A fluorophore ligase for site-specific protein labeling inside living cells (original) (raw)
Abstract
Biological microscopy would benefit from smaller alternatives to green fluorescent protein for imaging specific proteins in living cells. Here we introduce PRIME (PRobe Incorporation Mediated by Enzymes), a method for fluorescent labeling of peptide-fused recombinant proteins in living cells with high specificity. PRIME uses an engineered fluorophore ligase, which is derived from the natural Escherichia coli enzyme lipoic acid ligase (LplA). Through structure-guided mutagenesis, we created a mutant ligase capable of recognizing a 7-hydroxycoumarin substrate and catalyzing its covalent conjugation to a transposable 13-amino acid peptide called LAP (LplA Acceptor Peptide). We showed that this fluorophore ligation occurs in cells in 10 min and that it is highly specific for LAP fusion proteins over all endogenous mammalian proteins. By genetically targeting the PRIME ligase to specific subcellular compartments, we were able to selectively label spatially distinct subsets of proteins, such as the surface pool of neurexin and the nuclear pool of actin.
Keywords: fluorescence microscopy, biotechnology, enzyme engineering
Techniques for posttranslational labeling of proteins in living cells address some of the shortcomings of green fluorescent protein (GFP) by expanding the repertoire of chemical probes available for protein visualization (1). However, most of these methods, such as HaloTag (2), SNAP/CLIP (3), and DHFR (4), still use large protein tags that sterically interfere with protein trafficking and function, as GFP is known to do (5, 6). FlAsH (7) is the only peptide-based posttranslational labeling method that currently works inside living cells. At 12 amino acids, the FlAsH tag is much smaller than GFP, but poor labeling specificity (7–9), cellular toxicity, and undesired palmitoylation (7) and oxidation (8) of the Cys4 recognition motif limit its utility.
Here we introduce a method for protein labeling that utilizes a peptide tag while preserving high sequence specificity inside living cells. Our method, called PRIME (PRobe Incorporation Mediated by Enzymes), is based on a “fluorophore ligase” that is engineered from the Escherichia coli enzyme lipoic acid ligase (LplA). LplA’s natural function is to ligate lipoic acid onto three E. coli proteins involved in oxidative metabolism (10). We previously used LplA for labeling of cell-surface proteins by demonstrating that the wild-type enzyme can ligate an azidoalkanoic acid instead of lipoic acid (11) (Fig. 1A, Middle). Ligated azide could then be chemoselectively derivatized using cyclooctyne-fluorophore conjugates.
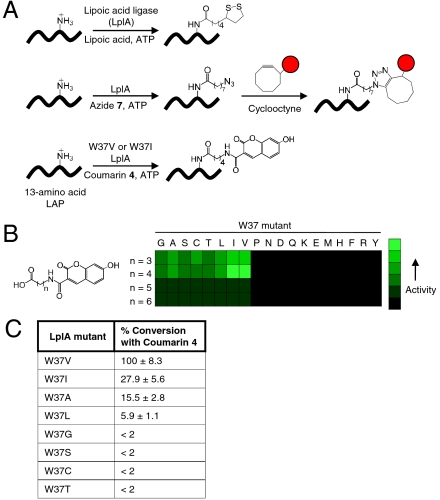
Fig. 1.
Engineering a coumarin ligase. (A) Natural and engineered ligation reactions catalyzed by lipoic acid ligase (LplA). The middle row shows two-step probe targeting via alkyl azide 7 ligation followed by [3 + 2] cycloaddition (12), and the bottom row shows direct fluorophore ligation by an LplA mutant. LAP = LplA Acceptor Peptide (20). The red circle represents any probe. (B) Relative activities of W37 LplA mutants with 7-hydroxycoumarin derivatives. Assays were performed under two conditions: 12 hours with LplA’s natural protein substrate E2p (15) or 40 min with LAP4.3D (20) (see SI Methods). No product was detected for the black-shaded quadrants. (C) Percent of LAP4.3D peptide converted to the coumarin (n = 4) conjugate for the eight most active W37 LplA mutants. Enzymes were used at 1 μM and reaction times were 40 min. Values are normalized to the percent conversion obtained with the best mutant, W37V, which is set to 100%. Measurements were performed in triplicate. Errors, ± 1 s.d.
In this work, we wished to extend LplA-mediated labeling to intracellular proteins but recognized the challenges associated with our two-step labeling scheme. First, labeling sensitivity is limited by the kinetics of strain-promoted [3 + 2] cycloaddition, which has a rate constant of 4.3 × 10-3 M-1 sec-1 (12). Second, for intracellular applications, two washout steps would be needed: first to remove excess azide, and second to remove excess fluorophore. We sought to create a simpler, one-step fluorophore ligation strategy as an alternative.
