Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention (original) (raw)
Abstract
The availability of both the mouse and human genome sequences allows for the systematic discovery of human gene function through the use of the mouse as a model system. To accelerate the genetic determination of gene function, we have developed a sequence-tagged gene-trap library of >270,000 mouse embryonic stem cell clones representing mutations in ≈60% of mammalian genes. Through the generation and phenotypic analysis of knockout mice from this resource, we are undertaking a functional screen to identify genes regulating physiological parameters such as blood pressure. As part of this screen, mice deficient for the Wnk1 kinase gene were generated and analyzed. Genetic studies in humans have shown that large intronic deletions in WNK1 lead to its overexpression and are responsible for pseudohypoaldosteronism type II, an autosomal dominant disorder characterized by hypertension, increased renal salt reabsorption, and impaired K+ and H+ excretion. Consistent with the human genetic studies, Wnk1 heterozygous mice displayed a significant decrease in blood pressure. Mice homozygous for the Wnk1 mutation died during embryonic development before day 13 of gestation. These results demonstrate that Wnk1 is a regulator of blood pressure critical for development and illustrate the utility of a functional screen driven by a sequence-based mutagenesis approach.
The sequencing of the mouse (1) and human (2) genomes is providing an unprecedented opportunity for the biomedical community to better understand and treat human disease. To realize this goal, efficient methods are required to determine mammalian gene function. Genetic screens combining efficient approaches to mutate genes with assays designed to identify phenotypic changes of interest have been carried out in the fly (3), worm (4), and other model organisms (5, 6). Such screens have been valuable for the identification of genes that play a role in multiple processes of interest. Insertional mutagenesis using transposons revolutionized Drosophila genetic screens by expediting the correlation of phenotypes with the identification of the causative genes. An insertional mutagenesis screen in the mouse provides gene function information in the context of mammalian physiology (7, 8). The potential utility of this approach for drug discovery is supported by the correlation between the knockout phenotypes and the pharmacological effects of drugs against the major protein targets of the pharmaceutical industry (9). Here we describe a large-scale mammalian reverse genetics operation in mouse enabled by efficient DNA insertional mutagenesis, sequencing, and phenotypic screening.
There are three widely used methods of mutagenesis in the mouse: gene targeting by homologous recombination (10), genetrapping by insertional mutagenesis (11–13), and chemical mutagenesis (14). Of these, gene-trapping offers the balance in scalability, throughput, mutagenicity, and mutation identification required for determination of gene function on a genomic scale (15). Gene-trapping is a method of random mutagenesis in which the insertion of a DNA element into endogenous genes leads to their transcriptional disruption. Unlike gene targeting by homologous recombination, a single gene-trap vector can be used to mutate thousands of individual genes as well as to efficiently produce sequence tags for the rapid identification of altered alleles (15). By contrast, random chemical mutagenesis by agents such as _N_-ethyl-_N_-nitrosourea (ENU) produces thousands of base pair mutations that can be difficult to identify. Genomewide gene-trapping in embryonic stem (ES) cells allows for the preselection of genes in specific gene families or members of a common biochemical pathway, enabling a reverse genetic screen in mammals, focused on candidate genes.
We previously have described the development and automation of a high-throughput 3′ gene-trapping technology (15). In this study, we describe improvements to the original gene-trapping vectors and analyze the expanded ES cell gene-trap library with regard to efficiency of gene identification, genome coverage, precise genomic mapping of gene-trap mutations, and mutagenicity. As an example of our approach, we describe the generation and analysis of mice carrying a gene-trap mutation at the Wnk1 locus. WNK1 [with no lysine (K)] is a member of a recently identified family of serine/threonine kinases that lacks a key catalytic lysine (16). Despite the lack of this lysine, WNK1 has been reported to display kinase activity, phosphorylating myelin basic protein and itself (16, 17). WNK1 protein localizes to diverse chloride-transporting epithelia (18), and large intronic deletions that lead to overexpression of the WNK1 gene recently have been shown to cause pseudohypoaldosteronism type II, an autosomal dominant disorder characterized by hypertension, increased renal salt reabsorption, and impaired K+ and H+ excretion (19). Analysis of _Wnk1_-deficient mice demonstrates a corresponding role for Wnk1 in blood pressure regulation in mice.
