Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation (original) (raw)
Abstract
When transplanted into Xenopus oocytes, the nuclei of mammalian somatic cells are reprogrammed to express stem cell genes such as Oct4, Nanog, and Sox2. We now describe an experimental system in which the pluripotency genes Sox2 and Oct4 are repressed in retinoic acid-treated ES cells but are reprogrammed up to 100% within 24 h by injection of nuclei into the germinal vesicle (GV) of growing Xenopus oocytes. The isolation of GVs in nonaqueous medium allows the reprogramming of individual injected nuclei to be seen in real time. Analysis using fluorescence recovery after photobleaching shows that nuclear transfer is associated with an increase in linker histone mobility. A simultaneous loss of somatic H1 linker histone and incorporation of the oocyte-specific linker histone B4 precede transcriptional reprogramming. The loss of H1 is not required for gene reprogramming. We demonstrate both by antibody injection experiments and by dominant negative interference that the incorporation of B4 linker histone is required for pluripotency gene reactivation during nuclear reprogramming. We suggest that the binding of oocyte-specific B4 linker histone to chromatin is a key primary event in the reprogramming of somatic nuclei transplanted to amphibian oocytes.
Keywords: chromatin, nuclear transfer, Xenopus
The transfer of somatic cell nuclei to eggs is an experimental means of reversing the process of cell differentiation in which cells become progressively restricted in the developmental pathways open to them (1–3). By comparison with other procedures (4, 5), nuclear transfer (NT) is relatively efficient (6), and it makes use of natural components of eggs without any accompanying change to the genome. A variant of this technique is NT to Xenopus oocytes (7). Although no new cell types are generated in this type of NT, reactivation of pluripotency genes takes place within a day after NT and in the absence of cell division. The oocyte (M1 prophase I) is the immediate progenitor of an egg (M2 metaphase) and is believed to reprogram transcription in the same way that an egg reprograms the sperm nucleus after fertilization. Therefore the direct and efficient transcriptional reprogramming activity of the Xenopus oocyte makes it a favorable cell in which to analyze an important part of the mechanism of nuclear reprogramming.
Our aim is to understand the mechanism of reprogramming by NT in eggs and oocytes. The substrate for this reprogramming activity is the chromatin of transplanted nuclei. The chromatin of eukaryotes contains DNA wrapped around the four core histones arranged as a nucleosome. The linker DNA joining two nucleosomes is also bound by chromatin proteins such as linker histones, high mobility group proteins (8, 9), and poly (ADP-ribose) polymerase 1 (10). Several linker histone variants are present in somatic cells, and the ratio of the various forms varies from one cell type to another (11). Linker histones initially were thought to have a general function in repressing gene activity. Recent work has demonstrated that linker histones also are involved in a more specific regulation of gene activity (12). The oocytes and eggs of both frogs and mammals contain an oocyte-specific linker histone (B4 in Xenopus, H1foo in mouse) (13–16). This oocyte variant is associated with the phase of development when the embryo is dependent on maternal transcripts, i.e., before zygotic genome activation. In Xenopus embryos, the transition from an oocyte to a somatic type of linker histone modulates cell fate in response to a morphogen (17). In nuclear transplantation in mouse and Xenopus, a loss of somatic H1 and incorporation of oocyte linker histone have been described (18–22). However, it remains unclear to what extent these changes are causally connected with the transcriptional reactivation of pluripotent genes.
Here we analyze acquisition of linker histones by transplanted nuclei, using a variant of a NT experiment in which differentiated ES nuclei are reprogrammed by oocytes in first meiotic prophase. This approach enables us to monitor linker histone exchanges in single transplanted nuclei in real time. The changes undergone by the transplanted nuclei are related to transcriptional activation but not to DNA replication. We find that gene activation in transplanted nuclei is related to an increase in chromosomal protein mobility. Most importantly, we show that the uptake of linker histone B4 is required for efficient pluripotency gene activation.
Results
Nuclei of Differentiated ES Cells Are Reprogrammed with High Efficiency Following NT to Xenopus Oocytes.
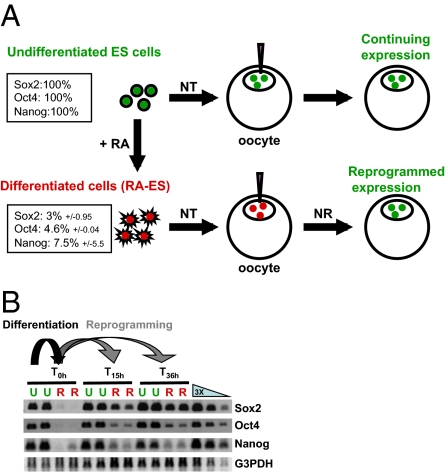
To quantify transcriptional reprogramming by oocytes, we measured transcription in nuclei whose pluripotency genes are in either an active or a repressed state before transplantation (Fig.1_A_). Such nuclei are obtained from undifferentiated ES cells or retinoic acid-differentiated ES cells (RA-ES). Incubation of RA-ES nuclei for 24 h in an oocyte shows reactivation up to 100% of the ES cell level for Sox2 and to a lesser extent for Oct4 and Nanog (Fig.1_B_ and Fig. S1). Therefore we have a system in which nuclear reprogramming takes place efficiently and in a short time (24 h at 14 °C). It allows the extent of reactivation of the three pluripotency genes to be analyzed and so provides a quantitative assay to test the importance of oocyte reprogramming components.