Through LplA active site mutagenesis, we created a ligase capable of directly attaching the blue fluorophore 7-hydroxycoumarin to proteins fused to a 13-amino acid LplA recognition sequence (Fig. 1A, Bottom). We characterized the kinetics and specificity of this fluorophore ligation in vitro. We then demonstrated coumarin PRIME labeling on different proteins in the cytosol and nucleus of various mammalian cell types. A side-by-side comparison with FlAsH showed that PRIME labeling is faster, more specific, and less toxic. We also used PRIME to label protein subpopulations in specific cellular compartments and to probe the biology of nuclear actin.
Results
Structure-Guided Mutagenesis and Screening for a Coumarin Fluorophore Ligase.
We selected 7-hydroxycoumarin as our fluorophore for this study because its excitation and emission wavelengths are suitable for live-cell imaging (excitation 387–405 nm; emission 448 nm) (13), it is brighter than Enhanced Blue Fluorescent Protein (EBFP) [_ε_ = 36,700 M-1 cm-1 and QY 0.7 for the anionic form (13), compared to ε = 31,500 M-1 cm-1 and QY 0.2 for EBFP (14)], and it is small and hydrophobic, which increases the likelihood that it will be accepted by LplA. We synthesized four 7-hydroxycoumarin structures (Fig. 1B) with varying linkers separating the fluorophore and the carboxylic acid that is activated by LplA to the adenylate ester.
Using a high-performance liquid chromatography (HPLC) readout of ligation onto E2p, a domain from LplA’s natural protein substrate pyruvate dehydrogenase (15), we found that wild-type LplA, not surprisingly, does not use any of the coumarin substrates. We then examined the crystal structure of E. coli LplA bound to lipoic acid (16), and the related structure of Thermoplasma acidophilum LplA bound to lipoyl-AMP (17), to identify candidate positions for mutagenesis. Thirteen residues lie within 7.5 Å of the dithiolane moiety of lipoic acid in either LplA structure: N16, L17, V19, E20, F35, W37, S71, S72, H79, T87, R140, F147, H149 (E. coli numbering; Fig. S1_A_ and B). We mutated each of these positions to alanine one by one and tested for uptake of the two smaller coumarin probes (n = 3,4). Only W37ALplA gave detectable ligation product.
Fig. S1_B_ shows that W37 is 8.5 Å from the dithiolane ring in the E. coli structure, whereas the analogous residue in T. acidophilum LplA, Y39, is much closer, at 3.9 Å (Fig. S1_A_). A new structure of E. coli LplA with lipoyl-AMP bound (18) instead of lipoic acid suggests that the enzyme undergoes a conformational change upon formation of the adenylate ester. Interestingly, in this new structure, the W37 position is much closer to the dithiolane ring (3.6 Å, Fig. S1_C_). We note that our previous work to engineer an aryl azide ligase also identified W37 as the position most important (out of nine positions tested) for expanding LplA substrate specificity (19).
Encouraged by the coumarin ligation activity of W37ALplA, we prepared a panel of all possible W37 LplA mutants and screened them against all four coumarin probes (Fig. 1B). We found that 11 mutants were inactive, although most of these were still able to ligate lipoic acid. Of the eight active mutants, little product was detected with the longer coumarin probes (n = 5,6), whereas the most product was seen with coumarin 4 (n = 4). We therefore reassayed these eight mutants under milder conditions with coumarin 4 to differentiate among them. Instead of E2p, the HPLC assay was performed with an “LplA acceptor peptide” (LAP). Specifically, we used the yeast display-evolved sequence “LAP4.3D” (20) due to its superior chromatographic properties on HPLC when fused to the carrier protein HP1 (11).
Fig. 1C compares the amount of coumarin 4-LAP4.3D conjugate obtained with the eight active W37 mutants. Intermediate-sized hydrophobic sidechains—such as valine and isoleucine—were best able to complement the shape and size of coumarin 4. The preference for hydrophobic residues suggests that the ligase binds the neutral form of 7-hydroxycoumarin, rather than its anionic form (pKa = 7.5) (13). This idea is supported by the observation that a fluorinated isostere of coumarin 4 called Pacific Blue, with a pKa of 3.7 (13), is ligated by W37VLplA and W37ILplA with much lower efficiency than coumarin 4.
Coumarin Ligation Kinetics and Sequence Specificity.
We proceeded to characterize the two most active coumarin ligases identified in our screen: W37VLplA and W37ILplA. First, HPLC assays were performed, this time using our most kinetically efficient LAP called “LAP2” (20). Controls in Fig. 2A show that coumarin ligation to LAP2 requires ATP and is not catalyzed by wild-type LplA. The HPLC product peaks from the reactions with both ligases were collected and analyzed by mass spectrometry, confirming the identity of the covalent coumarin 4-LAP2 adducts (Fig. S2).