Methods
Gene-Trapping in ES Cells and Generation of OmniBank Sequence Tags (OSTs). The procedures for infecting, selecting, and growing mouse ES cells have been described (15, 20). ES clone DNA was obtained as follows. Confluent 96-well plates were treated with lysis/binding buffer [0.2 M Tris·HCl/0.5 M LiCl/125 mM EDTA/10% lithium dodecyl sulfate (LiDS)], and each well was mixed thoroughly to fully lyse the cells. The cell lysate then was transferred to a binding plate treated with oligo(dT) (5′-NH2-TTTTTTTTTTTTTTTTTTTT-3′). After a 30-min incubation, the supernatant was removed, and the bound RNA was washed twice with wash buffer 1 (0.01 M Tris·HCl, pH 8/0.15 M LiCl/1 mM EDTA, pH 8/0.1% LiDS) and three times with wash buffer 2 (0.01 M Tris·HCl, pH 8/0.15 M LiCl/1 mM EDTA, pH 8). RNA then was eluted with the addition of elution buffer (1 mM EDTA, pH 8), followed by incubation at 70°C for 6 min. The supernatant then was transferred to a fresh 96-well plate containing reverse transcription (RT) buffer as described (15). The RT primer used was 5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT-3′.
The OST cDNA was then amplified with two rounds of nested PCR. In the first round, primers complementary to the 5′ tail of the RT primer and to the retroviral vector (5′-CCAGTGAGCAGAGTGACGAGGAC-3′ and 5′-GCCGCCGCCATGGCTCCGGTAGGTCCAGAGTCTTCAGA-3′, respectively) were used (94°C for 60 sec, 67°C for 10 min, 1 cycle; 94°C for 60 sec, 65°C for 5 min, 5 cycles; 94°C for 60 sec, 54°C for 60 sec, 65°C for 5 min, 15 cycles). In the second round, nested primers (5′-GAGGACTCGAGCTCAAGC-3′ and 5′-CCAGAGTCTTCAGAGATCAAGTC-3′) were used (94°C for 60 sec, 54°C for 60 sec, 65°C for 5 min, 35 cycles). The amplified cDNA was then cleaned by centrifugation through Sephacryl S-300 and Sephadex G-50 columns and cycle-sequenced by using standard methods with an Applied Biosystems 3700 capillary sequencer (sequencing primer, 5′-GGAGCTACCTGCATTAAGTC-3′).
Determination of Genome Coverage. An E value of _E_–4 was used as the cutoff for significant matches (21). A reciprocal blast (22) analysis of the entire Ensembl database was performed by using the best OST matches. Only those OSTs with reciprocal best matches were scored as confirmed hits.
Determination of Gene-Trap Location Along Mouse Chromosomes. Genes annotated in the Ensembl database were matched to OmniBank and plotted along their chromosomal location. To be counted as trapped, an Ensembl gene had to be matched by an OST with a minimum of 92% sequence identity and an E value of <_E_–30 (21).
Generation of OmniBank Mouse Lines. Mouse lines were generated by microinjection of OmniBank ES cell clones into host blastocysts by using standard methods (20). ES cell clones were chosen for microinjection based on confirmation of exonic or intronic insertion by the cloning of genomic insertion sites using inverse PCR (23). Gene-trap mutations were generated in ES cells derived from the 129/SvEvBrd strain. The chimeric mice were bred to C57BL/6J albino mice. All mice analyzed were of mixed genetic background (129/SvEvBrd and C57BL/6J).
Analysis of OmniBank Mutations by RT-PCR. RNA was extracted from spleen and thymus by using a bead homogenizer and RNAzol (Ambion, Austin, TX) according to the manufacturer's instructions. RT was performed with SuperScript II (Invitrogen) and random hexamer primers, according to the manufacturer's instructions. PCR amplification (95°C for 30 sec, 59°C for 45 sec, 70°C for 60 sec) was performed for 30 cycles with oligonucleotide primers (5′-CTACCTACTCGGATCTGTGTTATCA-3′ and 5′-ATGAGGATTTTGCCGCTGTTCCACTTGTAC-3′) complementary to the PolH mRNA (GenBank accession no. NM_030715) flanking the gene-trap insertion site. Control primers to the murine β actin gene were used (GenBank accession no. M12481). PCR products were verified by sequencing.
Genotyping of OmniBank Mice. Oligonucleotide primers (LTR reverse, 5′-ATAAACCCTCTTGCAGTTGCATC-3′; A, 5′-AAAATACTCTGTCAGGCTTAAGTGT-3′; and B, 5′-TGAAGCCAGGCATTAAGCACTC-3′) were used in a multiplex reaction to amplify corresponding Wnk1 (representative mouse EST AA239582) alleles on mouse chromosome 6. Approximately 125 ng of purified mouse tail genomic DNA was used as a template for PCR in a 50-μl reaction volume. Cycling conditions were 96°C for 15 sec, 62°C for 30 sec, and 72°C for 30 sec (35 cycles). Amplified products were separated on 2% agarose gels.
