellow square). Frames displayed here are separated by 4.5 min, but actual time resolution is higher. c,d Two single cell time traces showing Crz1 localization behavior of the two cells in b. Note that there is a synchronized initial burst of nuclear localization followed by subsequent unsynchronized isolated and cluster bursts of localization. Individual burst duration, τburst, and cluster duration, τcluster, as well as the delay between calcium addition and the initial response, τdelay, are defined on the traces. e, Averaged localization trace shows how single-cell burst dynamics yield partial adaptation across a population of cells.
Frequency modulated bursts of nuclear localization
In the absence of calcium, Crz1 was cytoplasmic in all cells (Supp Movie 1). Upon the addition of calcium, individual cells exhibited a rapid, synchronized burst of Crz1 nuclear localization, similar to behavior observed by the yeast osmosensor Hog17,8. However, unlike Hog1, this initial burst was followed by sporadic unsynchronized localization bursts, typically lasting about 2 minutes (Fig. 1 b-d) and persisting throughout the course of the movie (up to 10 hours, see Supplementary Movie 1). Moreover, these single-cell Crz1 dynamics are consistent with micro-array studies performed on cell populations5: After a step change in calcium, an initial overshoot in mRNA levels of Crz1 target genes results from the initial synchronous burst of Crz1, while the subsequent elevated average expression levels are due to sustained unsynchronized bursts in individual cells (Fig. 1c-e).
We next addressed how the amount of calcium affects the dynamics of nuclear localization. We observed that the fraction of cells with nuclear-localized Crz1 increased with calcium concentration. Because Crz1 localizes in bursts, this calcium-dependence could in principle result from increases in burst frequency or duration. Strikingly, analysis of movies revealed that only the burst frequency increased (Fig. 2a), while the distribution of burst durations remained constant at all calcium concentrations (Fig. 2b). This distribution was consistent with two rate-limiting stochastic steps, each with a timescale of ∼70 sec (Fig. 2b). Thus, cells use stereotyped Crz1 localization bursts in a frequency-modulated fashion to encode and respond to extracellular calcium. This contrasts with amplitude- modulation control, in which the fraction of Crz1 molecules found in the nucleus would change with calcium, but remain constant over time.
Figure 2.
Calcium modulates the frequency, but not the duration, of Crz1 nuclear localization bursts. a, Frequency of bursts increases with calcium concentration (error bars calculated by using different thresholds for burst determination, see Supplementary Information). b, Burst duration is independent of calcium concentration. Normalized histograms, h(t), of total burst duration at two calcium concentrations are both well-fit by h(t) = t e_−_t/τ, with τ = 70 sec (black line). c. The proportion of cells that respond rapidly to extracellular calcium increases with the calcium concentration. Bars represent the fraction of cells with nuclear-localized Crz1 within 15 minutes of addition of calcium. d, Average auto-correlation functions of localization trajectories (N=58 and 85 cells respectively) from a population of cells at two calcium concentrations. At low Ca2+ concentrations (blue, 50mM), the autocorrelation can be well-fit by a single exponential with timescale τ_burst_≈60s, whereas at high Ca2+ concentrations (red, 200mM), two time scales of fluctuations emerge, τ_burst_≈ 60s and τ_cluster≈_ 720s, corresponding to isolated and clustered bursts, respectively. Inset shows the relative weight of the clustered bursts, which appear at Ca2+ concentrations greater than 100mM and increase in frequency as calcium increases. Error bars are estimated from bootstrap.
Movies also revealed two modes of nuclear localization bursts: isolated individual bursts and clusters of bursts, analogous to spike trains in neurons (Fig. 1c,d). At calcium concentrations less than 100 mM, only isolated bursts were observed and the averaged autocorrelation function of the localization trajectories from individual cells was well-fit by a single exponential. However, as clustered bursts emerged at calcium concentrations greater than 100mM (Fig. 1c,d), the averaged autocorrelation function was better fit by a sum of two exponentials, whose timescales matched the typical durations of isolated bursts and burst clusters, respectively (Fig. 2d). Higher levels of calcium led to an increasing proportion of bursts occurring in clusters (Fig. 2d inset). Eventually, at the highest calcium levels, Crz1 nuclear localization trajectories appeared more similar to sustained oscillations9 than to the isolated stochastic bursts seen at lower calcium concentrations.