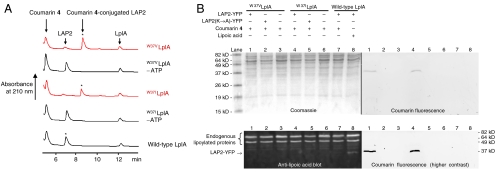
Fig. 2.
In vitro characterization of coumarin ligase. (A) HPLC traces showing conversion of LAP2 peptide (20) to the coumarin 4 conjugate by W37VLplA or W37ILplA (red traces). Negative controls with wild-type LplA, or ATP omitted, are shown in black. Starred peaks were collected and analyzed by mass spectrometry (Fig. S2). (B) Coumarin labeling of mammalian cell lysate to determine sequence-specificity. Lanes 1 and 4 show coumarin 4 ligation to LAP2-YFP catalyzed by W37VLplA and W37ILplA, respectively. Negative controls are shown with wild-type LplA (lane 7), an alanine mutation in LAP2 (lanes 2 and 5), and untransfected lysate (lanes 3 and 6). The same samples were also analyzed by blotting with anti-lipoic acid antibody. Lysate in lane 8 was subjected to in vitro lipoylation with exogenous wild-type LplA.
Second, we used HPLC to measure the kinetics of coumarin ligation onto LAP2 (Fig. S3_A_). For W37VLplA, we obtained a _k_cat of 0.019 ± 0.004 sec-1 and a K m of 56 ± 20 μM. W37ILplA had a similar _k_cat (0.016 ± 0.002 sec-1) but a much higher K m of 261 ± 69 μM. These values can be compared to previously determined kinetic constants for natural and unnatural LplA-catalyzed ligations (Fig. S3_B_). Whereas the _k_cat of coumarin ligation is 10–30-fold poorer than previous _k_cat values determined for lipoic acid (11, 15, 16), azidoalkanoic acid (11), and aryl azide (19), the coumarin K m for W37VLplA is better than previous K m values for unnatural substrates. In contrast, the coumarin K m for W37ILplA is much poorer. For live-cell labeling, we were uncertain whether low coumarin K m would confer an advantage, because intracellular coumarin concentrations following probe loading may exceed 300 μM (vide infra). We therefore tested both LplA mutants for coumarin ligation activity in living cells.
Third, we characterized the sequence specificity of both coumarin ligases. It was unclear if remodeling of the lipoic acid binding pocket would affect LplA’s recognition of peptide and protein substrates. In addition, mammalian cells express protein substrates for their own mitochondrial lipoic acid ligase that might cross-react with E. coli LplA (15). To test for these possibilities, we performed coumarin labeling of LAP2-YFP (yellow fluorescent protein) in mammalian cell lysate and analyzed the samples by in-gel coumarin fluorescence imaging and anti-lipoic acid Western blotting. Fig. 2B shows that the endogenous protein substrates of mammalian lipoic acid ligase are present in the lysate (see anti-lipoic acid blot) but are not labeled by coumarin. In addition, no other endogenous proteins are labeled by coumarin, even though many are expressed at much higher levels than LAP2-YFP according to the Coomassie stain. It is possible that mitochondrial proteins are not labeled by coumarin because they are already saturated with lipoic acid. This idea is supported by the observation that in vitro lipoylation of lysate does not increase the lipoylation level of these endogenous proteins (lane 8 of Fig. 2B).
We also found that LAP2-YFP proteins were not detectably lipoylated, even though free lipoic acid may be present in the cytosol (lanes 1, 4, and 7). To confirm site-specificity, we mutated the central lysine in LAP2 to alanine and found that this abolished labeling by both W37VLplA and W37ILplA (lanes 2 and 5). Thus, both coumarin ligases retain the high sequence-specificity of wild-type LplA.
Characterization of PRIME Labeling Inside Living Mammalian Cells.
To test coumarin ligation inside cells, we first addressed the issue of probe delivery. Coumarin 4 has 1–2 negative charges at physiological pH, making it membrane-impermeant. We protected either the carboxylic acid alone or both the acid and the 7-hydroxyl group with acetoxymethyl (AM) groups (coumarin-AM2; structure shown in Fig. 3A), which are known to be cleaved inside mammalian cells by endogenous esterases (21). Over time, excess anionic fluorophore is removed from cells via the action of organic anionic transporters (22). We found that one AM protecting group was not sufficient for membrane permeability; however, coumarin-AM2 entered cells readily (Fig. S4_A_).