Fig. 1.
Transplantation of nuclei from growing and differentiated ES cells allows quantification of gene reactivation. (A) Experimental design. NT, nuclear transfer; NR, nuclear reprogramming; RA, retinoic acid. Percentages in boxes show the level of gene expression as measured by real-time PCR on cultured cells before nuclear transfer (three experiments). (B) Example of RT-PCR analysis of the experiment depicted in A. U, undifferentiated ES nuclei; R, RA-differentiated ES nuclei.
Real-Time Monitoring of Nuclear Reprogramming.
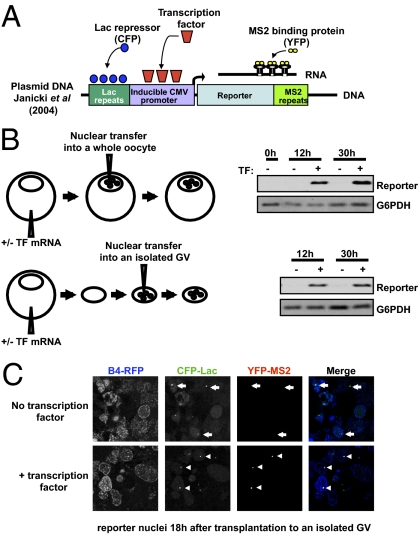
To analyze the reprogramming events leading to transcriptional reprogramming, we adapted an experimental system that allows us to monitor nuclear reprogramming in real time, and in individual nuclei, following NT. As described in Fig. 1, NT allows an accurate measurement of gene reactivation by RT-PCR, but the opacity of the Xenopus oocyte does not allow the transcriptional activation of individual transplanted nuclei to be seen in real time. We therefore isolated the germinal vesicle (GV) in a nonaqueous medium (23), because the reprogramming activity of the oocyte is located in its nucleus (the GV) (7). To facilitate the recognition of reprogramming in real time, we used nuclei from a human cell line (U2OS) containing an integrated series of numerous copies of the Lac repressor binding site (Fig. 2_A_) (24, 25). The binding of CFP-Lac protein identifies the position of this construct in a nucleus. The integrated construct also contains a cytomegalovirus (CMV) promoter with numerous binding sites for a transcription factor. Reporter transcripts from this construct can be seen by the binding of MS2-YFP protein to multiple sites on the mRNA (Fig. 2_A_). As shown in Fig. 2_B_, U2OS nuclei transplanted into the GV of an intact oocyte or into an isolated GV are able to activate the reporter as long as the transcription factor binding is provided by mRNA injected into oocytes. Therefore, gene activation takes place following NT into the isolated GV of a Xenopus oocyte.
Fig. 2.
Real-time monitoring of gene activation in nuclei transplanted to isolated Xenopus oocyte GVs. (A) The diagram shows an inducible CMV promoter (24) flanked by lacR binding site repeats thereby allowing visualization of the reporter gene in cells (by binding of CFP-LacR chimeric protein). The promoter drives the expression of a reporter mRNA containing MS2 binding site repeats, so that the transcribed mRNA at the transcription site can be visualized using a MS2-YFP chimeric protein. Nuclei containing this reporter gene for real-time imaging of transcription (24) were used for transplantation into isolated GVs. (B) Genes are reactivated in nuclei transplanted into a GV that already has been isolated from a Xenopus oocyte. Nuclei containing a reporter gene (diagram in A) were transplanted into the GV of a whole oocyte or into an isolated GV. Transplantation was carried out into GVs expressing or not expressing a transcription factor (TF) that is required to activate the reporter gene. RT-PCR analysis shows transcription factor-dependent activation of the reporter gene from both sets of transplanted nuclei. (C) Expression of the B4-RFP protein in the GV allows visualization of transplanted nuclei (Fig. 3), whereas expression of CFP-LacR shows the position of the reporter genes in these nuclei (dots in CFP-Lac channel). The MS2-YFP protein also expressed in the GV shows newly synthesized mRNA at the gene loci, but only when the GV contains the transcription factor specific to the reporter gene (Compare arrows in Upper with arrowheads on Lower Merge image).
The transparency of the isolated GV enables gene activation to be seen in individual nuclei in real time. Fig. 2_C_ (accompanied by Movie S1) shows the production of mRNA at the transcription sites. Therefore this procedure allows the real-time monitoring of early steps of nuclear reprogramming in experimental conditions compatible with the transcriptional activation of a gene. Using this experimental setting, we can test the importance of oocyte reprogramming factors, including B4, in real time.
