How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? (original) (raw)
Abstract
Chromosome replication occurs precisely once during the cell cycle of almost all eukaryotic cells, and is a highly complex process that is still understood relatively poorly. Two conserved kinases called Cdc7 (cell division cycle 7) and cyclin-dependent kinase (CDK) are required to establish replication forks during the initiation of chromosome replication, and a key feature of this process is the activation of the replicative DNA helicase in situ at each origin of DNA replication. A series of recent studies has shed new light on the targets of Cdc7 and CDK, indicating that chromosome replication probably initiates by a fundamentally similar mechanism in all eukaryotes.
Keywords: Cdc7, CDK, Cdc45–MCM–GINS, Treslin/Ticrr, GEMC1, DUE-B
With very few exceptions, eukaryotic cells make a single precise copy of their chromosomes in each round of the cell cycle. This means that each individual origin must fire just once per cell cycle, and is achieved by ensuring that the replicative DNA helicase can be loaded only at origins during G1 phase, but can be activated only in situ at origins during the subsequent S phase. In all eukaryotes, the replicative DNA helicase comprises a heterohexamer of six related MCM2–7 (minichromosome maintenance 2–7) proteins, originally identified via mutations in the budding yeast Saccharomyces cerevisiae that caused defects in maintaining minichromosomes. The MCM2–7 complex is loaded at origins in an inactive form during G1 phase, and the loaded helicase is then activated during S phase, in a manner that requires both Cdc7 (cell division cycle 7) kinase and CDK (cyclin-dependent kinase) (Bochman and Schwacha 2009; Remus and Diffley 2009; Araki 2010). Activation of MCM2–7 is associated with the recruitment of many other factors to the origin, and initial unwinding of the duplex DNA allows the establishment of two DNA replication forks with opposite polarity. After initiation, replication proceeds bidirectionally away from the origin, until each fork meets another from a neighboring origin, at which point replication of that part of the chromosome is terminated.
The mechanism and regulation of the MCM2–7 helicase remain unclear, although two recent studies used an in vitro loading system with purified budding yeast proteins to show that the inactive helicase is loaded onto dsDNA as a double hexamer (Evrin et al. 2009; Remus et al. 2009). In principle, the activated helicase could remain as a double hexamer, or else the inactive double hexamer could be separated into active single hexamers encircling dsDNA, or even into single hexamers around ssDNA at each replication fork (Bochman and Schwacha 2009). Precedents for each of these active forms exist among viral or bacterial helicases, and so the mechanism by which the MCM2–7 helicase acts at forks remains to be elucidated experimentally. What does seem clear, however, is that one key aspect of the activation mechanism is the stable association of the MCM2–7 complex with other proteins that are also important for helicase function in vivo. The recruitment of these factors to the MCM2–7 complex occurs only at origins, and appears to require the action of both Cdc7 and CDK in all species that have been examined.
The first such factor to be identified was the budding yeast protein Cdc45, which, together with MCM2–7, is required for the progression of DNA replication forks (Aparicio et al. 1997; Labib et al. 2000; Tercero et al. 2000), as are the orthologous factors in Xenopus (Pacek and Walter 2004; Shechter et al. 2004). A physical interaction between Cdc45 and MCM2–7 proteins was initially suggested by the fact that a mutation in budding yeast Cdc45 can be suppressed by compensating mutations in Mcm5 or Mcm7, and a mutation in fission yeast Cdc45/Sna41 was subsequently found to suppress a particular mutation in fission yeast Mcm5/Nda4 (Moir et al. 1982; Miyake and Yamashita 1998; Yamada et al. 2004). Cdc45 is recruited to early origins of DNA replication during G1 phase as discussed below, but a stable Cdc45–MCM complex is formed only during S phase in a manner dependent on CDK and Cdc7, and it appears that the assembly process can occur only in situ at nascent replication forks (Zou and Stillman 2000; Masai et al. 2006; Sheu and Stillman 2006; Im et al. 2009).
It is now clear that Cdc45 binds stably to MCM2–7 only as part of a larger complex of proteins built at replication origins, the existence of which is dependent on an additional component known as GINS (Go-Ichi-Ni-San, or 5-1-2-3, referring to the latter part of the names of each of the four subunits), which itself is a complex of four small proteins that are distantly related to each other. The GINS complex was first identified in budding yeast and extracts of Xenopus eggs, and GINS is currently the last replication factor conserved in all eukaryotes to be identified (Kubota et al. 2003; Takayama et al. 2003). The components of budding yeast GINS were also identified in a separate screen, and were shown to be important for the progression of DNA replication forks, just like MCM2–7 and Cdc45 (Kanemaki et al. 2003). Subsequently, GINS was found to associate at replication forks with MCM2–7 and Cdc45 as part of a large replisome progression complex (RPC) together with other factors, and GINS is required to maintain the association of MCM2–7 and Cdc45 within this complex (Gambus et al. 2006). In addition, an analogous CMG complex of Cdc45–MCM–GINS was isolated from extracts of early Drosophila embryos and found to contain DNA helicase activity, and the components of this complex were found to be part of the replicative helicase or “unwindosome” complex on replicating DNA in extracts of Xenopus eggs (Moyer et al. 2006; Pacek et al. 2006). It now seems clear that CMG interacts on replicating DNA during chromosome replication in human cells too (Bauerschmidt et al. 2007; Aparicio et al. 2009; Im et al. 2009). It thus appears that Cdc45 and GINS are essential components of the active MCM2–7 complex during chromosome replication in all eukaryotes, although the precise contribution of Cdc45 and GINS to helicase activity remains to be elucidated. Assembly of CMG and the RPC are key steps during activation of the MCM2–7 helicase at origins, and appear to be at the heart of the mechanism by which Cdc7 and CDK lead to the initiation of chromosome replication (Fig. 1), as discussed in the following sections.
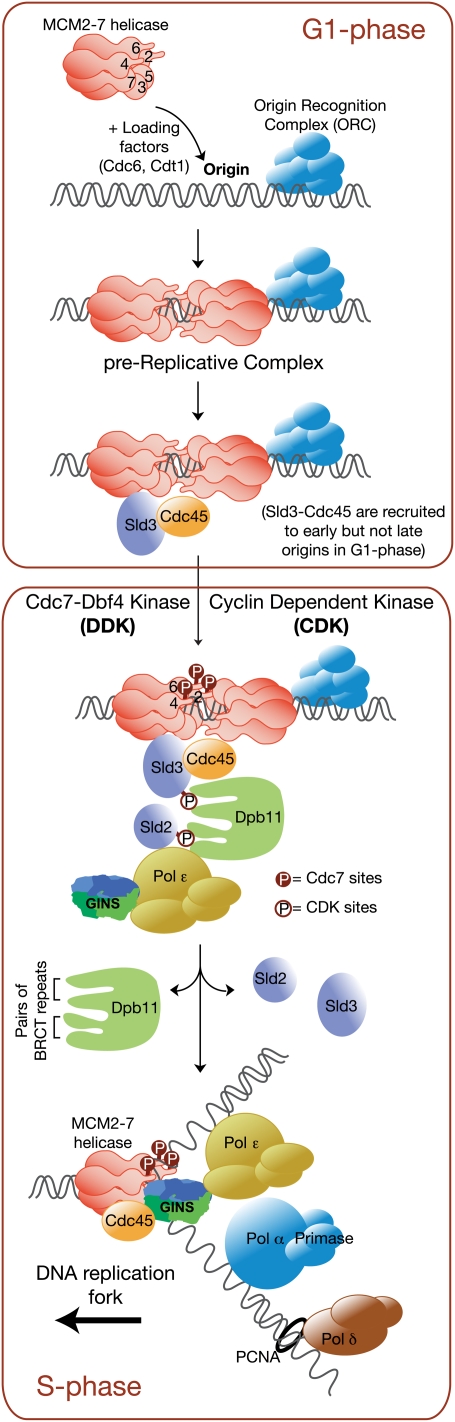
Figure 1.