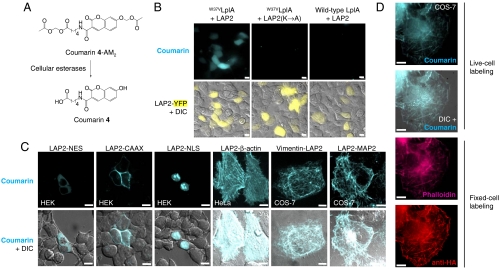
Fig. 3.
Coumarin PRIME labeling in living mammalian cells. (A) Structure of membrane-permeant coumarin 4-AM2 probe used for intracellular labeling. Reaction catalyzed by endogenous cellular esterases produces the parent coumarin 4 structure. (B) PRIME labeling of LAP2-YFP in HEK cells. Negative controls are shown with a Lys → Ala mutation in LAP2, and with W37VLplA replaced by wild-type LplA. (C) Labeling of different LAP2 fusion proteins. NES = nuclear export sequence. CAAX = prenylation tag. NLS = nuclear localization sequence. MAP2 = microtubule-associated protein 2. (D) Comparison of methods for actin visualization. Coumarin PRIME labeling was performed on live cells expressing HA-LAP2-β-actin. After fixation, actin was detected by staining with phalloidin-Alexa Fluor 647 and anti-HA antibody. For B_–_D, coumarin labeling was performed for 10 min with W37VLplA, followed by 1-hour washout of excess probe. All scale bars, 10 μm. Epifluorescence images are shown in B and D; confocal images are shown in C.
We used the “wedge method” (see SI Methods) to measure the intracellular concentration of coumarin probe immediately after loading. With 20 μM coumarin-AM2, the intracellular coumarin concentration was ∼469 μM after 10 min of loading (Fig. S4_B_). This concentration exceeds the coumarin K m for both W37VLplA and W37ILplA. Empirical optimization showed that coumarin probe could be thoroughly “washed out” of cells by replacing the media several times over 30 min (Fig. S4_C_).
Next, we evaluated LplA expression in mammalian cells. The wild-type E. coli LplA gene was found to express poorly in Human Embryonic Kidney (HEK) and HeLa cells. Fusions to fluorescent proteins helped, but we obtained the best results by resynthesizing the LplA gene with human-preferred codons. Humanized genes for both W37VLplA and W37ILplA gave robust and reproducible expression in a variety of cell lines, and their levels could be titrated by modulating the plasmid concentration used for liposome-mediated transfection (Fig. S5_A_).
Combining the optimized coumarin loading protocol, humanized LplA gene, and a LAP2-YFP fusion construct, we performed coumarin labeling with both W37VLplA and W37ILplA in live HEK cells (Fig. S5_B_). We observed specific labeling with both coumarin ligases, evident from comparing the coumarin signal for LAP2-YFP and LAP2(K → A)-YFP. W37VLplA gave higher coumarin signal/background ratios on average, consistent with its superior k cat/K m compared to W37ILplA. However, when each ligase was overexpressed by using 200 ng plasmid instead of 20 ng (per 0.95 cm2 of cells), some nonspecific labeling was seen with W37VLplA (Fig. S5 B and C). Analysis using targeted ligase constructs showed that this background correlated with W37VLplA localization and expression level. We hypothesize that W37VLplA may retain the coumarin-AMP ester intermediate within its active site, as the homologous enzyme biotin ligase is known to do with biotin-AMP ester (23). W37ILplA does not exhibit this behavior, likely due to its much higher K m for coumarin.
From these observations, we conclude that W37VLplA is the best coumarin ligase for PRIME labeling, but W37ILplA is preferred when ligase overexpression cannot be avoided. We did not observe background labeling with W37ILplA under any experimental condition that we tested, including six-hour-long incubation with 20 μM coumarin-AM2.
PRIME labeling with W37VLplA was characterized in HEK cells along with negative controls (Fig. 3B). After 10-min incubation with probe, followed by 60-min washout, only cells expressing LAP2-YFP retained coumarin fluorescence, whereas neighboring untransfected cells were not labeled. Mutation of LAP2 or replacement of W37VLplA with wild-type LplA eliminated coumarin labeling. PRIME labeling was performed successfully in a variety of cell lines, including HEK, HeLa, and COS-7 (Fig. 3C).
We also tested labeling of various LAP2 fusion proteins (Fig. 3C). LAP2-YFP targeted to the nucleus, cytosol, or plasma membrane gave the expected coumarin labeling patterns. Similarly, LAP2 fusions to the cytoskeletal proteins β-actin and vimentin or to MAP2 (microtubule-associated protein 2) could be specifically labeled with coumarin. Fig. 3D shows a comparison of PRIME labeling of β-actin in live cells to actin visualization by phalloidin or antibody staining after cell fixation. The coumarin signal overlaps well with the other stains but has the benefit of compatibility with live cells. Actin can also be visualized via fusion to fluorescent proteins. Fig. S9_A_ shows mApple-actin imaged in live COS-7 cells; here, actin is clearly excluded from the nucleus, an artifact that probably arises from the mApple tag interfering with nuclear import. We note that LAP2 was recognized by LplA at N-terminal, C-terminal, and internal fusion sites (see Table S1 for LAP2 placement in each construct).