Linker Histone Exchange in Transplanted Nuclei Is an Early Remod-eling Event.
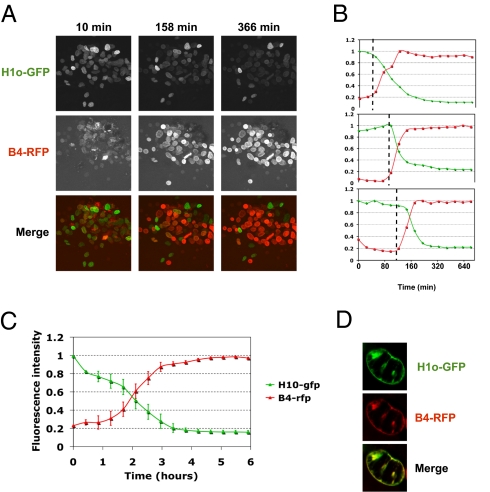
The Xenopus oocyte contains a large stock of maternal histone proteins, enough for the assembly of chromatin on the thousands of embryonic nuclei that are generated following fertilization and before zygotic activation of transcription (26). Using our real-time monitoring assay, we have tested whether chromatin components are exchanged between transplanted nuclei and the surrounding nucleoplasm. Linker histones are exchanged when Xenopus erythrocyte nuclei are incubated in Xenopus egg extracts (19, 20). We investigated whether such a global exchange occurs during transcriptional reprogramming. We transplanted nuclei containing chromatin-associated somatic linker histone variants H1c- or H1o-GFP 27 into GVs containing the Xenopus oocyte-specific linker histone variant B4-RFP. Real-time monitoring of individual transplanted nuclei by confocal microscopy shows a rapid loss of somatic linker histone from chromatin and the incorporation of oocyte linker histone (Fig. 3_A_ and Movie S2). Quantitative analysis of somatic- and oocyte-linker histone levels shows that linker histone movements take place simultaneously in individual nuclei (Fig. 3_B_) and are completed within 3 h in almost all injected nuclei (Fig. 3_C_). Moreover, B4 is incorporated in the general position previously occupied by H1, namely into chromatin, as shown by the colocalization of the two proteins at time points when B4 and H1 coexist in the same transplanted nucleus (Fig. 3_D_). These observations suggest an exchange mechanism in which somatic linker histone is competitively removed from transplanted nuclei by oocyte linker histone.
Fig. 3.
Real-time monitoring of linker histone exchange during nuclear reprogramming, (A) Xenopus oocyte linker histone (B4) replaces somatic linker histone (H1) following NT. Real-time monitoring of H1o-GFP (present in NIH 3T3 nuclei before transplantation) (27) and B4-RFP (expressed in the oocyte by mRNA injection) was carried out during the first 6 h of reprogramming. (A complete series of images is shown in Video S1). (B) Three individual nuclei from the sequence depicted in A were quantified for fluorescence intensity of H1 (green line) and B4 (red line). (C) Average change in fluorescence intensity with time in the experiment shown in A. Errors bars indicate ± SEM (n = 11 nuclei). (D) Confocal images of a nucleus at a time point following transplantation into an oocyte when B4 starts to be incorporated into chromatin and H1 is not yet completely lost from the nucleus.
By contrast, when core histone loss from transplanted nuclei is monitored within the same time frame, only a small loss, if any, is observed. Nuclei expressing H2B-GFP show only a minimal loss of this protein for at least 6 h following NT (Fig. S2 and Movie S3). Similarly, nuclei transplanted into a GV containing a fluorescent version of mammalian core histone H3.1 or H3.3 do not show any incorporation of these proteins in less than 1 day of incubation (Fig. S3).
These results demonstrate that core histones remain stably associated with transplanted nuclei, whereas linker histones are exchanged rapidly. This latter global-scale exchange of a chromatin component following NT takes place before any gene reactivation can be detected in transplanted nuclei (new transcripts are first detected 6 h after NT). We therefore conclude that the loss of the somatic linker histone and the incorporation of its oocyte-specific counterpart precede the transcriptional reactivation of transplanted nuclei.
Loss of Somatic Linker Histone H1 Is Not Required for Gene Reactivation.