Quantitative PCR Analysis of Wnk1 mRNA Levels. TaqMan assay. Quantitative RT-PCR was performed on a TaqMan 7900 as described in Applied Biosystems User Bulletin no. 2 by using the following primers: forward, 5′-AGGTCTGGACACCGAAACCA-3′; reverse, 5′-GACCCTTTAACATTTCAGCCTCTTC-3′; and a 6-carboxy-fluorescein-labeled probe of 5′-TGTGAATTGCAGGATCGA-3′ (oligos were obtained from Applied Biosystems). RNA was extracted from mouse tissues and quantitated by the RiboGreen method (Molecular Probes); 200 ng was used for RT with the TaqMan RT kit (Applied Biosystems). Relative Wnk1 mRNA levels were calculated with 18S rRNA as the internal control (Applied Biosystems kit) by using the standard curve method. Student's t test (one-tailed, assuming unequal variances) was performed to analyze the statistical significance of the data.
SYBR Green assay. We performed SYBR Green quantitative PCR on an Applied Biosystems PRISM 7700, using the standard curve method of Applied Biosystems User Bulletin no. 2. The primer sequences used for Wnk1 were 5′-GACAGTCTACAAAGGTCTGGACAC-3′ and 5′-GACCCTTTAACATTTCAGCCTCTTC-3′. Control primers were Actin (5′-GGGACCTGACGGACTACCT-3′ and 5′-GCTCGT TGCCA ATAGTGATG-3′) and the TaqMan rodent Gapd control primers (Applied Biosystems). Three heterozygous animals were compared with three WT animals in triplicate.
Blood Pressure Measurements. Systolic blood pressure was measured with tail cuff in untrained conscious mice by using the Visitech BP-2000 system (Visitech Systems, Apex, NC). Blood pressures were measured 10 times per day for 4 consecutive days, and a mean value was generated for each individual mouse.
Results
Optimizing Gene-Trapping Vectors. To maximize the number of genes that can be trapped, vectors were modified to contain the first exon, splice donor sequence, and partial first intron of the murine Bruton's tyrosine kinase (Btk) gene as the 3′ trapping component, rather than a selectable marker (Fig. 1). Because of more efficient 3′ splicing, likely due to exon size, a larger group of genes can be trapped and identified by using a noncoding exon as the 3′ trapping component. Because the selectable marker in the 5′ trapping component lacks a promoter and requires transcription from the endogenous gene to select for trapping events, one might expect this strategy to produce a strong bias against trapping genes that are not expressed in ES cells. However, the data do not support this assumption. RT-PCR expression analysis was carried out on 211 genes trapped in OmniBank versus 110 genes from the mouse EST databases; both sets of genes were randomly chosen. Sixteen of 211 trapped genes (7.6%) were not expressed in ES cells, compared with 10 of 110 randomly selected genes (9.1%). The overall percentage of genes trapped, discussed below, also demonstrates that the use of selection for 5′ trapping events is not a major limiting factor in mouse ES cells.
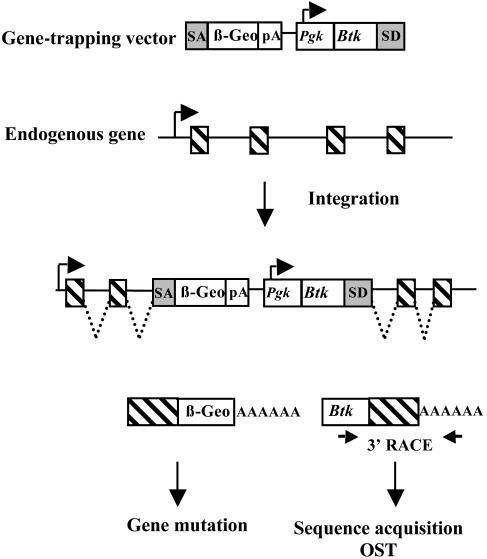
Fig. 1.