To estimate the intracellular coumarin ligation yield, we used the “wedge method” (see SI Methods) to determine the single-cell concentrations of LAP2-YFP and attached coumarin probe after PRIME labeling. We found that 24 ± 13% of LAP2-YFP was labeled with coumarin after 10 min by W37VLplA (Fig. S6). The yield increased to 53 ± 25% after 1 h incubation with coumarin probe.
We also performed complete cellular characterization with W37ILplA, the less sensitive but more specific coumarin ligase. PRIME labeling with this mutant is shown with specificity controls, various LAP2 fusion proteins, and in various mammalian cell lines in Fig. S7 A and B. Intracellular labeling yield is reported in Fig. S6.
Comparison of PRIME and FlAsH Labeling.
We quantitatively analyzed the specificity, sensitivity, and toxicity of PRIME labeling in conjunction with a side-by-side comparison to FlAsH methodology. For the comparison, we prepared a single nuclear-targeted construct containing LAP2, mCherry, and the optimized 12-amino acid FlAsH recognition motif (7). In live HEK cells, PRIME labeling was performed first with W37ILplA for 10 min, followed by FlAsH for 30 min (Fig. S8_A_). To assess labeling specificity, we compared the nuclear coumarin or FlAsH signal (representing specific labeling, as the substrate is nuclear-localized) to the cytosolic signal (representing nonspecific labeling) in single cells. Fig. S8_B_ shows that FlAsH gives more cytosolic background than PRIME. We also observed nonspecific FlAsH labeling in untransfected cells. A similar analysis of labeling sensitivity revealed that both methods can detect as little as 10 μM target protein with signal/background ratio > 2∶1 (Fig. S8_C_). An assay for mitochondrial respiration showed that FlAsH is more toxic than PRIME (Fig. S8_D_).
Imaging Nuclear Actin with PRIME.
We used PRIME methodology to explore the biology of nuclear actin. Cytosolic β-actin has a well-recognized role in controlling cell migration and morphology, but biochemical studies have also suggested several roles for β-actin in the nucleus, including involvement in transcriptional regulation (24) and chromatin remodeling (25). Studies of nuclear actin have been hampered by the lack of methods to noninvasively detect all forms of actin in living cells. For example, antibody detection and phalloidin staining require cell fixation. Fluorescent proteins disrupt nuclear entry of actin, as shown in Fig. S9_A_. Phalloidin and Lifeact (26) are specific for polymeric actin over monomeric actin. Rhodamine-labeled actin can be visualized by fluorescent speckle microscopy, but this requires invasive microinjection (27).
We tested PRIME for labeling of nuclear actin in living cells. Fig. S9_A_ (Left) shows confocal images of coumarin-labeled LAP2-β-actin in COS-7 cells also expressing a nuclear marker (histone 2B fused to YFP, or H2B-YFP). Coumarin signal is clearly present in the nucleus, which contrasts with confocal images of mApple-β-actin (Fig. S9_A_, Right). Though actin was overexpressed in these experiments, the diffuse coumarin labeling pattern suggests that nuclear actin may exist as monomers or short oligomers, consistent with previous studies (28).
Previous work has shown that cell stress, such as heat shock, can increase the abundance of nuclear actin (29). We therefore imaged actin after 30–90 min of heat shock (43 °C) and found that coumarin-labeled actin accumulates in the nucleus within 30 min, whereas mApple-actin accumulation is much slower (Fig. S9_A_). In a small fraction (< 10%) of heat-shocked cells, we observed interesting coumarin-labeled nuclear actin structures (Fig. S9_B_), such as nucleolar clusters and filaments (which also stained with phalloidin, confirming F-actin morphology; Fig. S9_C_). This heterogeneity contrasted with the uniformly diffuse distribution of nuclear actin in nonstressed cells as well as mApple-actin in the nuclei of stressed cells. Perhaps filamentous actin, which has not previously been observed in the nuclei of living somatic cells, plays a role in transcriptional regulation (30) or nuclear stabilization (31).
The ability of PRIME methodology to detect nuclear pools of polymeric and nonpolymeric actin in living cells differentiates it from antibody, phalloidin, Lifeact, and fluorescent protein visualization techniques. PRIME may prove to be a useful tool for studying the biological function of nuclear actin.