The initiation of chromosome replication in budding yeast. The MCM2–7 helicase is recruited to origins during the G1 phase of the cell cycle, and is loaded around dsDNA by the Cdc6 and Cdt1 proteins together with the origin recognition complex. Studies of the loading reaction in vitro indicate that the MCM2–7 helicase is loaded as a double hexamer, in contrast to the single-hexameric form in solution, and multiple double hexamers are probably loaded at each origin. This produces the prereplicative complex at origins, and is equivalent to the step referred to in Xenopus as the “licensing reaction.” At the earliest budding yeast origins, the Sld3 protein is recruited in a weakly bound form together with Cdc45, perhaps by direct interaction with the MCM2–7 complex. The MCM2–7 helicase remains inactive until cells enter S phase and activate Cdc7 and CDKs. The Cdc7 kinase phosphorylates the N-terminal tails of Mcm2/4/6 and probably induces a structural change in the MCM2–7 complex. It seems likely that CDK also phosphorylates MCM2–7, but the major targets are Sld2 and Sld3, the phosphorylated forms of which appear to be bridged by Dpb11 (the N-terminal pair of BRCT repeats of Dpb11 binds Sld3, whereas the C-terminal pair of BRCT repeats binds Sld2), allowing the recruitment of GINS and DNA polymerase ɛ to origins. This leads to the stable association of CMG together with a variety of other factors (not shown for simplicity) to form the RPC. Activation of the MCM2–7 helicase unwinds the origin and allows the priming of leading and lagging strands by DNA polymerase α. At present, the nature of the active CMG helicase within the RPC is unclear, as discussed in the text. DNA polymerase ɛ extends the leading strand at each DNA replication fork, and DNA polymerase δ extends each Okazaki fragment on the lagging strand. Many other factors also act at DNA replication forks, but these have been omitted, as they remain beyond the scope of this review. Similar events are thought to occur subsequently at later origins, to which the various initiation factors including Sld3 and Sld2 are recruited only around the time that initiation occurs at each particular origin. At present it is not clear whether CDK phosphorylates Sld3 only in situ at origins, or whether CDK can also phosphorylate Sld3 away from origins.
Cdc7 and CDK are required throughout S phase to activate origins of replication
Studies with budding yeast first showed that both Cdc7 and CDK are required throughout S phase for the firing of early and later origins of replication. Rather like CDK, the Cdc7 kinase acts in association with an essential regulatory subunit called Dbf4 (Dumbbell-forming 4, as yeast cells lacking Dbf4 arrest with a large bud; vertebrate species have a Dbf4 protein as well as a second Dbf4-related factor, called Drf1), and Cdc7 is also referred to as DDK (Dbf4-dependent kinase). It seems very likely that the Cdc7–Dbf4 kinase acts locally at each origin, initially suggested by the isolation of the budding yeast DBF4 gene in a one-hybrid screen that used an origin of DNA replication as bait (Dowell et al. 1994). Subsequently, two studies showed that yeast Cdc7 is needed early in S phase for the firing of early origins, but is then still required subsequently for the activation of later origins of replication (Bousset and Diffley 1998; Donaldson et al. 1998a). The Cdc7–Dbf4 kinase is probably recruited to origins of DNA replication by interaction with the MCM2–7 complex in both yeast and Xenopus (Lei et al. 1997; Jares and Blow 2000; Walter 2000). As discussed in more detail below, it seems that the targets of Cdc7 are phosphorylated preferentially on chromatin, presumably at origins of DNA replication.
CDK is also needed throughout S phase for the activation of replication origins. Budding yeast cells lacking the major S-phase cyclin Clb5 activate early but not later origins, and subsequent studies showed that this was simply because the minor S-phase cyclin Clb6 is present during early S phase but disappears later, so that it is no longer present to activate later origins (Donaldson et al. 1998b; Gibson et al. 2004; Jackson et al. 2006). In vertebrate cells, CDK also appears to be required throughout S phase—in this case, to activate clusters of origins (probably equivalent to “replication factories”) throughout each chromosome (Thomson et al. 2010).
Several studies using budding yeast and Xenopus eggs tried to establish whether Cdc7 and CDK must act in a particular order to promote the activation of replication origins. In extracts of Xenopus eggs, Cdc7 can complete its role in the absence of CDK activity, but the converse is not true (Jares and Blow 2000; Walter 2000). In yeast, however, it was found that Cdc7 could not complete its role in the absence of CDK activity (Nougarede et al. 2000), in apparent disagreement with the studies involving frog eggs. A trivial explanation for this discrepancy could simply be that the relevant phosphatases for the targets of Cdc7 and CDK in yeast and frog eggs have different activities, as this would produce different half-lives for the phosphorylated substrates. Whatever the explanation for the observed differences, however, a deeper understanding of how Cdc7 and CDK activate replication origins clearly required the identification of the physiologically relevant substrates of these kinases.
The MCM2–7 helicase appears to be the major target for Cdc7–Dbf4 in budding yeast
A wealth of genetic and biochemical data indicate that Cdc7 promotes the initiation of chromosome replication in budding yeast by binding to the MCM2–7 complex at origins of DNA replication and phosphorylating several subunits of the helicase, although the precise contribution of such phosphorylation events to the initiation mechanism still remains to be established. A mutation in the DBF4 gene was isolated as an allele-specific suppressor of the temperature-sensitive mcm2-1 allele, indicating that a specific alteration in Dbf4 could compensate for a particular defect in MCM2–7 function (Lei et al. 1997). The same study found that Dbf4–Cdc7 interacts with Mcm2, phosphorylates Mcm2 in vitro, and is required for Mcm2 phosphorylation in vivo. Furthermore, Mcm3, Mcm4, and Mcm6 were also phosphorylated by Cdc7–Dbf4 in vitro, and a later study showed that the same was true for Mcm7 (Weinreich and Stillman 1999). In contrast, Mcm5 has not been found to be a substrate of Cdc7–Dbf4, in budding yeast or any other species.