Simultaneous gene mutation and identification by gene-trapping in mouse ES cells. A retroviral vector contains a splice acceptor sequence (SA) followed by a promoterless selectable marker such as β-Geo, a functional fusion between the β-galactosidase and neomycin resistance genes, with a polyadenylation signal (pA). Insertion of the retroviral vector into an expressed gene leads to the splicing of the endogenous upstream exons (hatched boxes) into this cassette to generate a fusion transcript. The vector also contains a promoter that is active in ES cells [such as that of the mouse phosphoglycerate kinase (Pgk) gene] followed by a first exon (such as that of the Btk gene) upstream of a splice donor (SD) signal. Splicing from this signal to the exons downstream of the insertion gives rise to a fusion transcript that can be used to generate a sequence tag (OST) of the trapped gene by RACE (15). The Btk exon contains termination codons in all reading frames to prevent translation of downstream fusion transcripts.
Generation of the OmniBank Gene-Trap Library. We have used high-throughput gene-trapping with retroviral vectors in mouse ES cells to generate OmniBank, a library of 522,666 mutated ES cell clones. Each clone is frozen in duplicate in liquid nitrogen. We generated a tractable sequence tag from 271,860 clones (52%), using a third replicate of the ES cell clones that was subjected to an automated RT-PCR-based direct-sequencing protocol (15). We refer to such sequence as OSTs, which represent cDNA sequence of the mutated genes downstream of the genomic insertion site (Fig. 1).† Analysis of >3,500 Omni-Bank clones by RT-PCR using vector-specific primers in combination with primers to exons located 5′ and 3′ of the insertion demonstrates that the OST sequence is a reliable indicator of genomic location of gene-trap inserts. OSTs from 185,211 of the 271,860 sequence-tagged clones are at least 60 contiguous nucleotides in length with an average length of 319 nucleotides after screening against repetitive elements and low-complexity regions. These OSTs average 2.4% sequence ambiguities with an upper limit of 11%. When using blast-based algorithms (22), the 185,211 OSTs condensed into 42,279 nonoverlapping sequence clusters with a distribution of OST sequences per cluster (Fig. 2_A_) that suggests that the integration events are largely random. An additional 86,614 ES cell clones are identified by OSTs of 24–60 nucleotides with <15% ambiguity. In our experience, such OSTs are often sufficient to identify a trapped gene or provide a strong indication of gene identity that can be verified once a clone is thawed.
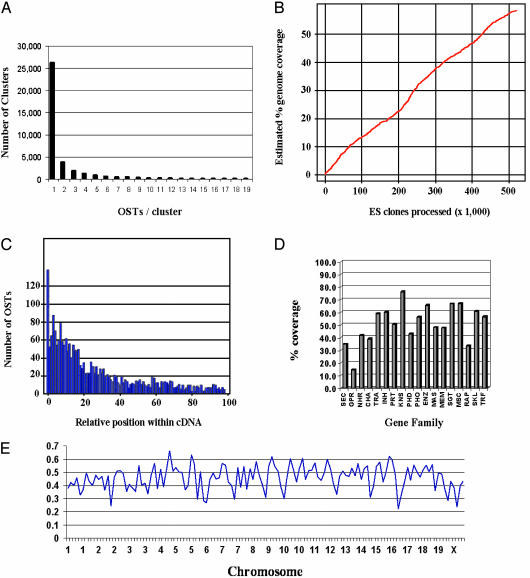
Fig. 2.
(A) Distribution of OSTs per OmniBank cluster is shown. (B) Historic progression of the estimated genome coverage of OmniBank is shown. (C) Distribution of OSTs relative to the cDNA sequence within the 2,170 genes analyzed is shown. (D) Estimated OmniBank gene coverage by gene family is shown. SEC, secreted; GPR, G protein-coupled receptor; NHR, nuclear hormone receptor; CHA, channel; TRA, transporter; INH, inhibitor; PRT, protease; KNS, kinase; PHD, phosphodiesterase; PHO, phosphatase; ENZ, enzyme; MAS, membrane and secreted; MEM, membrane; SGT, signal transduction; MSC, miscellaneous; RAP, receptor-associated protein; SKL, cytoskeletal; TRF, transcription factor. (E) Distribution of OmniBank gene-trap events throughout the mouse genome. Units on the x axis represent 20-Mbp genomic intervals. The y axis shows the percentage of genes trapped within each 20-Mbp interval. Additional detail on chromosomewide coverage is available in Fig. 6.