Labeling Spatially Defined Protein Subpopulations Using Genetically Targeted Ligase.
A unique feature of PRIME labeling is its ability to selectively highlight protein subpopulations via genetic targeting of the fluorophore ligase. To test this concept, we transfected HEK cells with both nuclear LAP2 and cytosolic LAP2. When cytosolic W37VLplA was coexpressed, we observed specific labeling of the cytosolic subpopulation of LAP2 (Fig. 4A, Left). Conversely, when nuclear-localized W37VLplA was coexpressed, we observed specific labeling of the nuclear subpopulation of LAP2 (Fig. 4A, Right). When enzyme and substrate were not colocalized, no coumarin labeling was observed. Similar results were obtained with genetically targeted W37ILplA (Fig. S7_C_).
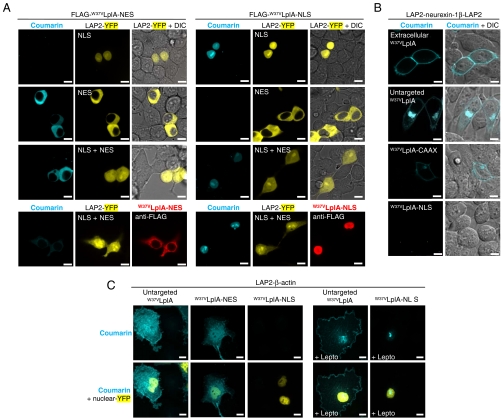
Fig. 4.
Compartmentalized PRIME labeling with genetically targeted coumarin ligase. (A) Compartmentalized labeling of cytosolic or nuclear LAP2-YFP. HEK cells expressing both LAP2-YFP-NES and LAP2-YFP-NLS (third row), or either construct alone (first and second rows), were labeled with coumarin using nuclear-excluded (left) or nuclear-targeted (right) ligase. On the bottom, anti-FLAG staining of fixed cells shows the expression pattern of FLAG-W37VLplA. (B) Compartmentalized labeling of the membrane protein neurexin-1β. HEK cells expressing double-LAP2 tagged neurexin-1β were labeled with W37VLplA added to the media (first row), expressed in the cytoplasm (second row), anchored to the inner plasma membrane (third row), or targeted to the nucleus (fourth row). (C) Compartmentalized labeling of actin. COS-7 cells expressing LAP2-β-actin and untargeted, nuclear-excluded, or nuclear-localized W37VLplA were labeled with coumarin for 30 min. The right panels show labeling in the presence of 2 ng/ml leptomycin B (Lepto) to inhibit nuclear export. All images in A_–_C are confocal. Scale bars, 10 μm.
We also tested this concept on neurexin-1β, a transmembrane adhesion protein that functions in synapse development and plasticity (32). We prepared a LAP2-neurexin-LAP2 double fusion construct to provide access to labeling reagents on both sides of the membrane. Fig. 4B shows PRIME labeling of the cell surface neurexin population by either purified W37VLplA applied to the cell medium or W37VLplA targeted to the inner leaflet of the plasma membrane. In contrast, untargeted W37VLplA labels the total intracellular pool of LAP2-neurexin-LAP2, including LAP2 present on the cytosolic face of the endoplasmic reticulum (ER) and the Golgi apparatus. Nuclear-targeted W37VLplA, as expected, does not label neurexin. Similar results were obtained using targeted forms of W37ILplA (Fig. S7_D_).
We also experimented with W37VLplA and W37ILplA targeted to the ER using a KDEL sequence. We did not detect coumarin ligation onto LAP2-neurexin-LAP2 or a number of other LAP2 fusion proteins. This may be a consequence of high calcium concentration in the ER (33), which we found inhibits LplA activity.
To provide further evidence for nuclear actin, we performed compartmentalized labeling of LAP2-β-actin using W37VLplA-NLS (Fig. 4C). We initially did not detect any nuclear coumarin signal, perhaps because nuclear actin is bound within protein complexes and not sterically accessible to LplA. The nuclear actin signal seen when labeling with untargeted W37VLplA (Fig. S9_A_ and Fig. 4C, Top) probably results from translocation of coumarin-labeled cytosolic actin to the nucleus, as suggested by a labeling experiment with W37VLplA-NES (Fig. 4C).
Therefore, we repeated the experiment with W37VLplA-NLS in the presence of leptomycin B to inhibit nuclear export of actin. In this case, we observed nuclear labeling, including actin structures similar to those seen in the nuclei of heat-shocked cells (Fig. 4C). Leptomycin B may cause nuclear actin levels to rise, saturating actin-binding proteins and producing free actin that is accessible to nuclear LplA.