In principle, phosphorylation of MCM2–7 by Cdc7–Dbf4 could create direct binding sites for other factors such as Cdc45 and GINS, or could cause a structural change in the MCM2–7 complex that is important for activation of the helicase. Evidence for the latter possibility came with the identification of the bob1 (bypass of block 1) mutation, which had been shown previously to suppress the lethality of cdc7Δ or dbf4Δ (Jackson et al. 1993; Hardy et al. 1997). Ironically, bob1 was found to be a single point mutation (P83L) in an N-terminal region of the Mcm5 protein, the only MCM2–7 subunit not phosphorylated by Cdc7–Dbf4 in vitro. The mode of action of the bob1 mutation is still not understood very well, but studies of an archaeal MCM complex with or without the analogous mutation suggest that the mcm5-P83L allele might cause a subtle structural change in the MCM2–7 complex (Fletcher et al. 2003), perhaps mimicking what normally happens when the other subunits are phosphorylated by Cdc7–Dbf4.
Many archaeal species possess a single ortholog of the eukaryotic MCM2–7 proteins, and several archaeal MCM proteins have been shown to form homohexameric complexes that have been useful for structural studies of the MCM helicase. The isolated N terminus of two species of archaeal MCM were found to be sufficient to form a hexameric ring, and it now seems likely that both archaeal and eukaryotic MCM hexamers have the form of a dumbbell containing two rings formed by N-terminal and C-terminal domains, the latter of which contains the ATPase region and thus the motor domain of the helicase (Sclafani et al. 2004). The double-hexameric forms of eukaryotic and archaeal MCM helicase appear to be head-to-head complexes, in which the N-terminal domains of each hexamer interact to form the double hexamer.
Interestingly, Mcm2, Mcm4, and Mcm6 in all eukaryotic species possess relatively unstructured extensions at the N terminus of up to a couple of hundred amino acids, preceding the structured N-terminal and C-terminal domains that are shared with archaeal MCM proteins. It now seems that these N-terminal tails of eukaryotic Mcm2–4–6 represent the major substrates for Cdc7–Dbf4, as well as containing sites for other kinases, including CDK.
The best characterized of these tails comprises the first 174 amino acids of budding yeast Mcm4 (Sheu and Stillman 2006, 2010; Devault et al. 2008). Although the N-terminal tails of eukaryotic Mcm4 proteins appear to lack primary sequence homology with each other, in contrast to the rest of the protein, this is probably misleading, as the tails contain conserved features that underlie their function. Most strikingly, the N-terminal tails of eukaryotic Mcm4 proteins are very rich in serine and threonine (29% of the first 174 amino acids of budding yeast Mcm4, and 24% of the first 148 amino acids of human Mcm4), and contain multiple motifs that are now known to be targets for Cdc7 kinase. These include motifs in which CDK appears to prime phosphorylation by Cdc7, such as “SSP,” in which CDK phosphorylates the second serine, thus helping Cdc7 to phosphorylate the first (Masai et al. 2000; Montagnoli et al. 2006; Devault et al. 2008). In addition, Cdc7 appears to target runs of consecutive Ser/Thr, as well as Ser/Thr embedded within stretches of acidic amino acids.
Two elegant studies from Bruce Stillman's group (Sheu and Stillman 2006, 2007) have analyzed the N-terminal tail of budding yeast Mcm4 (called NSD [N-terminal Ser/Thr-rich domain]) in great detail. The first study showed that the NSD contains redundant sites for Cdc7, and is followed by a “docking region” for Cdc7–Dbf4 within the structured N-terminal domain of Mcm4 (Sheu and Stillman 2006). The precise sequence context of the Cdc7 sites does not seem critical, as the NSD can be replaced by either the equivalent tail of Mcm2 or even a short stretch of exogenous sequence, as long as it contains Cdc7 target sites. The hyperphosphorylated form of Mcm4 is dependent on Cdc7 kinase activity in vivo, and is greatly enriched in the Cdc45–MCM complex that probably represents the fraction of the MCM2–7 helicase at replication forks within the RPC. In a mcm5-bob1 cdc7Δ strain, cells are alive but S phase is slow, and the Cdc45–MCM complex either is formed much less efficiently, or else is much less stable than in cells containing Cdc7 (Sheu and Stillman 2006). It thus appears that the phosphorylation of MCM2–7 by Cdc7 promotes the stable formation of the Cdc45–MCM complex.
In a recent second study from the same group (Sheu and Stillman 2010), the region of Mcm4 between residues 74 and 174 was found to contain an inhibitory activity that is relieved when Cdc7 phosphorylates the NSD. In cells lacking this inhibitory region but containing the rest of the NSD, Cdc7–Dbf4 becomes dispensable, as in mcm5-bob1, although once again cells grow slowly, and the formation or stability of the Cdc45–MCM complex is defective. Removing the whole NSD also allows proliferation in the absence of Cdc7, but cells are even sicker, indicating that phosphorylation of the NSD by Cdc7 normally has two roles: first, relieving an inhibitory activity in the Mcm4 NSD, and second, stimulating replication in some other way. The molecular details of these inhibitory and stimulatory activities remain to be determined.
At present it is unclear whether the N-terminal tails of Mcm2 and Mcm6 also have inhibitory regions analogous to that of Mcm4, although removal of the tails of either Mcm2 or Mcm6 is not sufficient to bypass the requirement of Cdc7 for proliferation (Sheu and Stillman 2010). Another recent study found that the structured N-terminal domain of Mcm2 probably contains a docking region analogous to that of Mcm4, and also reported that phosphorylation of S170 of Mcm2 by Cdc7–Dbf4 is essential for replication (Bruck and Kaplan 2009). As deletion of a region containing S170 is not lethal (Sheu and Stillman 2006), it still remains possible that the Mcm2 tail does contain an inhibitory activity that is relieved by Cdc7 phosphorylation of S170, but this idea requires further investigation.
In summary, it seems very clear that the MCM2–7 helicase is a major target of the Cdc7 kinase. Phosphorylation of the N-terminal tails of Mcm2–4–6 is probably the main way by which Cdc7 promotes the initiation of chromosome replication in budding yeast, and this allows the stable formation of the Cdc45–MCM complex. The mechanism by which phosphorylation of the tails of Mcm2–4–6 contributes to activation of the MCM2–7 helicase at origins remains unclear, but an attractive idea is that it causes a structural change within the MCM2–7 complex that is needed for stable formation of the RPC. If this is true, it is not yet clear whether the phosphorylated tails still play an important role after initiation, and whether dephosphorylation of the MCM2–7 complex forms any part in the mechanism of termination, when two neighboring forks meet each other.
It is still possible that other important targets of Cdc7 also remain to be characterized. It is worth noting that S phase is markedly defective in the absence of Cdc7, even in cells that have the mcm5-bob1 mutation, or in cells that lack the inhibitory region of the N-terminal tail of Mcm4. Although this could just be because the bypass mutations do not fully reproduce the normal effects of MCM2–7 phosphorylation by Cdc7, it might also reflect the importance of other targets of Cdc7. Previous studies have shown that Cdc45 is also a target for Cdc7 in vitro (Nougarede et al. 2000), as is DNA polymerase α, which primes DNA synthesis on the leading and lagging strands (Weinreich and Stillman 1999), but the importance of such potential targets in vivo remains to be addressed in the future.