Genome Coverage in OmniBank. Extrapolating from a set of 3,904 full-length mouse genes used in blast searches against the OST collection, we estimate that the OmniBank library contains sequence-tagged clones representing gene-trap mutations in ≈60% of mouse genes. The sentinel set of 3,904 full-length mouse cDNAs was generated from the Ensembl database (www.ensembl.org) by using three criteria: The mouse gene must have an identified human ortholog, it must be mapped to a specific chromosomal location in the mouse genome, and it must be represented in the RefSeq database of the National Center for Biotechnology Information (www.ncbi.nih.gov/RefSeq/). We believe these criteria include sequences that are most representative of genes confirmed experimentally and supported by expression data. blast analysis of OmniBank with this reference gene list as the query demonstrated that 2,170 (55.6%) genes were represented by OSTs. Although OSTs generally represent exonic sequence, a minority represent intronic sequence, presumably because of splicing into cryptic splice acceptor sites in the genome. An additional 148 genes were represented by OSTs when the full genomic sequences within the boundaries of the genes' transcribed regions were used as queries, bringing the OmniBank coverage to 59.4%. The historic progression of OmniBank coverage indicates a relatively constant growth rate of ≈10% genome coverage per 100,000 ES cell clones processed (Fig. 2_B_).
Further examination of gene-trap events sheds light on some characteristics of retroviral insertions. To determine the integration location of gene-trap vectors within genes, we examined the OST sequence relative to the cDNA for the 2,170 genes in our reference list with exonic matches (Fig. 2_C_). There is a clear bias for integration at the 5′ end of genes. Analysis of OmniBank coverage by gene family within the reference list (Fig. 2_D_) reveals differences, from 76.0% coverage for kinases to 14.4% for G protein-coupled receptors. This variation is likely due to differences in the predominant genomic organization of genes in various families. Supporting this view, a large proportion of G protein-coupled receptors is known to be encoded by single-exon genes that are not amenable to gene-trapping.
Distribution of Gene-Trap Events. The distribution of gene-trap events is generally homogeneous across all mouse chromosomes, as depicted in Figs. 2_E_ and 6, which is published as supporting information on the PNAS web site. The percent coverage shown in Figs. 2_E_ and 6 underestimates actual percent coverage because of the automated nature of the genomewide analysis.
Generation and Analysis of OmniBank Mouse Lines. The OmniBank ES clones of interest were chosen for mouse production based on their corresponding OST. After the thawing of each OmniBank clone, the precise genomic insertion site of the vector was determined by using inverse genomic PCR (23). This insertional information confirmed the exact genomic insertion site, facilitating the genotyping of mice derived from OmniBank (Fig. 3). Importantly, this procedure also allowed for the quality control of clones before mouse production to maximize the probability of producing a null mutation. Although the majority of gene-trapping events are within the exons or introns of the trapped gene, some trapping events occur in the promoter region at the 5′ end of the first exon and are unlikely to be mutagenic. Of >1,000 clones analyzed by inverse genomic PCR to date, 16.2% fall within this category of upstream insertions and are not used for mouse production.
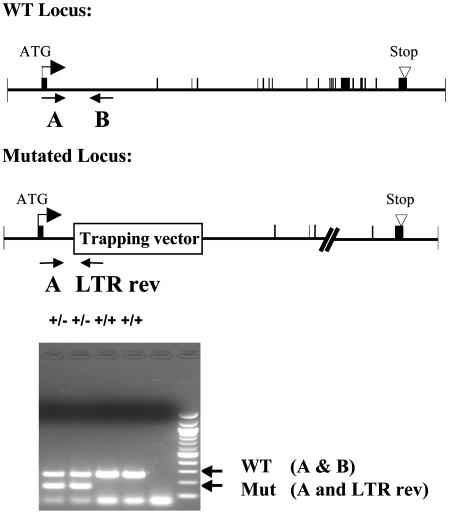
Fig. 3.
Generation of _Wnk1_-deficient mice. Primers A and B flank the genomic insertion site of the gene-trap vector in intron 1 of the Wnk1 gene and amplify a product for the WT allele. The LTR reverse primer, complementary to OmniBank vectors, was used in conjunction with primer B to specifically amplify the mutated allele. Rev, reverse; Mut, mutated.