Discussion
Using structure-guided mutagenesis of E. coli LplA, we have engineered a coumarin fluorophore ligase for PRIME labeling in living cells. Compared to other protein labeling methods, PRIME uniquely offers the combination of a peptide tag, high labeling specificity, and compatibility with the interior of living mammalian cells.
Aside from FlAsH, other peptide-based protein labeling methods are restricted to the cell surface either because the labeling reagents are not membrane-permeable [e.g., AcpS/Sfp (34) and sortase (35)] or because the chemistry is not orthogonal to intracellular functional groups [e.g., aldehyde tag (36) and ketone biotin (37)]. Unnatural amino acid mutagenesis stands apart as a tagless protein labeling method. Though demonstrations of fluorophore incorporation in mammalian cells are promising (38), the prevalence of native amber stop codons in mammalian cells (38) and potential dominant negative effects from truncated protein by-products (39) could limit the utility of this method.
Several other protein labeling methods, like PRIME, use enzymes to catalyze probe conjugation to proteins (40). Here we explored genetic targeting of the labeling enzyme as a method to spatially restrict labeling to protein subpopulations of interest. With suitable sensitivity and labeling speeds, one might be able to use this strategy to study protein trafficking between subcellular compartments and organelles. The utility would be greatest for compartments that are too small to selectively illuminate with focused light, such as neuronal synapses.
Materials and Methods
In Vitro Coumarin Ligation.
General reaction conditions: 100–150 μM LAP or E2p substrate was incubated with 1–2 μM LplA, 500 μM coumarin probe, 5 mM ATP, and 5 mM Mg(OAc)2 in 25 mM Na2HPO4 pH 7.2 at 30 °C for various lengths of time. Reactions were quenched with EDTA (C f 100 mM) and analyzed on a Varian Prostar HPLC using a reverse-phase C18 Microsorb-MV 100 column (250 × 4.6 mm). Chromatograms were recorded at 210 nm. For analysis of LAP2 peptide (GFEIDKVWYDLDA), we used a 10-min gradient of 30–60% acetonitrile in water with 0.1% trifluoroacetic acid at 1 ml/ min flow rate. Percent conversions were calculated by dividing the product peak area by the sum of (product + starting material) peak areas.
Coumarin Labeling of Mammalian Cell Lysate.
HEK cells expressing LAP2-YFP (or LAP2(K → A)-YFP) were lysed under hypotonic conditions in 1 mM HEPES pH 7.5 with 5 mM MgCl2, 1 mM phenylmethylsulphonyl fluoride, and protease inhibitor cocktail (Calbiochem). After three cycles of freeze-thaw, cells were mixed by vortexing for 2 min. Lysate was cleared by centrifugation and stored in aliquots at -80 °C. To label with coumarin or lipoic acid, lysates were incubated with 200 nM LplA, 500 μM coumarin 4 or 250 μM lipoic acid, 1.5 mM ATP, and 5 mM Mg(OAc)2 in 25 mM Na2HPO4 pH 7.2. After overnight incubation at 30 °C, reactions were boiled in protein loading buffer containing 2-mercaptoethanol for 7 min, then separated on a 12% SDS/PAGE gel. Coumarin fluorescence was visualized on an Alpha Innotech ChemiImager 5500 instrument using 365 nm UV light for excitation.
General Protocol for Coumarin PRIME Labeling in Living Cells.
HEK cells were transfected at ∼70% confluency with expression plasmids for W37VLplA (20 ng for a 0.95 cm2 dish) and the LAP2 fusion protein of interest (400–600 ng) using Lipofectamine 2000 (Invitrogen). 8–24 h after transfection, cells were incubated with 20 μM coumarin-AM2 in serum-free DMEM (Dulbecco’s modified Eagle medium, Cellgro) for 10 min at 37 °C. 0.1% w/v Pluronic F-127 (Invitrogen) can be optionally added to the labeling solution to give more even coumarin distribution in cells. The media was replaced 3–4 times over 30–60 min at 37 °C, with DMEM with 10% FBS (Fetal Bovine Serum), to wash out excess coumarin. Cells were imaged as described in SI Methods.