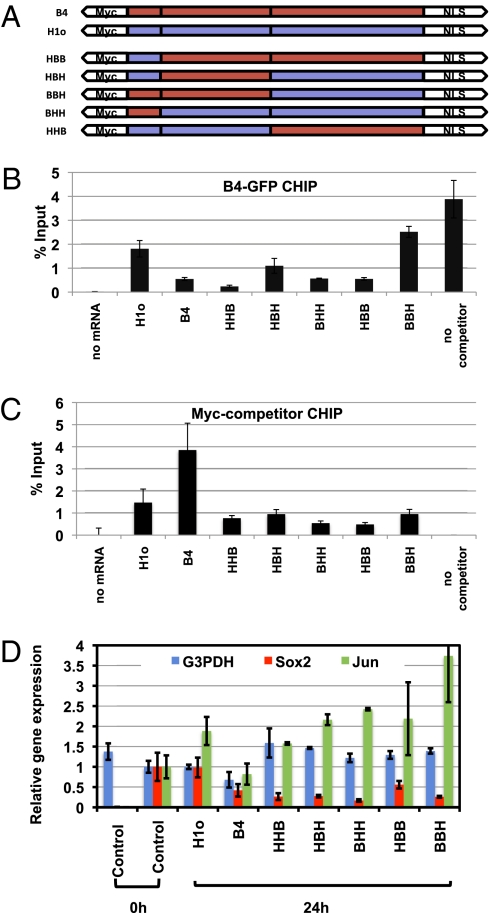
To prevent the loss of somatic linker histone from transplanted nuclei, we overexpressed H1 in oocytes by mRNA injection. Through such overexpression we obtained oocytes whose GVs contained a level of H1 more than five times greater than that of the endogenous B4 Fig S4_D_ (H1o). The overexpressed H1 is found associated to chromatinized plasmid (Fig. 4_C_, H1o). When transplanted into H1-containing oocytes, nuclei retain the overexpressed H1 on their chromatin (Fig. 5_A_). Therefore H1 overexpression in the oocyte is sufficient to prevent the loss of H1 from the chromatin of transplanted nuclei. Surprisingly, a global analysis shows that B4 is incorporated into transplanted nuclei whether H1 is lost or not (Fig. 5_A_). Therefore, it seems that a large number of B4 binding sites on chromatin are unaffected by H1 binding. Similarly, ChIP analysis on plasmid DNA shows that H1o competes only partially for B4 binding sites (see ref. 28 and Fig. 4 B and C).
Fig. 4.
Reactivation of a pluripotent gene in transplanted nuclei is prevented by expression of chimeric B4/H1 linker histone. (A) Structure of chimeric linker histones. Amino-terminal, globular, and carboxyl-terminal domains of oocyte- and somatic-linker histones were swapped to generate chimeric linker histones. (B) Chimeric linker histones interfere with B4 binding to chromatin. ChIP analysis of B4-GFP binding to chromatin was performed in the presence of chimeric linker histone as in Fig S4_C_ (B4-GFP, 0.2-fold endogenous B4; Myc-tagged chimeric linker histone, 6- to 9-fold endogenous B4). Error bars indicate the mean ± SEM of triplicate samples. (C) All chimeric linker histones are found associated with chromatin. ChIP analyses with an Myc antibody were performed in same experimental conditions as in B. Error bars represent the mean ± SEM of triplicate samples. (D) Pluripotency gene reactivation is inhibited by chimeric linker histones. RA-ES cells were transplanted into oocytes overexpressing the chimeric linker histones described in A, and gene expression was analyzed by RT-PCR 24 h after NT. The gene reactivation level from RA-ES nuclei 24 h after transplantation into control oocytes is set to 1. Gene expression was averaged from triplicate samples and normalized to the G3PDH level. Error bars indicate the mean ± SEM.
Fig. 5.
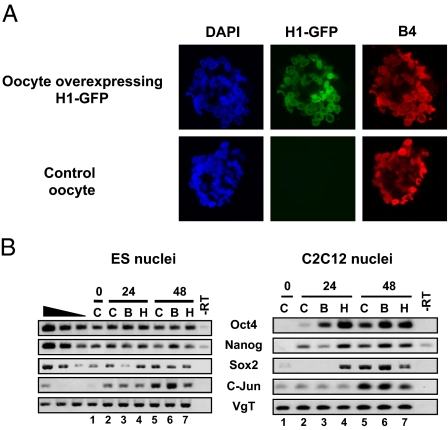
Loss of somatic H1 from transplanted nuclei is not required for gene reactivation. (A) H1 overexpression in the oocyte maintains H1 presence in transplanted nuclei but does not inhibit B4 incorporation. C2C12 nuclei were transplanted into control or H1-GFP-expressing oocytes. B4-RFP was detected by immunolabeling 24 h after NT. Loss of H1 from transplanted nuclei (Fig. 3A) can be prevented by expressing H1-GFP in the oocyte before NT (Upper Row, Center). B4 is loaded onto chromatin (Right), whereas H1-GFP is maintained (Upper Row, Center) or not (Bottom Row, Center). (B) Pluripotency genes are reactivated even when H1 is maintained in transplanted ES or C2C12 nuclei. RT-PCR analysis was carried out under conditions depicted in A. Pluripotency genes are reactivated whether H1 is maintained or not in transplanted nuclei (compare columns 5 and 7). Data are from three experiments. B, oocyte injected with B4 mRNA; C, control oocyte; H, oocyte injected with H1 mRNA.