To date, 741 lines of mice from the OmniBank resource have been produced and studied to determine the mutagenic effects of gene-trap insertions. Gene-trap insertions mutate genes by forcing exons at the 5′ end of the insertion to splice with the splice acceptor of the gene-trap vector and by preventing them from splicing into exons at the 3′ end of the insertion, thereby preventing production of the endogenous transcript. To systematically evaluate this ability of gene-trap insertions to prevent the production of the endogenous transcript in homozygous mutants, RNA is isolated from selected tissues known to normally express the transcript and subjected to RT-PCR using primers complementary to exons flanking the insertion site. An example of our standard RT-PCR quality control is depicted in Fig. 4 for the mouse PolH gene. Analysis of non-embryonic lethal mouse lines demonstrates that gene-trap insertions within both exons and introns of the gene of interest lead to the disruption of the endogenous mRNA transcript in all cases. Of these, >96% show complete absence of WT message, with the remaining lines showing an average reduction in mRNA levels of 91.6% as measured by quantitative PCR. These data demonstrate that intragenic insertion efficiently disrupts gene transcription in vivo and can be used to reliably predict mutagenicity before mouse production.
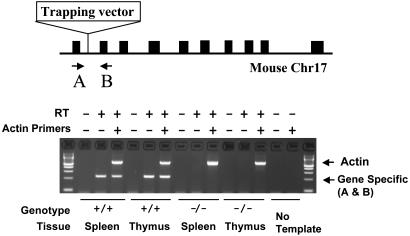
Fig. 4.
Assessing the mutagenicity of OmniBank gene-trap insertions by RT-PCR. Primers A and B are complementary to exons flanking the insertion site in the PolH gene in mouse chromosome 17 (Chr17) (GenBank accession no. NM_030715). RT-PCR using primers A and B shows the absence of endogenous message in the spleen and thymus of homozygous animals. Control primers to the murine β actin gene were used (GenBank accession no. M12481).
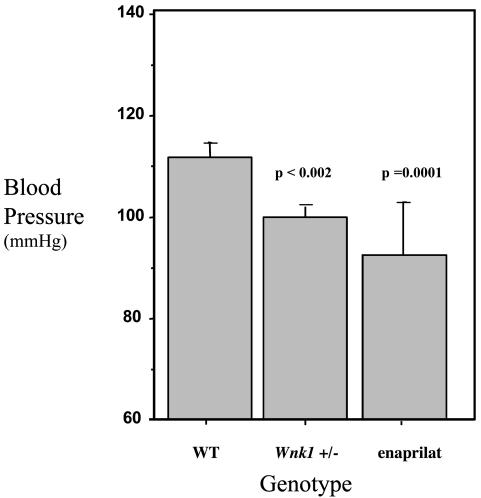
Generation and Analysis of Wnk1-Deficient Mice. The ES cell clone represented by OST 38262 was chosen for the generation of _Wnk1_-deficient mice based on sequence identity to the published mouse Wnk1 EST sequences. Inverse genomic PCR (23) of DNA isolated from OST 38262 cells confirmed that the retroviral gene-trap vector had inserted in intron 1 of the mouse Wnk1 gene on chromosome 6, downstream of the exon encoding the initiation methionine (Fig. 3). Mice heterozygous for the mutation were generated by using standard methods of host embryo microinjection, chimera production, and germ-line transmission (20). Heterozygous intercrosses yielded no homozygous animals (17 WT and 44 heterozygous pups, from a total of 61), demonstrating that Wnk1 is required for embryonic development. Analysis of timed matings revealed that no homozygous embryos survived past day 13 of gestation. Mice heterozygous for the Wnk1 gene-trap mutation were subjected to our standard phenotypic analysis protocol, which is applied to all lines generated and includes tests of behavior, immunology/inflammation, obesity/diabetes, bone, cell proliferation, and cardiovascular function (described in detail in ref. 24). Wnk1 heterozygotes displayed a reduction in blood pressure of ≈12 mmHg (1 mmHg = 133 Pa) (Fig. 5) and no notable phenotype in all other areas examined (9, 24). This decrease in blood pressure was not related to any pathological changes in the kidney, because no notable differences in kidney histology were observed (data not shown).
Fig. 5.
Lower blood pressure in heterozygous Wnk1 mice. Blood pressure was measured in untreated WT (n = 20), Wnk1 heterozygotes (n = 11), and WT animals treated with 1.25 mg of the antihypertensive drug enaprilat per kg (n = 3). [Enaprilat is an inhibitor or angiotensin-converting enzyme (ACE), a key regulator of blood pressure in mouse and man.] Blood pressures were measured 10 times per day for 4 consecutive days, and a mean value was generated for each individual mouse. n, Number of mice per cohort.