An additional level of quantization emerged in the initial response to a change in calcium, which exhibited an all-or-none response at the single-cell level. At calcium concentrations ≥ 100 mM, most cells displayed a synchronous initial burst of Crz1 nuclear localization. This resulted in a sharp histogram of values of τ_delay_, defined as the time interval between calcium addition and the first burst of Crz1 nuclear localization (Fig. 1c). In contrast, at lower calcium levels of about 10mM, no synchronous initial response occurs, as reflected in the much broader distribution and larger mean of τ_delay_. Intermediate calcium concentrations produced a mixture of the two discrete behaviors (Supplementary Fig. 2). Thus, rather than controlling the amplitude of the initial response, calcium modulates the proportion of fast-responding cells (Fig. 2c). Critically, in both the initial response and the subsequent bursts, we never observed persistent intermediate levels of localization. Taken together, these results reveal that Crz1 burst activity is ‘quantized’ at multiple levels in cells and that only the frequencies of burst events are modulated by extracellular calcium.
Active Generation of Bursts
Since nuclear localization bursts occurred stochastically in single cells and were unsynchronized within a population on the same slide, they could not be driven by fluctuations in external conditions. These bursts also cannot be explained simply by independent fluctuations in the phosphorylation or localization state of individual Crz1 molecules because each burst involves coherent translocation of a large fraction of the ∼1000 copies of Crz1 present per cell10. In order to gain insight into these dynamics, we next asked whether Crz1 bursts were related to, or driven by, other dynamic cellular phenomena.
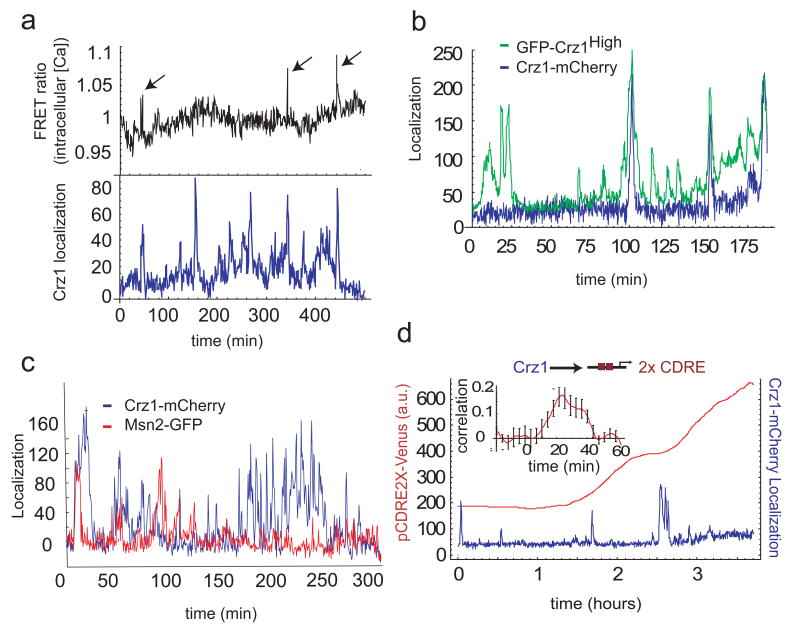
First, we acquired movies of cells expressing both the G1 cell cycle phase marker Whi5-GFP11 and Crz1-mCherry. We found no evidence for cell cycle regulation of Crz1 localization bursts, nor were bursts in daughter cells correlated with those in corresponding mother cells (Supplementary Fig. 5).
Second, we examined the role of intracellular calcium, which in other cell types has been shown to exhibit spike-like dynamics12. We acquired movies of yeast cells expressing both a FRET-based calcium sensor13 and Crz1-mCherry. We observed sporadic transient spikes in intracellular calcium lasting about 38 s (Supplementary Fig. 7). These spikes coincided with some Crz1 localization bursts. (Fig. 3a and Supplementary Fig. 6). However, Crz1 localization bursts also occurred without corresponding calcium spikes, indicating that localization bursts may be stimulated by calcium spikes, but are not exclusively determined by them.
Figure 3.