Sld2 and Sld3 are the major targets for CDK during the initiation of chromosome replication in budding yeast
Whereas the MCM2–7 helicase itself is the major target of Cdc7, and the N-terminal tail of Mcm4 is also phosphorylated by CDK (Devault et al. 2008), the principal targets of CDK during the initiation of chromosome replication in budding yeast appear to be factors that are required specifically for initiation, and that are not incorporated subsequently into the replisome (Fig. 1). The molecular function of these factors is not understood, but they are required for the recruitment of factors such as Cdc45 and GINS to the MCM2–7 complex at origins, probably contributing thereby to assembly of the CMG helicase within the RPC.
A key player in this process, called Dpb11 (DNA polymerase B 11), was originally found as a high-copy suppressor of a mutation in the Dpb2 subunit of DNA polymerase ɛ (Araki et al. 1995), which is responsible for synthesis of the leading strand at DNA replication forks. All groups of eukaryotes have a single ortholog of Dpb11 that is required for the initiation of chromosome replication, known in fission yeast as Rad4/Cut5, and in metazoans as TopBP1/Mus101/Cut5 (Garcia et al. 2005). All of these orthologs are built from a series of BRCT repeats, which often function as phosphoprotein-binding modules (Manke et al. 2003; Yu et al. 2003), although the number of repeats varies between species, and the TopBP family of proteins can clearly have other roles in addition to the initiation of chromosome replication (Garcia et al. 2005). In a seminal screen for mutations that cause synthetic lethality in combination with the dpb11-1 allele, Hiro Araki's group (Kamimura et al. 1998) isolated a series of SLD (synthetic lethal with Dpb11) genes that have subsequently been found to include several key players in the initiation process. The first of these to be characterized was SLD2 (Kamimura et al. 1998; Wang and Elledge 1999), which encodes a factor of unknown molecular function that binds to the C-terminal pair of the four BRCT repeats of Dpb11, in a manner now known to require phosphorylation of Sld2 by CDK (Masumoto et al. 2002; Tak et al. 2006).
Subsequently, the Sld3 protein was also shown to be required for initiation (Kamimura et al. 2001). A screen for multicopy suppressors of the temperature-sensitive phenotype of a mutated allele of SLD3 identified CDC45, and high-copy SLD3 was also found to suppress mutations in CDC45. Moreover, both Sld3 and Cdc45 were found by chromatin immunoprecipitation to be recruited in a mutually dependent manner to early origins of replication in G1 phase before the initiation of chromosome replication, despite the fact that stable association of Cdc45 with the MCM2–7 complex can be detected only during S phase, when replication forks are established. This led to the idea that a complex of Sld3–Cdc45 might associate weakly with the earliest origins in G1 phase, ready to act once CDK and Cdc7 trigger initiation subsequently (Kamimura et al. 2001).
As Cdc45 is required for both initiation and elongation, and Sld3 seemed to behave in a similar manner to Cdc45, these data also led to the notion that Sld3 might function in association with Cdc45 in both of these processes (Kamimura et al. 2001). Subsequently, however, it was found that Sld3 is displaced from origins during initiation and is dispensable for elongation (Kanemaki and Labib 2006), in contrast to Cdc45. Consistent with this fact, Sld3 was not found to be a component of the RPC, in contrast to Cdc45 (Gambus et al. 2006). This indicated that Sld3 is an initiation factor that helps to recruit Cdc45 to origins, perhaps contributing thereby to assembly of the RPC following activation of Cdc7 and CDK.
In 2007, two studies (Tanaka et al. 2007; Zegerman and Diffley 2007) showed that Sld3 is an essential target of CDK for the initiation of chromosome replication in budding yeast. As with Sld2, phosphorylation of Sld3 by CDK promotes its association with Dpb11—in this case, with the N-terminal pair of BRCT repeats. These data led to a model for initiation whereby Sld3 associates with some factor at origins (perhaps the MCM2–7 helicase itself) and helps to recruit Cdc45, whereas Dpb11 bridges phosphorylated Sld3 to phosphorylated Sld2, apparently leading to recruitment of GINS and the assembly of the RPC (Araki 2010).
Until very recently, it remained unclear how GINS might be recruited to origins during the initiation process. Previous work involving chromatin immunoprecipitation experiments had indicated that GINS might be recruited together with Dpb11 and, more surprisingly, together with DNA polymerase ɛ (Masumoto et al. 2002; Takayama et al. 2003). A very recent study has shed new light on this aspect of the initiation process in budding yeast (Muramatsu et al. 2010). By treating cells with a cross-linking agent to preserve weak interactions between proteins, it was found that GINS, DNA polymerase ɛ, Dpb11, and phosphorylated Sld2 could all form a so-called preloading complex as cells enter S phase. This required CDK but not Cdc7–Dbf4, and the complex formed even if the loading of the MCM2–7 helicase at origins was blocked in order to prevent initiation from occurring.
Studies with purified proteins in vitro indicated that phosphorylation of Sld2 by CDK is not absolutely required for the individual components to associate with each other, but strengthens their ability to interact and assemble together into the preloading complex. Moreover, DNA polymerase ɛ was found to be essential for GINS to interact with Dpb11–Sld2 in vitro. In addition, high-copy expression of the four GINS proteins in vivo was found to suppress temperature-sensitive mutations in Dpb11, Pol2, and Sld2. Overall, it appears that GINS is recruited to origins together with DNA polymerase ɛ in budding yeast, and this requires Dpb11 and probably CDK phosphorylation of Sld2 (Muramatsu et al. 2010).
Importantly, chromosome replication can proceed in the absence of CDK activity in budding yeast cells expressing a phosphomimetic mutant of Sld2 combined with a fusion of Sld3 to the C-terminal pair of BRCT repeats of Dpb11 (Zegerman and Diffley 2007). A particular mutated allele of Cdc45 can also substitute in a similar assay for the fusion of Sld3 to Dpb11 (Tanaka et al. 2007). These findings led to the proposal that Sld2 and Sld3 are the “minimal set” of CDK targets for the initiation of chromosome replication in budding yeast (Tanaka et al. 2007; Zegerman and Diffley 2007). But chromosome replication proceeds very slowly in the bypass strains upon inhibition of CDK (Tanaka et al. 2007; Zegerman and Diffley 2007), and assembly of the RPC is highly defective (H Morohashi and K Labib, unpubl.), as seen with the mutations that allow cells to proliferate in the absence of Cdc7. So it is still possible that CDK contributes in other ways to the normal progression of chromosome replication, and has other important targets. The N-terminal tail of Mcm4 is one such example, as it can be phosphorylated by both Cdc7 and CDK, and the CDK sites become particularly important if the number of Cdc7 sites is reduced (Devault et al. 2008; Sheu and Stillman 2010).
The initiation of chromosome replication in fission yeast: fundamentally similar, but slightly different?