To demonstrate that the decrease in blood pressure in heterozygotes was due to a decrease in Wnk1 mRNA expression, Wnk1 mRNA was quantitated by using both SYBR Green and TaqMan quantitative PCR assays. Wnk1 heterozygotes exhibited a 41% and a 57% reduction in Wnk1 mRNA levels in the kidney and the heart, respectively, by the SYBR Green method. A reduction in expression of Wnk1 of 49% and 45% was observed in kidney and thymus, respectively, by the TaqMan method. Recent DNA microarray and proteomics studies have demonstrated a general correlation between mRNA and protein levels (25, 26).
To further evaluate kidney function, blood chemistry and urinalysis were carried out on Wnk1 heterozygotes. Plasma blood urea nitrogen and creatinine levels were within the normal parameters, as were levels of urinary protein, blood, urobilinogen, glucose, nitrite, ketone, and leukocytes, suggesting normal kidney function. Expression studies in Xenopus oocytes suggest a role of WNK kinases in the regulation of thiazide-sensitive Na–Cl cotransport (27). Urinary electrolytes were measured, and chloride, potassium, calcium, and phosphorus levels were all normal, indicating no abnormalities in electrolyte excretion. To determine whether Wnk1 heterozygotes could retain electrolytes normally when challenged with a low-salt diet, mice were fed a low-salt diet (0.01% NaCl) for 3 weeks. During the last week, the animals were placed in metabolic cages to measure water and food intake, as well as urinary output. There were no significant differences in water and food intake or urinary output between animals fed a low-salt diet and those fed a normal diet, nor were there any differences in these parameters between Wnk1 heterozygotes and WT animals. Although both the heterozygotes and WT animals showed reduced urinary electrolyte output to compensate for the lack of salt intake, there was no significant difference in urine sodium, potassium, or chloride levels, indicating that Wnk1 haploinsufficiency does not affect the ability of mice to conserve electrolytes.
Discussion
The OmniBank library contains ES cell clones carrying mutations in ≈60% of all genes and can be used for functional annotation of the genome through reverse genetics. Our data indicate that it should be possible to further increase genome coverage by collecting and sequencing more gene-trapped clones, although gene structure and other factors would pose an upper limit to the percentage of genes that can be trapped. A genomewide reverse genetic functional screen is likely to require a combination of technological approaches. We are using the OmniBank gene-trap resource in combination with gene targeting by homologous recombination to produce knockout mice that are subjected to a broad phenotypic screen to understand gene function. The Wnk1 gene is one example of a gene that was analyzed as part of this process.
The role of WNK1 in regulating human blood pressure has been corroborated through the generation and analysis of mutant mice; activating human mutations lead to hypertension, whereas deficiency in the mouse leads to hypotension. Lethality of Wnk1 homozygotes demonstrates the essential role of Wnk1 in embryonic development. The decrease in blood pressure displayed by Wnk1 heterozygotes (mean = 12.2 mmHg) is analogous to that observed in mice heterozygous for a targeted mutation in the type 1A angiotensin II receptor gene (12 mmHg), a known regulator of blood pressure in mouse and man (28, 29) and the target of antagonist drugs widely used for the treatment of hypertension (30). A heterozygous effect (a decrease of 15–20 mmHg) has also been reported for mice carrying a targeted mutation in the angiotensin-converting enzyme (ACE) gene (31), another target of antihypertensive drugs such as enaprilat (Fig. 5). The fact that heterozygous mice display a significant decrease in blood pressure while still carrying a functional copy of the gene suggests that even incomplete inhibition of WNK1 in humans could lead to significant antihypertensive effects.
Supplementary Material
Supporting Figure
Abbreviations: ES, embryonic stem; OST, OmniBank sequence tag.
Data deposition: The sequences reported in this article have been deposited in the GenBank database (accession nos. CG472819–CG671551).