Crz1 localization bursts are partially independent of other cellular processes and affect downstream gene expression. a, Time traces of Crz1-mCherry localization (blue) in arbitrary units and FRET ratio changes (black), indicating intracellular calcium levels. Arrows indicate spontaneous calcium spikes coincident with Crz1 localization bursts. b, Single-cell time traces of Crz1-mCherry (blue) and the Crz1high-GFP mutant (green) with increased affinity to calcineurin. Both Crz1 proteins are expressed and measured simultaneously in the same cell. c, single-cell traces of Crz1-mCherry and Msn2-GFP in the same cell. Note that the two proteins exhibit statistically similar burst-like behavior but only weak correlation. d, Crz1-mCherry localization (blue) increases expression of the Crz1 target synthetic promoter (2X CDRE) (red). Transcriptional bursts in p2XCDRE-venus are preceded by corresponding Crz1 localization bursts. Inset shows positive cross-correlation (_n_=9 cells) between the promoter activity and Crz1 localization with a delay corresponding to target protein maturation. Error bars are estimated from bootstrap.
Third, we asked whether Crz1 bursts are driven by fluctuations in calcineurin, the upstream phosphatase that initiates Crz1 nuclear localization.14 We analyzed a Crz1 mutant15 with higher affinity to calcineurin, GFP-Crz1high, together with Crz1-mCherry simultaneously in the same cell. If Crz1 bursts were just passive readouts of spikes in calcineurin activity, one would expect both types of Crz1 to burst simultaneously, with amplitudes related to their relative calcineurin affinities. Instead, we observed that Crz1high exhibited an increased burst frequency. Wild-type Crz1 bursts were a subset of these Crz1high bursts (Fig. 3b). Similarly, sub-saturating concentrations of the calcineurin inhibitor FK50616 reduced the frequency of bursts, but did not affect their amplitude (Supplementary Fig 8). As the calcineurin-Crz1 interaction controls only the frequency of burst initiation, Crz1 localization dynamics do not simply follow upstream fluctuations in Calcineurin activity.
Finally, to investigate the generality of localization bursts, we examined Msn2, a general stress response transcription factor that was previously reported to exhibit nuclear localization oscillations.17,18,19 We found that Msn2-GFP localization is induced by calcium stress. Like Crz1, it localized in short bursts on the timescale of 1.5-2 minutes, and also exhibited clustered bursts (Fig. 3c and Supplementary Fig 9). However, despite these statistical similarities, bursts of the two proteins were largely uncorrelated when observed simultaneously in the same cells under calcium stress (Fig 3c and Supplementary Fig. 10). 18.1±2.0% of Msn2-GFP bursts coincided with Crz1-mCherry bursts, a fraction only slightly higher than the 14.0±1.2% of overlapping events expected by chance if the two proteins burst independently (Supplementary Materials). Furthermore, bursts of Msn2 nuclear localization can occur in the absence of calcium, while Crz1 bursts cannot. Similarly, we observed that the glucose-responsive repressor Mig1 exhibited bursts of nuclear localization at low glucose concentrations. These results suggest that cells operate multiple transcription factor localization burst systems in a largely independent manner.
The functional role of frequency modulation
What effect do FM-regulated Crz1 localization bursts have on downstream genes? We analysed the transcriptional activity of a synthetic Crz1-dependent promoter containing calcineurin-dependent response elements (2X CDRE),20 driving expression of the fluorescent protein Venus. We also monitored Crz1-mCherry localization simultaneously in the same cell (Fig. 3d). We found that the promoter was expressed in transcriptional bursts that followed Crz1 localization bursts (Fig. 3d). Interestingly, not all Crz1 localization bursts resulted in observable transcriptional bursts, suggesting that transcription initiation is probabilistic. Nevertheless, the rate of Venus production (time derivative of fluorescence) was correlated with Crz1-mCherry bursts with a time-delay comparable to the maturation time of the Venus fluorophore, as expected (Fig. 3d inset and Supplementary Fig 12). We observed similar results with a natural Crz1 target gene, Cmk2 (Supplementary Fig. 11).5 Thus, transcription factor localization bursts propagate to downstream targets, and represent a general mechanism for generating “transcriptional bursting” 21,22,23 in downstream gene expression24 25 26 27 28 29 30 31 32 33 34.