The initiation of chromosome replication has also been studied extensively in the fission yeast Schizosaccharomyces pombe, and it seems very likely that the underlying mechanism will be extremely similar to the situation in budding yeast. Indeed, orthologs of both Cdc7 and CDK are required for recruitment of Cdc45 to chromatin during S phase (Gregan et al. 2003; Dolan et al. 2004). Moreover, the fission yeast ortholog of Cdc7–Dbf4, known as Hsk1 (homolog of CDC seven kinase 1)–Dfp1 (DBF four in pombe 1), was found to phosphorylate the Mcm2 and Mcm4 components of the MCM2–7 complex, and Hsk1 activity is also important in vivo for hyperphosphorylation of Mcm6/Mis5 (Brown and Kelly 1998; Masai et al. 2006). Removal of the N-terminal tail of Mcm4 reduced viability, and putative target sites for Hsk1 in the N-terminal tail of Mcm4 were found to share a redundant role with sites in the tails of Mcm2/6 (Masai et al. 2006). Tethering of Hsk1 close to an origin increased the efficiency of that origin, suggesting that Hsk1 acts locally at origins as in budding yeast (Patel et al. 2008).
Fission yeast has a single ortholog of each of Dpb11, Sld2, and Sld3, all of which are essential for chromosome replication (Fenech et al. 1991; Nakajima and Masukata 2002; Noguchi et al. 2002). Sld3 can associate with origins in the absence of CDK activity as in budding yeast, and is required for the subsequent recruitment of GINS and Cdc45 (Yabuuchi et al. 2006). Moreover, the fission yeast ortholog of Sld2, known as Drc1 (DNA replication and checkpoint 1), is an essential target of CDK, and phosphorylation stimulates the interaction of Drc1 with the Rad4/Cut5 protein, which is the fission yeast ortholog of Dpb11 (Noguchi et al. 2002).
Nevertheless, several apparent differences have been found between the fission yeast Sld2 and Sld3 proteins and their budding yeast counterparts. First, recruitment of fission yeast Sld3 to origins of DNA replication requires Hsk1 activity, although the relevant target sites remain to be characterized (Nakajima and Masukata 2002). A role for budding yeast Cdc7 in recruiting Sld3 to origins has not yet been described and would not have been anticipated, as budding yeast Sld3 is recruited to early origins in G1 phase, when Cdc7 is thought to be inactive due to low levels of Dbf4. Second, mutation of consensus CDK sites in fission yeast Sld3 did not block chromosome replication (Nakajima and Masukata 2002), and at present it is unclear whether Sld3 is an essential target of CDK for chromosome replication in fission yeast. Nevertheless, interaction between Sld3 and Rad4/Cut5 does appear to be required for replication, indicating that the mode of action will be very similar to budding yeast Sld3, even if the regulation is slightly different (H Masukata, pers. comm.). Finally, fission yeast Sld2 was reported to interact with the N-terminal pair of BRCT repeats of Rad4/Cut4 (Noguchi et al. 2002), whereas the analogous repeats in budding yeast Dpb11 interact with Sld3 as described above. A more complete understanding of the interaction of fission yeast Rad4/Cut5, Sld2, and Sld3 clearly awaits further investigation.
In summary, it seems very likely that the MCM2–7 helicase is a major target of the fission yeast ortholog of Cdc7, analogous to the situation in budding yeast, although the details remain to be explored further. It also seems very likely that interaction of Rad4/Cut5 with Sld2 and Sld3 will be crucial for activation of the MCM2–7 helicase, and will promote recruitment of key factors such as GINS and Cdc45 to origins. CDK plays a key role in this process by phosphorylating Sld2, but the importance of Sld3 as a CDK target remains unclear. In contrast, the fission yeast equivalent of Cdc7 kinase is clearly required for the recruitment of Sld3 to origins, indicating that both CDK and Cdc7 allow Sld3–Rad4/Cut5–Sld2 to mediate recruitment to origins of factors such as GINS and Cdc45.
Cdc7 also phosphorylates the MCM2–7 helicase in vertebrate cells, and is an interesting potential target for new cancer therapies
A wealth of evidence indicates that the N termini of Mcm2 and Mcm4 are phosphorylated by Cdc7 in human cells, as well as during chromosome replication in extracts of Xenopus eggs. As with the yeast protein, vertebrate Cdc7 has a strong preference for Ser or Thr in an acidic context, either produced by adjacent amino acids that are acidic, or else due to an adjacent CDK site that appears to prime phosphorylation by Cdc7 (Masai et al. 2000, 2006; Cho et al. 2006; Montagnoli et al. 2006). In the case of Mcm4, isolated fragments from the N-terminal tail are phosphorylated less well than full-length protein, perhaps indicating the presence of a “docking domain” analogous to that of budding yeast Mcm4 (Masai et al. 2006). It also seems clear that Cdc7 phosphorylates the MCM2–7 complex preferentially on chromatin during S phase, and recruitment of Cdc7 to chromatin in extracts of Xenopus eggs appears to be via interaction with the MCM2–7 helicase itself, consistent with the studies of the yeast proteins (Jares and Blow 2000; Pereverzeva et al. 2000; Walter 2000).
The consequences of the phosphorylation of the MCM2–7 helicase by Cdc7 in human cells also seem likely to be similar to the situation in yeast, as a recent study found that Cdc7 (and CDK) is required for formation of the CMG helicase complex in human cells (Im et al. 2009). Interestingly, there is accumulating evidence to indicate that the process of chromosome replication is defective in many cancer cells, and this probably explains why such cells are especially sensitive to drugs that inhibit the initiation step of chromosome replication (Halazonetis et al. 2008). There is now considerable interest in inhibitors of Cdc7 as a potential anti-cancer therapy, and several studies have found that such drugs preferentially kill cancer cells compared with untransformed primary cells (Swords et al. 2010).
Clearly there is still much to learn regarding how the MCM2–7 helicase is regulated by Cdc7 phosphorylation in human cells, and indeed a recent study argues that Cdc7 might promote the assembly of the MCM2–7 complex onto chromatin during re-entry into the cell cycle from quiescence (Chuang et al. 2009). Nevertheless, it now seems very likely that the mechanism by which Cdc7 promotes activation of the MCM2–7 helicase and the initiation of chromosome replication will be fundamentally similar in all eukaryotes.
Is RecQ4 the vertebrate ortholog of yeast Sld2?
The major targets of CDK during the initiation of chromosome replication in the budding yeast S. cerevisiae appear to recruit essential replisome components such as Cdc45 and GINS to origins, but Sld3–Dpb11–Sld2 are not themselves part of the replication machinery at DNA replication forks. Perhaps it is not surprising that these factors have been free to evolve more rapidly than core replisome components, if they simply mediate protein–protein interactions during initiation. Whatever the reason, the primary sequence of Sld3, Dpb11, and Sld2 has diverged quite quickly during evolution, even in other budding yeast species. For this reason, finding orthologs of these factors in higher eukaryotes has been much more challenging than for many other replication factors.
Recognizing orthologs of Dpb11 is aided by the fact that such proteins are built from BRCT repeats, and it seems clear that each eukaryotic species has a single such protein, although the number of BRCT repeats varies from four in yeasts to eight in the human ortholog that is known as TopBP1 (Garcia et al. 2005). Predicted features for putative orthologs of Sld2 and Sld3 in higher eukaryotes might include some sequence similarity to the yeast proteins, association with TopBP1 (perhaps regulated by CDK), and an essential role in the initiation of chromosome replication.