Footnotes
References
- 1.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420**,** 520–562. [DOI] [PubMed] [Google Scholar]
- 2.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409**,** 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Adams, M. D. & Sekelsky, J. J. (2002) Nat. Rev. Genet. 3**,** 189–198. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen, E. M. & Mango, S. E. (2002) Nat. Rev. Genet. 3**,** 356–369. [DOI] [PubMed] [Google Scholar]
- 5.Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K., Farrington, S., et al. (2002) Nat. Genet. 31**,** 135–140. [DOI] [PubMed] [Google Scholar]
- 6.Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., et al. (2002) Plant Cell 14**,** 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, A. (2002) Nature 420**,** 512–514. [DOI] [PubMed] [Google Scholar]
- 8.Sands, A. T. (2003) Nat. Biotechnol. 21**,** 31–32. [DOI] [PubMed] [Google Scholar]
- 9.Zambrowicz, B. P. & Sands, A. T. (2003) Nat. Rev. Drug Discov. 2**,** 37–50. [DOI] [PubMed] [Google Scholar]
- 10.Capecchi, M. R. (1989) Science 244**,** 1288–1292. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich, G. & Soriano, P. (1991) Genes Dev. 5**,** 1513–1523. [DOI] [PubMed] [Google Scholar]
- 12.Skarnes, W. C., Auerbach, B. A. & Joyner, A. L. (1992) Genes Dev. 6**,** 903–918. [DOI] [PubMed] [Google Scholar]
- 13.Zambrowicz, B. P. & Friedrich, G. A. (1998) Int. J. Dev. Biol. 42**,** 1025–1036. [PubMed] [Google Scholar]
- 14.Balling, R. (2001) Annu. Rev. Genomics Hum. Genet. 2**,** 463–492. [DOI] [PubMed] [Google Scholar]
- 15.Zambrowicz, B. P., Friedrich, G. A., Buxton, E. C., Lilleberg, S. L., Person, C. & Sands, A. T. (1998) Nature 392**,** 608–611. [DOI] [PubMed] [Google Scholar]
- 16.Xu, B., English, J. M., Wilsbacher, J. L., Stippec, S., Goldsmith, E. J. & Cobb, M. H. (2000) J. Biol. Chem. 275**,** 16795–16801. [DOI] [PubMed] [Google Scholar]
- 17.Xu, B. E., Min, X., Stippec, S., Lee, B. H., Goldsmith, E. J. & Cobb, M. H. (2002) J. Biol. Chem. 277**,** 48456–48462. [DOI] [PubMed] [Google Scholar]
- 18.Choate, K. A., Kahle, K. T., Wilson, F. H., Nelson-Williams, C. & Lifton, R. P. (2003) Proc. Natl. Acad. Sci. USA 100**,** 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson, F. H., Disse-Nicodeme, S., Choate, K. A., Ishikawa, K., Nelson-Williams, C., Desitter, I., Gunel, M., Milford, D. V., Lipkin, G. W., Achard, J. M., et al. (2001) Science 293**,** 1107–1112. [DOI] [PubMed] [Google Scholar]
- 20.Joyner, A. L. (2000) Gene Targeting: A Practical Approach (Oxford Univ. Press, Oxford).
- 21.Karlin, S. & Altschul, S. F. (1990) Proc. Natl. Acad. Sci. USA 87**,** 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. (1997) Nucleic Acids Res. 25**,** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silver, J. & Keerikatte, V. (1989) J. Virol. 63**,** 1924–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.BeltrandelRio, H., Kern, F., Lanthorn, T., Oravecz, T., Piggott, J., Powell, D., Ramirez-Solis, R., Sands, A. T. & Zambrowicz, B. in Model Organisms in Drug Discovery, ed. Carroll, P. (Wiley & Sons, Chichester, U.K.), in press.
- 25.Lian, Z., Kluger, Y., Greenbaum, D. S., Tuck, D., Gerstein, M., Berliner, N., Weissman, S. M. & Newburger, P. E. (2002) Blood 100**,** 3209–3220. [DOI] [PubMed] [Google Scholar]
- 26.Kern, W., Kohlmann, A., Wuchter, C., Schnittger, S., Schoch, C., Mergenthaler, S., Ratei, R., Ludwig, W. D., Hiddemann, W. & Haferlach, T. (2003) Cytometry 55B**,** 29–36. [DOI] [PubMed] [Google Scholar]
- 27.Yang, C.-L., Angell, J., Mitchell, R. & Ellison, D. H. (2003) J. Clin. Invest. 111**,** 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, M., Oliverio, M. I., Mannon, P. J., Best, C. F., Maeda, N., Smithies, O. & Coffman, T. M. (1995) Proc. Natl. Acad. Sci. USA 92**,** 3521–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugaya, T., Nishimatsu, S., Tanimoto, K., Takimoto, E., Yamagishi, T., Imamura, K., Goto, S., Imaizumi, K., Hisada, Y., Otsuka, A., et al. (1995) J. Biol. Chem. 270**,** 18719–18722. [DOI] [PubMed] [Google Scholar]
- 30.Oliverio, M. I. & Coffman, T. M. (1997) Clin. Cardiol. 20**,** 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krege, J. H., John, S. W., Langenbach, L. L., Hodgin, J. B., Hagaman, J. R., Bachman, E. S., Jennette, J. C., O'Brien, D. A. & Smithies, O. (1995) Nature 375**,** 146–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure