The role of human bromodomains in chromatin biology and gene transcription (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 16.
Published in final edited form as: Curr Opin Drug Discov Devel. 2009 Sep;12(5):659–665.
Abstract
The acetylation of histone lysine is central to providing the dynamic regulation of chromatin-based gene transcription. The bromodomain (BRD), which is the conserved structural module in chromatin-associated proteins and histone acetyltranferases, is the sole protein domain known to recognize acetyl-lysine residues on proteins. Structural analyses of the recognition of lysine-acetylated peptides derived from histones and cellular proteins by BRDs have provided new insights into the differences between and unifying features of the selectivity that BRDs exhibit in binding biological ligands. Recent research has highlighted the importance of BRD/acetyl-lysine binding in orchestrating molecular interactions in chromatin biology and regulating gene transcription. These studies suggest that modulating BRD/acetyl-lysine interactions with small molecules may provide new opportunities for the control of gene expression in human health and disease.
Keywords: Acetyl-lysine recognition, bromodomain, gene transcription, lysine acetylation
Introduction
Changes in the structure of chromatin, which is a combination of histone proteins and DNA, are closely coupled to changes in gene transcription. This complex and tightly regulated relationship is possible because of the post-translational modification of DNA-packing histones present in the chromatin. Chromatin contains the entire genomic DNA in eukaryotic cells, and functions as the primary regulator controlling global dynamic changes in gene expression and silencing. Nucleosomes are primary components of chromatin and pack 147-bp lengths of DNA in two super-helical turns around a histone octamer, which consists of a histone-3-histone-4 (H3-H4) tetramer and two H2A-H2B dimers. These nucleosome core particles are connected by short lengths of DNA between the linker histones H1 and H5 to form a nucleosomal filament within the higher-order structure of the chromatin fiber. Within the chromatin structure, the N- and C-termini of the core histone octamers protrude out from the nucleosome particles and are subject to various post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination and small ubiquitin-like modification (SUMO)ylation. These site-specific modifications may act collectively in the cell nucleus to orchestrate genomic stability and gene expression or repression [1–3]. Of the various histone modifications, lysine acetylation [4] is the most dynamic as this modification directs both structural changes to chromatin and gene transcription [5–7].
The dynamic role of lysine acetylation in gene transcription is, to some extent, a result of the bromodomain (BRD), which is the only protein domain known to act as a acetyl-lysine binding domain [8]. BRD-containing proteins have also been implicated in disease processes, including cancer, inflammation and viral replication. This review describes the structural and functional features of human BRDs in chromatin biology and gene transcription.
The bromodomain fold and acetyl-lysine recognition
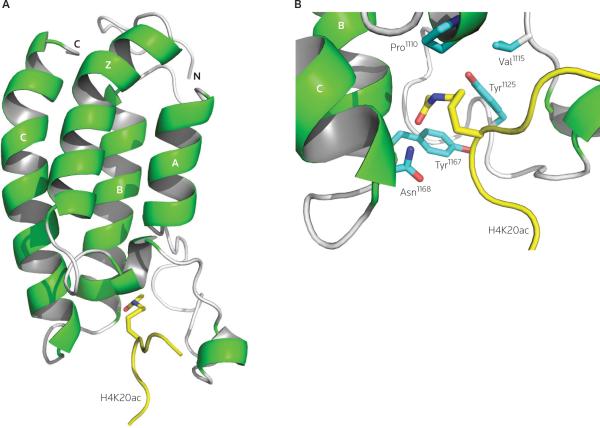
The available structures of the BRD from the human transcriptional co-activator PCAF (p300/CREB-binding protein-associated factor) [9,10] and the BRDs from the transcriptional protein TAF1 (transcription initiation factor TFIID subunit 1) [11] reveal that the BRDs of the histone acetyltransferase GCN5 [12,13], the co-activator CBP (CREB binding protein) [14], the BET family protein BRD2 [15], BPTF (the BRD and plant homeodomain [PHD] finger-containing transcription factor) [16] and the SNF2L4 (a SWI/SNF remodeling complex protein) [17] all adopt a distinct structural fold of a left-handed four-helix bundle (αZ, αA, αB and αC), termed the `BRD fold'. The inter-helical αZ-αA (ZA) and αB-αC (BC) loops constitute a hydrophobic pocket that recognizes the acetyl-lysine (Figure 1A). Notably, the structural features of BRD/acetyl-lysine binding are significantly different to those of chromodomain/methyl-lysine binding, in which a methyl-lysine sequence forms an anti-parallel β-strand to the β-barrel structure of the chromodomain [18,19]. The modular nature of the BRD fold enables the BRD to act as a functional unit within a protein, either individually or in combination with other modules.