Using these experimental conditions, we specifically tested whether the loss of H1 is required for transcriptional reprogramming, because the artificial maintenance of H1 in transplanted nuclei does not affect the bulk of B4 incorporation. Using pluripotent nuclei (from ES cells) and committed-cell nuclei (C2C12), we found that (i) pluripotent ES nuclei continue expressing Oct4, Nanog, and Sox2 for up to 48 h after NT in oocytes with or without H1 (Fig. 5_B_, Left, compare lanes 5–7) and (ii) committed C2C12 cell nuclei (Fig. 5_B_, Right, compare lanes 5–7) or RA-ES nuclei (Fig. 4_D_, compare Sox2 reactivation in control and H1o at 24 h) reactivate pluripotency genes to the same level, regardless of H1 removal. Therefore the maintenance of H1 in transplanted nuclei does not affect the continuing expression of pluripotency genes or their reactivation from transplanted nuclei.
Linker Histone Mobility Increases During Nuclear Reprogramming.
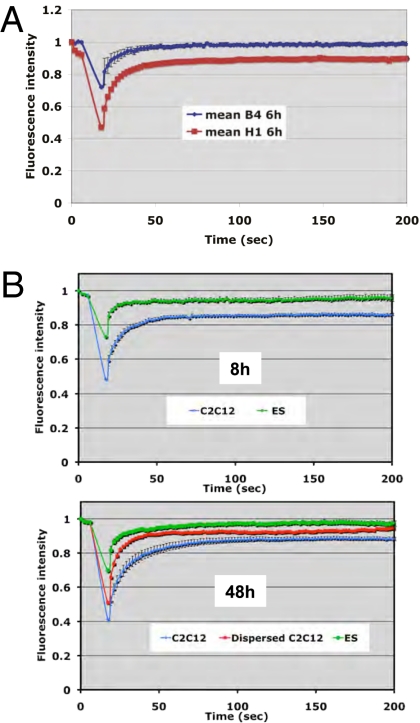
Recent work has shown that a characteristic of pluripotent cells is to exhibit a higher mobility than differentiated cells for a range of chromatin components, including core histone H3, heterochromatin protein 1, and linker histone H1 (29). We tested the possibility that the oocyte reverses gene transcription from a differentiated state by increasing the mobility of chromatin proteins, including linker histones. Fluorescence recovery after photobleaching (FRAP) analysis on nuclei 6 h after transplantation into oocytes expressing B4-GFP or H1o-GFP shows greater mobility of the oocyte linker histone than in the somatic type (Fig. 6_A_). We conclude that in NT experiments to Xenopus oocytes, linker histone mobility changes from a relatively immobile form (somatic H1) to a highly mobile form (oocyte B4). Therefore linker histone exchange following NT results in an increase in chromatin protein mobility that contrasts with the decrease in chromatin mobility observed during differentiation of ES cells (29).
Fig. 6.
Nuclear reprogramming is associated with an increase in linker histone mobility. (A) B4 is more mobile than H1 in transplanted nuclei. HeLa cell nuclei were transplanted into oocytes overexpressing B4-GFP or H1-GFP (mRNA injection). Linker histone mobility then was measured by FRAP 6 h after nuclear transplantation. The differences in bleach depth (B4 ∼30%, H1 ∼50%) as well as the extent of recovery (B4 ∼100%, H1 ∼90%) reflect the difference in mobility between B4 and H1. Error bars represent the mean ± SEM (n = 10 nuclei). (B) H1 mobility increases during nuclear reprogramming. Using oocytes overexpressing H1-GFP, we measured the mobility of H1 during nuclear reprogramming by FRAP analysis. ES cells (green lines) show high mobility 8 h and 48 h after NT. Committed cell nuclei (C2C12) show a fraction of H1 with low mobility (blue line) 8 h after NT. At 48 h after NR, H1 mobility in C2C12 nuclei (blue line) increases toward that observed in ES cell nuclei (red line). Error bars indicate mean ± SEM (n = 10 nuclei).
We then compared the mobility of H1 artificially maintained in pluripotent or committed cell nuclei over a longer period. We first observed that H1 on transplanted ES cell nuclei is highly mobile, with no immobile fraction at any time point tested (green line in Fig. 6_B_). By contrast we observed that C2C12 nuclei exhibit an immobile fraction at early time points after NT (blue line in Fig. 6_B_, Upper). But with longer incubation times (48 h), H1 mobility is greater in a subset of C2C12 nuclei showing dispersion of chromatin than in ES cells (Fig. 6_B Lower_, red line). In conclusion we observe the same difference in H1 mobility in ES versus C2C12 cells at early time points following NT as found by others in normal cultured cells (29). We also found that H1 artificially maintained in the chromatin of transplanted nuclei increases its mobility during nuclear reprogramming. The timing of the increase in H1 mobility in C2C12 nuclei correlates with the reactivation of pluripotency genes in these nuclei (Fig. 5_B_ and Fig. 6_B_). It therefore is possible that a change in the mobility of H1, rather than its mere presence/absence, may be a critical parameter for transcriptional reprogramming. The change in H1 mobility could result from modification of H1 itself or of H1 binding sites on chromatin.