Currently, the best candidate for Sld2 in higher eukaryotes is the RecQ4 DNA helicase that is mutated in a human genetic disease called Rothmund-Thomson syndrome, which predisposes patients to cancer (Sangrithi et al. 2005). The RecQ4 protein is essential for proliferation in mice and Drosophila, but the mutations causing human Rothmund-Thomson syndrome often cause truncations that leave the N-terminal portion of the protein intact. Strikingly, this part of the protein shows limited homology with yeast Sld2 (Sangrithi et al. 2005; Matsuno et al. 2006). Moreover, depletion of RecQ4 from extracts of Xenopus eggs blocked chromosome replication, and this could be rescued by addition of either full-length human or frog RecQ4 (Sangrithi et al. 2005). Importantly, an N-terminal fragment of frog RecQ4 could also rescue the defect in chromosome replication, and this fragment was able to interact with Cut5/TopBP1, the Xenopus ortholog of Dpb11 (Matsuno et al. 2006). It thus appears that the N-terminal domain of RecQ4 could play a related role to yeast Sld2 during the initiation of chromosome replication.
In addition to the surprising presence of a RecQ helicase domain in the same protein, there are several other features of RecQ4 that would not have been anticipated from studies of yeast Sld2. Most intriguingly, depletion of RecQ4 from Xenopus extracts blocked the initiation of chromosome replication but did not block the stable association of Cdc45 with chromatin (Sangrithi et al. 2005; Matsuno et al. 2006). This merits further investigation, but it seems likely that the CMG helicase complex can be assembled in the absence of RecQ4, although initiation is still blocked at some later step. At present it is unclear whether yeast Sld2 plays any role after recruitment of Cdc45 and GINS to the MCM2–7 complex, or whether RecQ4 and Sld2 truly promote the initiation of chromosome replication in different ways. As might have been expected, the N terminus of Xenopus RecQ4 binds directly to Cut5/TopBP1, and can be phosphorylated by CDK in vitro (Matsuno et al. 2006). But even the dephosphorylated N terminus of RecQ4 can bind Cut5/TopBP1 in vitro, so at present it is unclear whether CDK plays any role in stimulating the interaction between RecQ4 and Cut5/TopBP1 during the initiation of chromosome replication.
Finally, a recent study investigated the binding partners of human RecQ4 on chromatin, by expressing a tagged version of RecQ4 in human 293 cells (Xu et al. 2009). Surprisingly, RecQ4 was found to bind the human CMG complex on chromatin, as well as another replication factor called Mcm10 that is thought to recruit DNA polymerase α to origins during the initiation of chromosome replication. RecQ4 can bind directly to Mcm10 in vitro, and it was suggested that Mcm10 could mediate the association of RecQ4 with the CMG complex on chromatin (Xu et al. 2009). At present it is unclear whether these interactions occur at DNA replication forks or else represent intermediates in the initiation reaction, but they certainly challenge the predictions that might have been made for a putative ortholog of yeast Sld2. To complicate the situation still further, another recent study found that depletion of RecQ4 in HeLa cells blocked formation of the CMG complex (Im et al. 2009), although it is not clear at which stage of the cell cycle the cells became arrested (for example, a partial defect in replication could cause a subsequent problem in mitosis, which would also reduce the detectable CMG complex).
In summary, it seems clear that RecQ4 plays an important role during the initiation of chromosome replication in Xenopus and in human cells, and the N-terminal domain that is related to Sld2 is crucial for this process. Although it remains possible that the mode of action of Xenopus RecQ4 during initiation could differ from that of the human protein, this seems less likely given that human RecQ4 can rescue depletion of its frog ortholog from Xenopus egg extracts. Either way, there clearly is still a lot to learn about the molecular function of RecQ4, and the same could perhaps be said of yeast Sld2.
Searching for orthologs of yeast Sld3
Until very recently, it has not been possible to identify putative orthologs of Sld3 outside of fungi. This could either be due to the rapid divergence of Sld3 orthologs during evolution, making their identification by sequence searching extremely difficult, or else could reflect the fact that other proteins might have taken over the role of yeast Sld3 during the initiation of chromosome replication in other eukaryotic species. For example, TopBP1 itself has been reported to bind directly to Cdc45 (Schmidt et al. 2008), in part via the N-terminal pair of BRCT repeats that are required for chromosome replication as discussed below. Very interestingly, however, several recent studies have identified three new candidates for proteins that might play a role analogous to Sld3 during the initiation of chromosome replication in vertebrate cells.
Previous work showed that the first five BRCT repeats of TopBP1 are sufficient to support chromosome replication in extracts of Xenopus eggs (Hashimoto et al. 2006; Yan et al. 2006). To find proteins that might interact with TopBP1 during chromosome replication, Kumagai et al. (2010) used a tagged version of the first six BRCT repeats of Xenopus TopBP1 to pull down proteins extracted from replicating Xenopus nuclei. In this way, they found a novel 220-kDa protein of unknown function that they called Treslin (TopBP1-interacting, replication-stimulating protein). Depletion of Treslin from frog extracts blocked chromosome replication and prevented the association with chromatin of Cdc45 (and thus probably GINS) but did not block loading of the MCM2–7 helicase (Kumagai et al. 2010). These features would be consistent with Treslin having a role analogous to that of Sld3, although at present it is not clear how Treslin itself is recruited to chromatin, and unfortunately the depletion of Treslin could not be rescued by recombinant protein.
Despite lacking detectable motifs that could explain its mode of action, the sequence of frog Treslin is extremely rich in potential CDK sites, having 67 Ser–Pro or Thr–Pro sites, of which 35 have Lys/Arg two residues beyond the proline, as generally found in optimal CDK sites. Moreover, the interaction between Treslin and TopBP1 could be detected only in nuclear extracts in which Treslin becomes phosphorylated, and was inhibited by addition to the nuclear extracts of the CDK inhibitor p27 (Kumagai et al. 2010). By making further truncations of TopBP1, it was found that the first three BRCT repeats are sufficient to promote chromosome replication in extracts of Xenopus eggs, and phosphorylated Treslin binds to the first two BRCT repeats of TopBP1 (Kumagai et al. 2010). These features identify frog Treslin as a very interesting new replication factor, potentially analogous to yeast Sld3.
It now seems very likely that vertebrate orthologs of Treslin are generally required for chromosome replication. Depletion of Treslin in human cells blocked proliferation and appeared to inhibit chromosome replication (Kumagai et al. 2010). Moreover, an independent study identified the zebrafish ortholog of Treslin, called Ticrr (TopBP1-interacting checkpoint and replication regulator), as a novel factor that is crucial for chromosome replication and cell proliferation during early development (Sansam et al. 2010). In extracts of mutant ticrr early embryos, the MCM2–7 helicase can still be detected on chromatin, but association of GINS (and thus probably Cdc45) with chromatin is defective. The same study also showed that the first two BRCT repeats of human TopBP1 are required for interaction with human Ticrr/Treslin, although a role for CDK in promoting this interaction could not be detected in this case (Sansam et al. 2010). It thus seems clear that Treslin/Ticrr is an important new player in the initiation of chromosome replication, working together with TopBP1.
Interestingly, a single ortholog of Treslin can be detected not just in other vertebrate species, but also in simple invertebrate animals such as the Placazoan Trichoplax adhaerens. It remains to be determined whether distantly related homologs of Treslin occur in other eukaryotic taxa, and whether Treslin orthologs have an evolutionary origin in common with yeast Sld3 proteins, but this seems an intriguing possibility.
In addition to Treslin/Ticrr, another novel protein has just been shown to be an essential target of CDK during the initiation of chromosome replication in Xenopus. By searching for proteins that are related in sequence to Geminin, which inhibits loading at origins of the MCM2–7 helicase, Balestrini et al. (2010) found a novel factor that they call GEMC1 (Geminin coiled-coil-containing protein 1). In contrast to Geminin, however, GEMC1 is required for the initiation of chromosome replication (Balestrini et al. 2010). Depletion of GEMC1 from an extract of Xenopus eggs prevents Cdc45 and GINS from binding to chromatin, but does not prevent the loading of MCM2–7, TopBP1, or Cdc7. Moreover, these effects can be rescued by addition of recombinant GEMC1 protein to the depleted egg extracts (Balestrini et al. 2010).
The association of GEMC1 with chromatin requires the origin recognition complex and is greatly reduced in the absence of TopBP1 (in contrast, Treslin can bind to chromatin without TopBP1). Interestingly, TopBP1, Cdc45, and Cdk2–Cyclin E were found to coimmunoprecipitate with GEMC1 from extracts of interphase Xenopus eggs, although the region of TopBP1 that interacts with GEMC1 still remains to be characterized (Balestrini et al. 2010). Crucially, GEMC1 is phosphorylated in vitro by CDK, and mutation of the relevant sites prevents GEMC1 from supporting chromosome replication CDK phosphorylation. Nevertheless, CDK appears to regulate GEMC1 in a manner different from Treslin, as CDK phosphorylation does not appear to be essential for interaction of GEMC1 with TopBP1, although a phosphomimetic mutant of GEMC1 has enhanced binding to TopBP1 (Balestrini et al. 2010).
GEMC1 is required for proliferation during the development of fertilized frog eggs, and the mouse ortholog also appears to be required for chromosome replication in cultured cells. It thus appears that GEMC1 is another exciting new replication factor, required at a similar stage as Treslin, but with somewhat different properties. It seems clear that GEMC1 works together with TopBP1 in a manner regulated by CDK, at least in frog extracts, and the properties of GEMC1 are somewhat reminiscent of yeast Sld3 (or even Sld2 for that matter).
Finally, a third factor recently has been reported to be required for the association of Cdc45 with chromatin during chromosome replication in extracts of Xenopus eggs. The DUE-B (DNA unwinding element) protein was originally described as a factor that binds to the DNA unwinding element of the c-MYC origin of DNA replication in human cells (Casper et al. 2005). A new study has shown that depletion of DUE-B from extracts of Xenopus eggs not only inhibits the association of Cdc45 with chromatin, but also largely compromises the chromatin binding of TopBP1 too (the requirement of DUE-B for loading of TopBP1 contrasts with Treslin and GEMC1), and both effects can be rescued with recombinant DUE-B protein purified from HeLa cells (Chowdhury et al. 2010). Very surprisingly, the N-terminal 150 amino acids of DUE-B comprise a proofreading activity for tRNAs, known as D-aminoacyl-tRNA deacylase, which is conserved in all eukaryotes. In contrast, the C-terminal 59 amino acids are not found in the yeast orthologs and are required for DUE-B to associate with Cdc45 and TopBP1 (Chowdhury et al. 2010). Depletion of DUE-B in human cells reduces the association of Cdc45 but not MCM2–7 with chromatin, and a fraction of DUE-B can be seen in subnuclear foci (although this is not known to require the C-terminal 59 amino acids). In summary, the requirement of DUE-B for chromosome replication is very intriguing, but the role of DUE-B is apparently distinct from that of Treslin and GEMC1, as it appears to be required for the association of TopBP1 with chromatin. Clearly, further studies are required to understand in more detail the role of this factor during chromosome replication.
Is there a conserved mechanism for the initiation of chromosome replication in all eukaryotes?
It seems very likely that the machinery at DNA replication forks will be almost identical from yeast to human cells, as it is clear that most of the known replication factors have a single ortholog in all species. So it would be natural to think that the initiation mechanism will be highly conserved too, and in all eukaryotes, the underlying principles are likely to be the same: Cdc7 and CDK somehow activate the MCM2–7 helicase at origins, and an important and conserved feature of this process is the recruitment of Cdc45 and GINS, allowing the formation of the CMG helicase. Nevertheless, the manner by which Cdc45 and GINS are recruited could alter over time in different species, in response to different requirements for regulation, without changing the end product of the initiation reaction. It is clearly too soon to describe the initiation mechanism in great detail, but it seems very likely that it will follow a number of emerging principles.
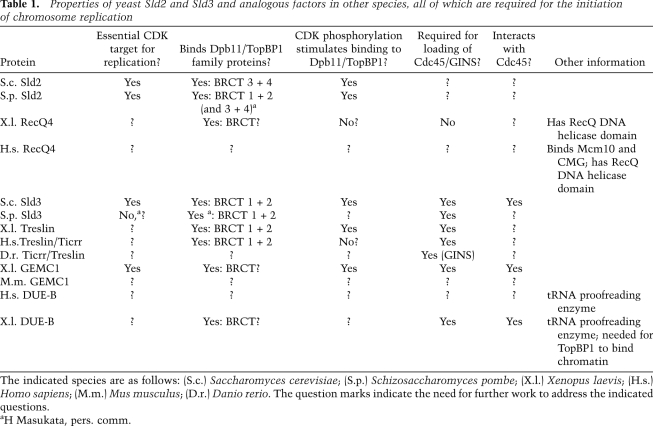
First, it appears that a major target of the Cdc7 kinase in most species will be the N-terminal tails of several subunits of the MCM2–7 helicase, perhaps inducing a structural change within the MCM2–7 complex that promotes activation and stable association with Cdc45 and GINS. Second, assembly factors promote the recruitment of GINS and Cdc45 to the MCM2–7 helicase at origins, and the ortholog of Dpb11/Rad4/Cut5/TopBP1 is likely to play a central role in mediating the interactions between these factors (Table 1). In budding yeast, this process is stimulated by CDK phosphorylation of Sld2 and Sld3. In fission yeast, it is clear that Sld2 is an important target of CDK, but Cdc7 also contributes to the same step by allowing recruitment of Sld3 to origins. It seems likely that CDK phosphorylation of analogous proteins such as Treslin/Ticrr and GEMC1 might be important for recruitment of Cdc45 to origins in vertebrate cells too. The mechanism by which GINS is recruited to origins remains unclear outside of budding yeast, and it is not known whether GINS is recruited together with Cdc45, or in association with other factors.
Table 1.