For further information on cloning, synthetic methods, cell culture, labeling, imaging, and other protocols, see SI Methods.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Amar Thyagarajan and Peng Zou for technical assistance. John Cronan (University of Illinois), Michael Davidson (Florida State University), Birka Hein (Max Planck, Göttingen), Daniel S. Liu (MIT), and Masahito Yamagata (Harvard) provided plasmids. Funding was provided by the National Institutes of Health (R01 GM072670), MIT, and the Dreyfus Foundation. S. T. was supported by an HHMI summer fellowship. M. F-S. was supported by a fellowship from La Caixa Foundation (Spain).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Lin MZ, Wang L. Selective labeling of proteins with chemical probes in living cells. Physiology. 2008;23:131–141. doi: 10.1152/physiol.00007.2008. [DOI] [PubMed] [Google Scholar]
- 2.Los GV, et al. HatoTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 3.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher SS, Sable JE, Sheetz MP, Cornish VW. An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem Biol. 2009;4:547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moritz OL, Tam BM, Papermaster DS, Nakayama T. A functional rhodopsin-green fluorescent protein fusion protein localizes correctly in transgenic Xenopus laevis retinal rods and is expressed in a time-dependent pattern. J Biol Chem. 2001;276:28242–28251. doi: 10.1074/jbc.M101476200. [DOI] [PubMed] [Google Scholar]
- 6.Marsh DR, Holmes KD, Dekaban GA, Weaver LC. Distribution of an NMDA receptor: GFP fusion protein in sensory neurons is altered by a C-terminal construct. J Neurochem. 2001;77:23–33. doi: 10.1046/j.1471-4159.2001.t01-1-00182.x. [DOI] [PubMed] [Google Scholar]
- 7.Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 8.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 9.Stroffekova K, Proenza C, Beam KG. The protein-labeling reagent FLASH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Arch Ges Phys. 2001;442:859–866. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- 10.Cronan JE, Zhao X, Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv Microb Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Suarez M, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agard NJ, et al. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 13.Sun WC, Gee KR, Haugland RP. Synthesis of novel fluorinated coumarins: Excellent UV-light excitable fluorescent dyes. Bioorg Med Chem Lett. 1998;8:3107–3110. doi: 10.1016/s0960-894x(98)00578-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang TT, et al. Improved fluorescence and dual color detection with enhanced blue and green variants of the green fluorescent protein. J Biol Chem. 1998;273:8212–8216. doi: 10.1074/jbc.273.14.8212. [DOI] [PubMed] [Google Scholar]
- 15.Green DE, et al. Purification and properties of the lipoate protein ligase of Escherichia-coli. Biochem J. 1995;309:853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara K, et al. Crystal structure of lipoate-protein ligase a from Escherichia coli—Determination of the lipoic acid-binding site. J Biol Chem. 2005;280:33645–33651. doi: 10.1074/jbc.M505010200. [DOI] [PubMed] [Google Scholar]
- 17.Kim DJ, et al. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: Insights into interaction with lipoyl domains. J Biol Chem. 2005;280:38081–38089. doi: 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara K, et al. Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A. J Biol Chem. 2010;285:9971–9980. doi: 10.1074/jbc.M109.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baruah H, et al. An engineered aryl azide ligase for site-specific mapping of protein-protein interactions through photo-cross-linking. Angew Chem Int Ed Engl. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puthenveetil S, et al. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J Am Chem Soc. 2009;131:16430–16438. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsien RY. Fluorescent probes of cell signaling. Annu Rev Neurosci. 1989;12:227–253. doi: 10.1146/annurev.ne.12.030189.001303. [DOI] [PubMed] [Google Scholar]
- 22.Oh YK, Straubinger RM. Cellular retention of liposome-delivered anionic compounds modulated by a probenecid-sensitive anion transporter. Pharm Res. 1997;14:1203–1209. doi: 10.1023/a:1012158924547. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Beckett D. Kinetics of biotinyl-5′-adenylate synthesis catalyzed by the Escherichia-coli repressor of biotin biosynthesis and the stability of the enzyme-product complex. Biochemistry. 1994;33:7354–7360. doi: 10.1021/bi00189a041. [DOI] [PubMed] [Google Scholar]
- 24.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 26.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 28.Vartiainen MK. Nuclear actin dynamics—From form to function. FEBS Lett. 2008;582:2033–2040. doi: 10.1016/j.febslet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann WA, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 31.Bohnsack MT, et al. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–263. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 32.Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashby MC, Tepikin AV. ER calcium and the functions of intracellular organelles. Semin Cell Dev Biol. 2001;12:11–17. doi: 10.1006/scdb.2000.0212. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, et al. Genetically encoded short peptide tags for orthogonal protein labeling by sfp and AcpS phosphopantetheinyl transferases. ACS Chem Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 35.Popp MW, et al. Sortagging: A versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 36.Wu P, et al. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc Natl Acad Sci USA. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen I, Howarth M, Lin WY, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 38.Liu WS, et al. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 39.Lu T, et al. Probing ion permeation and gating in a K+ channel with backbone mutations in the selectivity filter. Nat Neurosci. 2001;4:239–246. doi: 10.1038/85080. [DOI] [PubMed] [Google Scholar]
- 40.Sunbul M, Yin J. Site specific protein labeling by enzymatic posttranslational modification. Org Biomol Chem. 2009;7:3361–3371. doi: 10.1039/b908687k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information