B4 Is Enriched at the Promoter of Reactivated Pluripotency Genes.
Because the loss of H1 is not critical for gene reactivation, we next tested whether the incorporation of B4 into nuclei is a necessary step toward reprogramming. ChIP analysis demonstrates that B4 protein is accumulated at the promoter of all three pluripotency genes 6 h following NT (Fig. 7_A_). B4 is found more abundantly in RA-ES than in ES chromatin, presumably reflecting the heterochromatinization observed during RA-differentiation (30). When reprogramming is allowed to proceed for a longer time (25 h), B4 binding to pluripotency genes in RA-ES cells decreases to the levels seen in ES cells (Fig. 7_A_, 25 h). This decrease is seen in particularly Sox2, which is the gene that is reactivated most efficiently during this period (Discussion). However, B4 binding to heterochromatic regions such as the Major satellite, which represents 3.5% of the genome (30), is not affected (Fig. 7_A_, blue bars).
Fig. 7.
Incorporation of B4 into transplanted nuclei is necessary for pluripotency gene expression in oocytes. (A) ChIP analysis of B4 binding to pluripotency genes in transplanted nuclei. B4 binding to pluripotency gene promoters and to the major satellite region of ES or RA-ES nuclei was analyzed by ChIP 0.5 h, 6 h, and 25 h following transplantation. Error bars represent mean ± SEM. Data are from two experiments. No Ab, control chip performed in the absence of antibody. (B) Incorporation of B4 in transplanted nuclei (control Ab) can be prevented by injection of anti-B4 antibody into the oocyte GV (Lower Row, Center). C2C12 nuclei were transplanted with or without anti-B4 antibody into oocytes expressing B4-GFP. Then B4-GFP labeling of transplanted nuclei was detected by confocal microscopy. The graph on the right shows quantification of the B4-GFP signal. Error bars indicate mean ± SEM (n = 15 nuclei). (C) Pluripotency gene reactivation requires incorporation of B4 into transplanted nuclei. RT-PCR analysis was carried out in conditions depicted in B. Gene reactivation is inhibited in anti-B4 samples at 24 h in both ES and RA-ES transplanted nuclei. Data are from three experiments.
B4 Incorporation Is Required for Pluripotency Gene Reactivation Following NT.
We used a polyclonal antibody directed against B4, which, when injected in the GV of an oocyte containing fluorescently labeled B4, triggers the formation of large aggregates of the labeled B4. Nuclei transplanted into GVs containing the anti-B4 antibody failed to incorporate B4, whereas nuclei transplanted into control GVs contain a high level of B4 (Fig. 7_B_). Using anti-B4 injection, we then measured gene expression in NT experiments in which B4 incorporation was prevented. As shown in Fig. 7_C_, preventing the incorporation of B4 protein led to a decrease of pluripotency gene expression from ES cell nuclei and to a failure to reactivate these genes from differentiated cell nuclei. In both sets of nuclei the housekeeping genes G3PDH and C-jun are not affected. We conclude from these experiments that oocyte linker histone is required for pluripotency gene reactivation in the context of the oocyte.
To support this conclusion by an independent route, we generated and characterized dominant negative linker histone chimeric constructs, consisting of part H1 and part B4 sequences. We first set up a ChIP test that allows us to monitor B4 binding onto chromatin. Varying levels of nuclear GFP-B4 and Myc-B4 proteins are expressed by mRNA injection to the oocyte (Fig. S4 A and B). As shown by ChIP analysis, these tagged B4 proteins bind to chromatin assembled from ssDNA injected into the oocyte GV (Fig. S4_C_). When increasing amounts of nuclear B4-GFP (from 0 to 20% of endogenous B4) are expressed in the GV, a corresponding increase is found associated with chromatinized plasmid. This B4-GFP ChIP signal is reduced by coexpression in the oocyte of a Myc-B4 competitor (400–900% of endogenous B4) (Fig. S4_C_). Therefore, using B4-GFP ChIP, we can monitor the effect of linker histone overexpression on association of B4 with chromatin.
We then injected mRNAs encoding chimeric linker histones into the oocyte to obtain a nuclear level more than five times higher than that of endogenous B4 (Fig. S4_D_). B4-GFP ChIP indicates that most of these constructs interfere significantly with B4 binding (Fig. 4_B_), and can all be detected bound to chromatin (Myc-B4 ChIP, Fig. 4_C_). Therefore the expression of chimeric linker histone provides a way to interfere with endogenous B4 binding to chromatin injected to the oocyte nucleus. Using this interference approach, we then tested transcriptional reactivation from RA-ES nuclei transplanted into oocytes. The result shown in Fig. 4_D_ indicates a strong inhibition of Sox2 reactivation in the presence of the chimeric linker histone, whereas C-jun and G3PDH are not significantly affected.