Standard models of gene regulation involve amplitude modulation, or analogue, changes in transcription factor concentration in response to external signals. What biological functions could the frequency modulation form of regulation observed here provide for the cell? Crz1 regulates more than 100 different target genes,5 including Ca2+ pumps and structural proteins necessary for calcium adaptation. The target promoters of these genes may differ in their input functions, defined as the dependence of transcription rate on the concentration of transcription factor in the nucleus. Input functions vary widely in their minimal and maximal levels of expression, the concentration of transcription factor at which they reach half-maximal activity, and in the sharpness, or cooperativity, of their response.35 We used an analytic model to show that frequency-modulation regulation of nuclear localization bursts could allow transcription factors to modulate the expression of multiple target genes in concert, keeping their relative abundances fixed over a wide dynamic range, regardless of the shapes of their input functions.
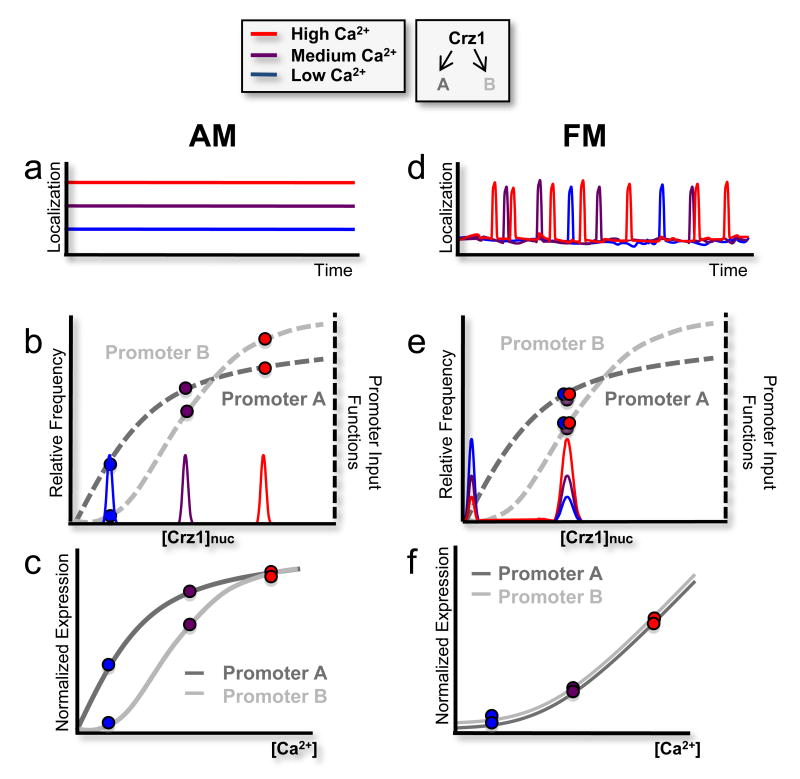
This hypothesis can be understood by comparing the effects that amplitude- and frequency-modulation regulation systems have on two hypothetical target promoters, labeled A and B, with different input functions (Fig. 4b, e, gray dashed curves). In the AM regulation system, the fraction of Crz1 molecules in the nucleus, Crz1nuc, would increase with calcium, but remain constant over time (Fig. 4a, b). Accordingly, histograms of Crz1nuc would exhibit a single peak whose position is calcium-dependent (Fig. 4b). The expression level of each gene would be proportional to its input function evaluated at this peak position. Because it is expected that these input functions would, in general, differ in shape, the normalized rates of A and B expression would vary differently with Crz1nuc, and hence their ratio would vary with calcium (Fig. 4c). Thus, in the amplitude modulation model, A and B would be expressed in an ‘uncoordinated’ fashion.
Figure 4.
Frequency- versus amplitude –modulation regulation of two hypothetical target genes, labeled A and B (schematic). a, In the amplitude-modulation regulation system, the fraction of nuclear Crz1 (Crz1nuc) changes with calcium, but remains constant over time. b, As such, the histogram of Crz1nuc yields single peaks at calcium-dependent positions. Target gene expression level is proportional to the input functions at these peak positions. c, Because their input functions differ, the normalized rates of A and B expression vary differently with nuclear Crz1, and hence with calcium, yielding different (uncoordinated) expression profiles as a function of calcium. d, In frequency modulation, where Crz1 molecules collectively move between nuclear and cytoplasmic compartments, Crz1nuc is either high or low, during or between bursts, respectively. This graph depicts the limiting case of rapid and complete transitions between two states, but results do not depend on this assumption (see Supplementary Information). e, This yields a bimodal histogram in which the height, but not the position, of the peak is calcium-dependent. f, Consequently, the expression levels A and B are each proportional to burst frequency, and hence to each other, yielding coordinated expression, as shown.
In contrast, frequency-modulation regulation (Fig. 4d) would control the fraction of time that Crz1 is nuclear, rather than the concentration of nuclear Crz1. In the limiting case of fast switching between two localization states, this would cause Crz1nuc to be either high, during a burst, or very low, between bursts, but rarely in between, resulting in a bimodal Crz1nuc histogram (Fig. 4e). The expression level of each target promoter would be determined mainly by the value of its input function near the location of the higher histogram peak, and by the fraction of time Crz1 spends in the localized state, which determines the height of the peak. In frequency modulation regulation, higher levels of calcium would increase the relative height of this peak, by increasing the frequency of bursts, but would not change its position (Fig. 4e). The expression levels of the two genes would therefore be individually proportional to the burst frequency, and consequently remain proportional to each other, as calcium is varied. Thus, A and B would be coordinated, expressed at a constant ratio at all calcium levels, regardless of the shapes of their input functions (Fig. 4f).
This regulatory strategy is general: It does not require bursts to saturate target promoter input functions, nor does it require that the Crz1nuc histogram be bimodal (see supplementary material for a more detailed treatment). Thus, it functions with bursts, such as those observed here, that span a range of localization amplitudes, giving rise to a non-bimodal histogram. Finally, this strategy is more general than FM regulation; it can also work by varying the duration, rather than the frequency, of localization bursts. We refer to this principle as FM-coordination.
Experimental validation of FM-coordination
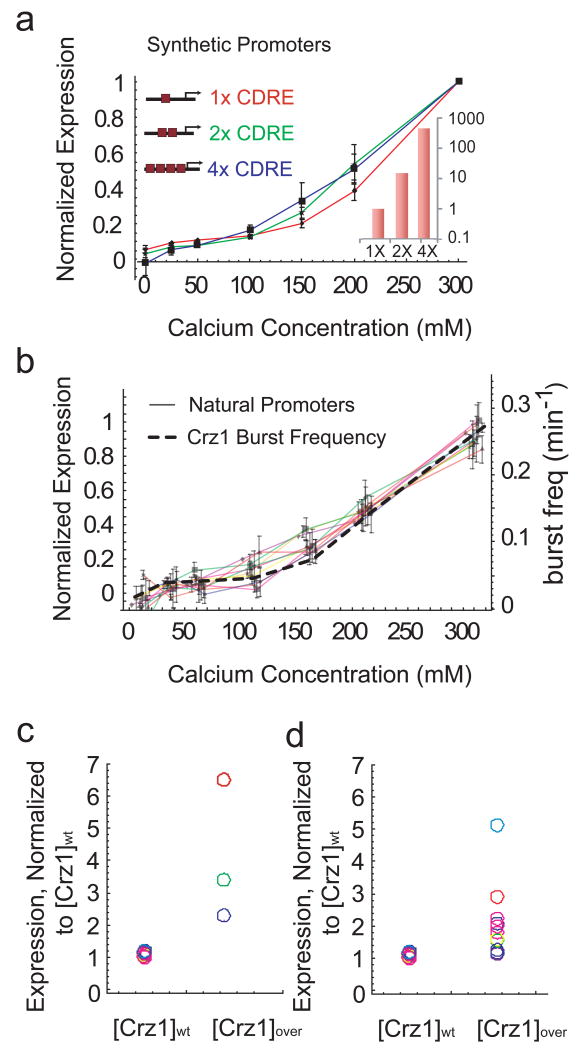
To test the FM-coordination hypothesis experimentally, we first analyzed three synthetic promoters containing one, two, or four CDREs. These promoters were previously shown to exhibit Crz1-dependent expression in response to calcium20. We used flow cytometry to analyze strains containing each of these three promoters fused to yellow fluorescent protein (YFP). The promoters varied in their expression strength over two orders of magnitude, in the ratio of 1:15:450, respectively (Fig. 5a inset). Nevertheless, they exhibited identical dependence on calcium, collapsing onto a single curve when normalized, as predicted by FM-coordination (Fig. 5a).
Figure 5.