Properties of yeast Sld2 and Sld3 and analogous factors in other species, all of which are required for the initiation of chromosome replication
It is still possible, however, that more parts of the puzzle remain to be identified even in budding yeast. For example, Sld3 is now thought to exist in a complex with a new factor called Sld7 (H Araki, pers. comm.), and it will be very interesting, for example, to see whether Treslin and GEMC1 interact in an analogous manner as part of the initiation mechanism. Finally, it is also possible that other kinases contribute to activation of the MCM2–7 helicase during the initiation of chromosome replication, in addition to Cdc7 and CDK. There is evidence that an unidentified kinase activity in budding yeast is able to phosphorylate preferentially the loaded MCM2–7 helicase at origins, and this appears to stimulate the subsequent phosphorylation of MCM2–7 by Cdc7 (Francis et al. 2009). One interesting candidate is Casein Kinase II, which is an acidophilic kinase that has been shown previously to phosphorylate human Mcm2 in vitro (Montagnoli et al. 2006). There is clearly still much to learn about the mechanism by which the MCM2–7 helicase is activated at origins during the initiation reaction in eukaryotic cells. But the recent progress in this area suggests that a more complete picture might not be too far away.
Acknowledgments
I am very grateful to Cancer Research UK, who funds the work in my group, and I also thank Hiro Araki, Vincenzo Costanzo, Bill Dunphy, Michael Leffak, and Hisao Masukata for generously sharing unpublished information during the writing of this review, and for comments on the manuscript.
Footnotes
References
- Aparicio OM, Weinstein DM, Bell SP 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM complexes and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J 2009. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res 37: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H 2010. Regulatory mechanism of the initiation step of DNA replication by CDK in budding yeast. Biochim Biophys Acta 1804: 520–523 [DOI] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A 1995. Dpb11, which interacts with DNApolymerase IIɛ in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell-cycle checkpoint. Proc Natl Acad Sci 92: 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V 2010. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol 12: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmidt C, Pollok S, Kremmer E, Nasheuer HP, Grosse F 2007. Interactions of human Cdc45 with the Mcm2–7 complex, the GINS complex, and DNA polymerases δ and ɛ during S phase. Genes Cells 12: 745–758 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A 2009. The Mcm complex: Unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73: 652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JFX 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ 1998. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem 273: 22083–22090 [DOI] [PubMed] [Google Scholar]
- Bruck I, Kaplan D 2009. Dbf4–Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem 284: 28823–28831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper JM, Kemp MG, Ghosh M, Randall GM, Vaillant A, Leffak M 2005. The c-myc DNA-unwinding element-binding protein modulates the assembly of DNA replication complexes in vitro. J Biol Chem 280: 13071–13083 [DOI] [PubMed] [Google Scholar]
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK 2006. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci 103: 11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Liu G, Kemp M, Chen X, Katrangi N, Myers S, Ghosh M, Yao J, Gao Y, Bubulya P, et al. 2010. The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol Cell Biol 30: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LC, Teixeira LK, Wohlschlegel JA, Henze M, Yates JR, Mendez J, Reed SI 2009. Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol Cell 35: 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A, Gueydon E, Schwob E 2008. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol Biol Cell 19: 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan WP, Sherman DA, Forsburg SL 2004. Schizosaccharomyces pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding. Chromosoma 113: 145–156 [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ 1998a. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev 12: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL 1998b. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell 2: 173–183 [DOI] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JFX 1994. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science 265: 1243–1246 [DOI] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C 2009. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci 106: 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M, Carr AM, Murray J, Watts FZ, Lehmann AR 1991. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res 19: 6737–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS 2003. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol 10: 160–167 [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP 2009. Incorporation into the prereplicative complex activates the Mcm2–7 helicase for Cdc7–Dbf4 phosphorylation. Genes Dev 23: 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Aparicio JG, Hu F, Aparicio OM 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol 24: 10208–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Lindner K, Brimage L, Franklin R, Namdar M, Hart EA, Aves SJ, Kearsey SE 2003. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol Biol Cell 14: 3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA 1997. Mcm5/Cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci 94: 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tsujimura T, Sugino A, Takisawa H 2006. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells 11: 993–1007 [DOI] [PubMed] [Google Scholar]
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK 2009. Assembly of the Cdc45–Mcm2–7–GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci 106: 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA 1993. Cell cycle regulation of the yeast CDC7 protein kinase by association with the DBF4 protein. Mol Cell Biol 13: 2899–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Reed SI, Haase SB 2006. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol 26: 2456–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Blow JJ 2000. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev 14: 1528–1540 [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak Y-S, Sugino A, Araki H 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 20: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K 2006. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–725 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JFX 2000. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK 1997. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev 11: 3365–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 bY Cdks. J Biol Chem 275: 29042–29052 [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. 2006. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 281: 39249–39261 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655 [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H 2006. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol Cell Biol 26: 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Yamashita S 1998. Identification of sna41 gene, which is the suppressor of nda4 mutation and is involved in DNA replication in Schizosaccharomyces pombe. Genes Cells 3: 157–166 [DOI] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D 1982. Cold-sensitive cell-division-cycle mutants of yeast: Isolation, properties, and pseudoreversion studies. Genetics 100: 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C 2006. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem 281: 10281–10290 [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR 2006. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ɛ, and GINS in budding yeast. Genes Dev 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R, Masukata H 2002. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol Biol Cell 13: 1462–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Shanahan P, Noguchi C, Russell P 2002. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Nougarede R, Della Seta F, Zarzov P, Schwob E 2000. Hierarchy of S-phase-promoting factors: Yeast Dbf4–cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol 20: 3795–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Walter JC 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J 23: 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Patel PK, Kommajosyula N, Rosebrock A, Bensimon A, Leatherwood J, Bechhoefer J, Rhind N 2008. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol Biol Cell 19: 5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereverzeva I, Whitmire E, Khan B, Coue M 2000. Distinct phosphoisoforms of the Xenopus Mcm4 protein regulate the function of the Mcm complex. Mol Cell Biol 20: 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JF 2009. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol 21: 771–777 [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF 2009. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121: 887–898 [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 24: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Wollmann Y, Franke C, Grosse F, Saluz HP, Hanel F 2008. Characterization of the interaction between the human DNA topoisomerase IIβ-binding protein 1 (TopBP1) and the cell division cycle 45 (Cdc45) protein. Biochem J 409: 169–177 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Fletcher RJ, Chen XS 2004. Two heads are better than one: Regulation of DNA replication by hexameric helicases. Genes Dev 18: 2039–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Ying CY, Gautier J 2004. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J Biol Chem 279: 45586–45593 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2006. Cdc7–Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords R, Mahalingam D, O'Dwyer M, Santocanale C, Kelly K, Carew J, Giles F 2010. Cdc7 kinase—A new target for drug development. Eur J Cancer 46: 33–40 [DOI] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H 2006. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2–Dpb11. EMBO J 25: 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JFX 2000. DNA synthesis at individual replication forks requires the essential initiation factor, Cdc45p. EMBO J 19: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM, Gillespie PJ, Blow JJ 2010. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol 188: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem 275: 39773–39778 [DOI] [PubMed] [Google Scholar]
- Wang H, Elledge SJ 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci 96: 3824–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Stillman B 1999. Cdc7p–Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J 18: 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y 2009. MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J 28: 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H 2006. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J 25: 4663–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Nakagawa T, Masukata H 2004. A novel intermediate in initiation complex assembly for fission yeast DNA replication. Mol Biol Cell 15: 3740–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Lindsay HD, Michael WM 2006. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J Cell Biol 173: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]