We conclude from these experiments that the binding of B4 to chromatin of nuclei transplanted into the GV of Xenopus oocyte is a necessary step for the reactivation of pluripotency genes.
Discussion
We suggest that the acquisition of linker histone B4 by transplanted nuclei in Xenopus is a key facilitating event in the reprogramming of somatic cell nuclei. We propose that it helps decondense the chromatin of specialized cells and so exposes genes that are repressed in somatic cells to the active transcriptional conditions of an oocyte.
The combination of real-time monitoring and comparative gene expression analysis of transplanted pluripotent and differentiated cell nuclei developed in this work facilitate the analysis of the mechanisms leading to transcriptional reprogramming. We believe that this approach is complementary to the induced pluripotent stem cell or cell fusion routes. It does not yield new cell types but promotes transcriptional reprogramming in a direct, rapid, and efficient way. Using the pluripotent Sox2 gene as reporter of reprogramming, we obtain up to 100% replication-independent reactivation of the gene within 24 h at 14 °C (equivalent to 6 h at 37 °C). The Oct4 gene shows more moderate reactivation, and Nanog appeared to be reactivated only from certain batches of oocytes, reminiscent of findings for the activation of oocyte ribosomal 5S genes (31). Alternatively a different epigenetic status of the repressed Oct4 and Nanog genes may explain the differential sensitivity to reactivation by the oocyte, as has been proposed for axolotl extract (32). The difference between the reactivation of the various pluripotency genes may depend on the methylation status of DNA on the repressed promoter. The unmethylated Sox2 promoter may be more prone to reactivation than the methylated promoters of Oct4 and Nanog (33).
We observe that reprogramming by oocytes reverses the decrease in linker histone mobility normally observed during ES cell differentiation (29). The unusually high content of acidic residues in the B4 C-terminus domain as compared with H1 could explain the high mobility of oocyte linker histone. In vitro studies have shown that B4-containing chromatin is more permissive to remodeling than H1 chromatin (34). Therefore B4 could enhance transcriptional reactivation indirectly by altering chromatin structure, possibly by allowing action of remodeling complexes. Such a global remodeling of chromatin structure/accessibility then could favor the action of gene-specific regulators already present in the oocyte. Our observation of a later decrease of B4 binding to the Sox2 promoter after its initial reactivation (Fig. 7_A_) suggests that B4 is required only at an early step of gene activation, perhaps when gene promoters are becoming derepressed. We also point out that B4 and H1 do not seem to compete for the same binding sites on chromatin and that the B4 effect on gene reactivation also occurs when both H1 and B4 are bound. Therefore, the enhancing effect of B4 observed on chromatin remodeling in vitro (34) probably results from the presence of B4 per se rather than from the replacement of H1 by B4.
A further question is: What causes B4 linker histone to become incorporated into chromatin of somatic nuclei transplanted to oocytes? We suggest the following as the simplest explanation: During its growth, the Xenopus oocyte accumulates an extraordinarily high concentration of certain proteins. We suggest that the very high concentration of the B4 protein in the oocyte GV enables it to form a sufficiently stable association with the chromatin of transplanted nuclei to initiate transcriptional activation. The loss of H1 from transplanted nuclei, although evidently not required for reprogramming, can be understood if the rapid exchange of H1 from chromatin results in its being diluted by dispersion in the large volume of the oocyte. We do not suppose that the binding of B4 to chromatin is sufficient, by itself, for reprogramming. Other abundant proteins in the GV probably also contribute. B4, nevertheless, is one of the components necessary for the oocyte, and subsequently for the egg, to achieve nuclear reprogramming by maternal components.
Materials and Methods
Oocytes and Cell Nuclei.
Nuclei and oocytes were prepared as previously described (7). When GV isolation was required, batches of 10–20 oocytes were transferred to mineral oil (23), and the excess modified Barth-Hepes saline (MBS) medium surrounding the oocyte was removed carefully. GVs were dissected free of yolk with forceps and transferred to imaging slides (ref 631–0469; WVR International) where they were flattened gently under a coverslip.
ChIP Analysis.
Experiments were carried out as described in ref. 35. Primers used are listed in Fig. S5. ChIP was carried out with anti-GFP (Abcam), anti Myc (9E10; Sigma), or anti-B4 antibodies. In Fig. 7_A_ the PCR signals (expressed as the per cent of input) from two independent experiments were normalized so that the signal from RA-ES nuclei at 6 h after NT was set to 1. The bar chart shows averaged values for the normalized ChIP signals from the two experiments.
Confocal Analysis.