Frequency-modulated bursts coordinate gene expression. a. Measured expression levels of synthetic Crz1 target promoters containing one, two and four CDREs. In each case, expression levels are normalized by their maximum. Note similarity of curves to each other and to burst frequency. Bottom inset shows expression at 200mM calcium normalized by expression of the 1× CDRE. b, Expression profiles of natural target promoters (solid lines) exhibit similar inductions as the Crz1 burst frequency (dashed line). Only ten are plotted here for clarity. Error bars indicate standard error from repeated experiments. Induction curves for 64 genes are shown in Supplementary Fig. 14. c, Expression level of synthetic target genes at wild-type versus overexpressed levels of Crz1. Each promoter is normalized by the maximum expression in wild-type Crz1 levels. Thus the normalized expression levels of one, two and four CDREs at [Crz1]wt each equal 1. If all promoter input functions were identical, then their normalized expression should increase by the same factor when Crz1 is overexpressed. However, the data show a range of fold changes in expression levels, excluding identical input function shapes as an explanation for proportional regulation. d. Same as c. for the natural promoters in b.
Natural eukaryotic promoters exhibit more diverse architectures than these pure synthetic promoters. Therefore, we also investigated whether FM-coordination could explain the behavior of natural Crz1 target genes. A previous microarray study identified 163 potential Crz1 target genes,5 83 of which were available as fluorescent protein fusions.6 Of these, 40 showed a measurable and predominantly Calcineurin-dependent response to calcium under our media conditions (Supplementary Fig. 15). These natural promoters, like their synthetic counterparts, exhibited a broad distribution of expression levels (Supplementary Figs 14 and 17). Notably, a large subset of these target genes (34 out of 40) exhibited normalized calcium response curves identical to each other and to those of the synthetic promoters. They were thus regulated in proportion to burst frequency, in accordance with FM-coordination (Fig. 5b and Supplementary Fig. 14a).
In principle, target promoters could respond proportionally because they share identical input functions. If so, increasing total Crz1 concentration would increase the expression of all promoters by the same factor (Fig, 5c, d). Conversely, if the expression levels of the promoters change by different factors when Crz1 concentration is increased, then their input functions must differ in shape. This is what we observed when we compared the expression of both the synthetic and natural promoters in wild-type strains to their expression in a Crz1-overexpressing strain.36 The increase in expression varied between onefold and sevenfold over this set of promoters (Fig. 5c, d), confirming that synthetic and natural promoters are proportionally regulated by the FM-coordination mechanism, despite non-identical input functions.
Discussion
Diverse perturbations including calcium, pharmacological inhibitors, and genetic mutations vary mean Crz1 activity across a broad dynamic range exclusively by affecting the frequency of localization bursts, rather than their duration or amplitude. Thus, Crz1 activity is quantized both in its initial response to a step change in calcium, which is all-or-none at the single-cell level, and also in its sustained response, which is composed of stereotyped bursts of localization. These data suggest that eukaryotic cells can encode information about the extracellular environment in the frequency of stochastic intracellular events, rather than in the concentrations of molecular species. This contrasts with canonical signal transduction pathways, in which concentrations of activated proteins are used to convey information to the nucleus. Using localization as a single-cell reporter for post-translational modifications, we observed of qualitatively similar, but largely uncorrelated, behavior in the Msn2 stress response transcription factor and the Mig1 glucose repressor, suggesting that localization bursts are broadly employed in the cell.
In engineering, frequency modulation appears in diverse signal processing and control applications such as broadcasting37 and rocket thruster control38, where transfer characteristics may be uncertain. Spike frequency control, or rate coding, is also fundamental to neural computation39, where it may overcome noise background for signal propagation, among other functions40. In genetic networks, frequency- modulation regulation solves a fundamental problem, allowing cells to co-regulate proportionally a large set of target genes with a diverse range of input functions. Note also that these results represent a complementary mode of regulation compared to previous observations of calcium oscillation frequency-dependent gene expression.41 FM-coordination connects the dynamic behavior of a single protein to the expression of large regulons, and may thus provide insight into genome-scale expression patterns. Furthermore, at the evolutionary level, FM-coordination enables promoter mutations to alter the level of expression of individual genes without disrupting their coordination. In contrast, to achieve coordinate regulation using amplitude modulation would require ‘fine-tuning’ of target promoter input functions, severely constraining promoter architecture and regulatory evolution. In light of previous observations of transcription factor pulsing in p5342, NFκB43 and SOS stress response systems44, and because of its potential utility in protein and metabolic, as well as transcriptional, networks, we anticipate that frequency-modulated regulation may represent a general principle by which cells coordinate their response to signals.
Methods
Strain construction
All GFP strains were obtained from the GFP C-terminal protein fusion library (Invitrogen).6 All other strains were constructed similarly to the library: Polymerase chain reaction (PCR) was used to amplify cassettes with yeast codon-optimized fluorescent proteins tagged to an auxotrophic marker.45 These amplified cassettes were made with primers containing 40 bp homology towards the C-terminus of the protein of interest. The cassettes were then transformed into the w303a lab strain of yeast using standard protocols.46 Transformants were selected using standard yeast media.47 Plasmid pGW845, a vector containing the FRET cameleon pair under the control of the ADH1 promoter, was provided by Jochen Stadler13 and p30, the Crz1high mutant with altered calcineurin affinity, was provided by Martha Cyert.15 Both were transformed directly into a yeast strain containing Crz1-mCherry (Fig 3b). A Crz1 overexpression plasmid, pLE66, provided by K. Cunningham,36 was transformed into each of 83 Crz1 target strains from the GFP library. A BamHI fragment of yeast codon-optimized Venus-YFP, cut from pKT103,45 was ligated into pAMS342, pAMS363 and pAMS366, provided by Martha Cyert, containing 1×,2×,and 4× CDRE's respectively.20
Media
Yeast were grown in synthetic complete or the appropriate drop-out media made using low-fluorescence yeast nitrogen, adapted from previous work.45 This media is yeast nitrogen base without riboflavin, folic acid, or calcium chloride: 2 g/l (NH4)2Cl2, 1 g/l KH2PO4, 0.5 g/l MgCl2, 0.05 g/l NaCl, 0.5 mg/l H3BO4, 0.04 mg/l CuSO4, 0.1 mg/l KI, 0.2 mg/l FeCl3, 0.4 mg/l MnCl2, 0.2 mg/l Na2MoO4, 0.4 mg/l ZnSO4, 2 μg/l biotin, 0.4 mg/l calcium pantothenate, 2 mg/l inositol, 0.4 mg/l niacin, 0.2 mg/l PABA, 0.4 mg/l pyridoxine HCl, 0.4 mg/l thiamine, 20 g/l dextrose.
Time-lapse microscopy
Cells were attached to glass bottom dishes (Willco) functionalized with Concanavalin-A (Sigma). Fluorescence images were taken at room temperature on an Olympus IX81 with the ZDC autofocus option and an Andor Ikon (DU-934) camera. CaCl2 was added during movies to nominal concentrations using a 2X stock solution. Images were acquired at 10 second intervals for single color movies and at 30 second intervals for two-color and FRET movies. Automation was controlled by Andor IQ software. Two-color movies of Crz1 and downstream target genes were taken at 30° to ensure fast maturation of the Venus fluorophore.
Flow cytometry
Yeast cells were grown overnight in 96-well plates and diluted. CaCl2 was added to nominal concentrations with a 2× stock solution. Cells were regrown in the calcium for 4-6 hours, diluted 5-fold, and gene expression was measured using a Beckman Coulter Cell Lab Quanta SC MPL. To identify cells, all particles were triggered and gated during acquisition. As shown in Figure S13, particles were triggered if their Coulter volume measurements were within a specified range. Either 5000 particles or 30 μl sample volume were acquired. These particles were then excited by a 22mW 488nm laser and side-scatter and fluorescence were measured. A polygon gate was drawn on the Coulter volume/Side-scatter two-parameter plot to identify cells. If more than 500 cells were found, mean fluorescence was calculated. Quanta Photomultiplier tube settings were 3.5 for Side-scatter, 8.00 for GFP measurements and from 4-6 for synthetic promoter-YFP measurements.
Image analysis
Fluorescence cell images were segmented using a Hough transformation algorithm in Matlab, provided by Sharad Ramanathan.48 Localization score was determined by the difference between the mean intensity of the 5 brightest pixels in the cell and mean intensity of the rest of the pixels in the cell. Bursts were identified by thresholding traces at >1 standard deviations above background noise, estimated from the lowest 20% of values. Subsequent data analysis used the resulting traces with standard routines in Matlab and Mathematica.