Confocal analysis was carried out on a Zeiss 510 META confocal LSM microscope equipped with argon (458/477/488/514 nm lines) and HeNe (543nm) lasers. For FRAP analysis, 1-μm2 dots within transplanted nuclei were photobleached with the argon laser at 100% intensity. Recovery of fluorescence within the bleached area was monitored immediately after photobleaching using 3-s intervals and low laser intensity (2%) to avoid additional photobleaching. Measurements were performed at 20 °C, and raw data were corrected for background and fluctuations of the laser power. For quantification of exchange experiments, Z-sections of the sample were projected on a single plane and quantified using the Zeiss LSM 510 software.
RT-PCR Analysis.
mRNA extraction was performed using a RNAeasy mini kit (Qiagen) with an on-column DNase step according to the manufacturer's specifications. cDNA synthesis was carried out with SSIII (Invitrogen) using gene-specific primers (Fig. S5). PCR analysis was done either with a standard PCR machine and Hot Start polymerase (Qiagen) or by qPCR, using primers described in Fig. S5.
Supplementary Material
Supporting Information
Acknowledgments
We are grateful to Dr. K. Ohsumi, Dr. K. Ura, and Dr. R. Heald for supplying anti-B4 antibody and cDNAs, to Dr. Ö. Wrange for providing the M13MMTV plasmid, and to Dr. D.T. Brown, Dr. T. Misteli, and Dr. D. Spector for providing cell lines. J.J., N.G., and J.B.G. are funded by the Wellcome Trust. C.A. is sponsored by a postdoctoral fellowship from the Swedish Research Council-Medicine. R.P.H-S. is supported by the National Research Foundation (South Africa), Cambridge Commonwealth Trust, and an Overseas Research Scholarship.
Footnotes
The authors declare no conflict of interest.
References
- 1.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurdon JB. The developmental capacity of nuclei taken from differentiating endoderm cells of Xenopus laevis. J Embryol Exp Morphol. 1960;8:505–526. [PubMed] [Google Scholar]
- 3.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 4.Pereira CF, et al. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Byrne JA, Simonsson S, Gurdon JB. From intestine to muscle: Nuclear repro-gramming through defective cloned embryos. Proc Natl Acad Sci USA. 2002;99:6059–6063. doi: 10.1073/pnas.082112099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne JA, Simonsson S, Western PS, Gurdon JB. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- 8.Catez F, et al. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ura K, Nightingale K, Wolffe AP. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: Structural transitions and transcriptional repression. EMBO J. 1996;15:4959–4969. [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 11.Fan Y, Skoultchi AI. Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 2004;377:85–107. doi: 10.1016/S0076-6879(03)77005-0. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Cho H, Wolffe AP. Xenopus laevis B4, an intron-containing oocyte-specific linker histone-encoding gene. Gene. 1994;143:233–238. doi: 10.1016/0378-1119(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: Homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- 15.Smith RC, Dworkin-Rastl E, Dworkin MB. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988;2:1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrov S, Almouzni G, Dasso M, Wolffe AP. Chromatin transitions during early Xenopus embryogenesis: Changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993;160:214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- 17.Steinbach OC, Wolffe AP, Rupp RA. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 18.Teranishi T, et al. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: Molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto K, et al. Reprogramming events of mammalian somatic cells induced by Xenopus laevis egg extracts. Mol Reprod Dev. 2007;74:1268–1277. doi: 10.1002/mrd.20691. [DOI] [PubMed] [Google Scholar]
- 21.Becker M, et al. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol Biol Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao S, et al. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: Evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Lund E, Paine PL. Nonaqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol. 1990;181:36–43. doi: 10.1016/0076-6879(90)81110-g. [DOI] [PubMed] [Google Scholar]
- 24.Janicki SM, et al. From silencing to gene expression: Real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinett CC, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodland HR, Adamson ED. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev Biol. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- 27.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 28.Belikov S, Astrand C, Wrange O. Mechanism of histone H1-stimulated glucocorticoid receptor DNA binding in vivo. Mol Cell Biol. 2007;27:2398–2410. doi: 10.1128/MCB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korn LJ, Gurdon JB. The reactivation of developmentally inert 5S genes in somatic nuclei injected into Xenopus oocytes. Nature. 1981;289:461–465. doi: 10.1038/289461a0. [DOI] [PubMed] [Google Scholar]
- 32.Bian Y, Alberio R, Allegrucci C, Campbell KH, Johnson AD. Epigenetic marks in somatic chromatin are remodelled to resemble pluripotent nuclei by amphibian oocyte extracts. Epigenetics. 2009;4:194–202. doi: 10.4161/epi.4.3.8787. [DOI] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saeki H, et al. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Astrand C, Belikov S, Wrange O. Histone acetylation characterizes chromatin presetting by NF1 and Oct1 and enhances glucocorticoid receptor binding to the MMTV promoter. Exp Cell Res. 2009;315:2604–2615. doi: 10.1016/j.yexcr.2009.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information