Supplementary Material
Movie
Supplementary
Acknowledgments
We thank M. Cyert for the CDRE and Crz1 mutant plasmids, K. Cunningham for the Crz1 overexpression plasmid pLE66, J. Stadler for the pGW845 FRET plasmid, S. Ramanathan for image analysis code, and K. Thorn and C-L. Guo for technical assistance. We thank U. Alon, M. Carlson, M. Cyert, H. Garcia, R. Kishony, G. Lahav, J. G-Ojalvo, I. Riedel-Kruse, B. Shraiman, G. Süel, members of the laboratory, and especially N. Friedman for discussions. L.C. is supported by the Beckman Fellows Program at Caltech. This work was supported by NIH grants R01GM079771 and P50 GM068763 for National Centers of Systems Biology, and the Packard Foundation.
Footnotes
Supplementary Information accompanies the paper.
References
- 1.Cyert MS. Regulation of nuclear localization during signaling. J Biol Chem. 2001;276(24):20805. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 2.Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24(4):469. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 3.Kyriakis JM. The integration of signaling by multiprotein complexes containing Raf kinases. Biochim Biophys Acta. 2007;1773(8):1238. doi: 10.1016/j.bbamcr.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999;13(7):798. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimoto H, et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277(34):31079. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- 6.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 7.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319(5862):482. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci U S A. 2008;105(20):7165. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suel GM, et al. Tunability and noise dependence in differentiation dynamics. Science. 2007;315(5819):1716. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 10.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 11.Di Talia S, et al. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448(7156):947. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 12.Fewtrell C. Ca2+ oscillations in non-excitable cells. Annu Rev Physiol. 1993;55:427. doi: 10.1146/annurev.ph.55.030193.002235. [DOI] [PubMed] [Google Scholar]
- 13.Wiesenberger G, et al. Mg2+ deprivation elicits rapid Ca2+ uptake and activates Ca2+/calcineurin signaling in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6(4):592. doi: 10.1128/EC.00382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boustany LM, Cyert MS. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 2002;16(5):608. doi: 10.1101/gad.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy J, Li H, Hogan PG, Cyert MS. A conserved docking site modulates substrate affinity for calcineurin, signaling output, and in vivo function. Mol Cell. 2007;25(6):889. doi: 10.1016/j.molcel.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breuder T, et al. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc Natl Acad Sci U S A. 1994;91(12):5372. doi: 10.1073/pnas.91.12.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquet M, et al. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J Cell Biol. 2003;161(3):497. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garmendia-Torres C, Goldbeter A, Jacquet M. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr Biol. 2007;17(12):1044. doi: 10.1016/j.cub.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 19.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 Link Calorie Restriction and TOR to Sirtuin-Mediated Lifespan Extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5(10):e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11(24):3432. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Raj A, et al. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175(1):67. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 25.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38(6):636. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 26.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440(7082):358. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 27.Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97(16):168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- 28.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: from single molecules to the proteome. Curr Opin Genet Dev. 2007;17(2):107. doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Maheshri N, O'Shea EK. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 31.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 32.Ozbudak EM, et al. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 33.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444(7119):643. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, et al. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311(5767):1600. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeld N, et al. Gene regulation at the single-cell level. Science. 2005;307(5717):1962. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 36.Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11(24):3445. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong EH. A method of reducing disturbances in radio signaling by a system of frequency modulation. P Ire. 1936;24(5):689. [Google Scholar]
- 38.Song GB, Buck NV, Agrawal BN. Spacecraft vibration reduction using pulse-width pulse-frequency modulated input shaper. J Guid Control Dynam. 1999;22(3):433. [Google Scholar]
- 39.Adrian ED, Zotterman Y. The impulses produced by sensory nerve-endings: Part II. The response of a Single End-Organ. J Physiol. 1926;61(2):151. doi: 10.1113/jphysiol.1926.sp002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarpeshkar R. Analog versus digital: extrapolating from electronics to neurobiology. Neural Comput. 1998;10(7):1601. doi: 10.1162/089976698300017052. [DOI] [PubMed] [Google Scholar]
- 41.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392(6679):933. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 42.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006 0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 44.Friedman N, et al. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3(7):e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21(8):661. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 46.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 47.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 48.Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131(3):544. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie
Supplementary