Figure 1. Structural basis of acetyl-lysine recognition by the bromodomain.
(A) The 3-D structure of the CREB binding protein (CBP) bromodomain (BRD) bound to a H4K20ac peptide (PDB code: 2RNY); and (B) the acetyl-lysine binding site, showing the key interactions between the CBP BRD and a H4K20ac peptide. The peptide is yellow and the side chains of the protein residues are color-coded by atom type.
Despite the structurally conserved BRD fold, the overall sequence similarity between members of the BRD family is not high, and there are significant variations in the sequences of the ZA and BC loops [20]. Nevertheless, the amino acid residues that are engaged in acetyl-lysine recognition are among the most conserved residues in the large BRD family, and correspond to Tyr1125, Tyr1167 and Asn1168 in CBP (Figure 1B) [14,21]. The crystal structure of the yeast GCN5 BRD bound to a histone H4 peptide containing acetylated-Lys16 identified that, in addition to binding to the conserved Tyr364 and Tyr406 residues (corresponding to Tyr1125 and Tyr1167 in CBP, respectively), the acetyl-lysine residue forms a specific hydrogen-bond between the oxygen of the acetyl carbonyl group and the side-chain amide nitrogen of the conserved asparagine residue, Asn407 (corresponding to Asn1168 in CBP) [13]. A network of water-mediated hydrogen bonds involving carbonyl groups from the protein backbone at the base of the binding pocket also contributes to acetyl-lysine binding. The critical role of these three conserved amino acid residues in acetyl-lysine recognition has been confirmed by mutagenesis studies [9,10,14], and data demonstrate that most of the BRD family members function as acetyl-lysine binding domains [9]. Significantly, the key Asn1168 residue in CBP (Asn407 in GCN5) is not present in a small subgroup of BRDs, such as that of the transcriptional corepressor TIF1β (transcription intermediate factor 1β) or the sixth BRD in the human Polybromo protein. The former BRD does not bind to lysine-acetylated histones [22], whereas the latter BRD does [Zhou MM: unpublished data], suggesting that there may be another mode of acetyl-lysine binding to the conserved BRD fold.
The human bromodomain family
The human genome encodes 42 BRD-containing proteins, each of which contains between one and six BRDs [23]. The total number of unique individual human BRDs is 56 (2 human proteins, which contain 2 BRDs each, are both annotated as BRD2 and share > 99% sequence identity; their corresponding BRDs are identical in sequence [23]). These numbers can be contrasted with those of BRDs in the yeast genome, which encodes only nine BRD-containing proteins with a total of 14 BRDs [23]. The structural diversity of the human BRD family can be examined indirectly by clustering the 56 BRD sequences into groups that share similar sequence length and at least 35% sequence identity [24]. This yields nine groups, each of which contains at least two BRDs and eight outliers (see Table 1).
Table 1.
Classification of the human bromodomain family.
| Group | Bromodomains | Reference |
|---|---|---|
| 1 | PCAF, hGCN5, TIF1α, TIF1γ, TRI66, Rack7, BAZ2A, BAZ2B, NURF (isoform 2), Sp100, Sp110, Sp140, BRD21, BRD22, BRD31, BRD32, BRD41, BRD42, TAFII2101, TAFII2102, TAF11, TAF12, BRDT1 and BRDT2 | [9,11,12,16,27]* |
| 2 | SNF2L2, SNF4L4, Polybromo2, Polybromo4 and Polybromo5 | [17,29]* |
| 3 | BRD1, BRD7, BRD9, HOTTL and BRD and PHD finger-containing protein 3 | [30]* |
| 4 | BRD 1 of BRD and WD-repeat-containing proteins 1, 2 and 3 | - |
| 5 | BRD 2 of BRD and WD-repeat-containing proteins 1, 2 and 3 | - |
| 6 | ATAD2 and ATAD2B | * |
| 7 | Polybromo1 and Polybromo3 | - |
| 8 | CBP and p300 | [14]* |
| 9 | BRD81 and BRD82 | - |
| Outliers | Polybromo6, HRX/ALL-1, ASH1L, BAZ1B, BAZ1A, MYND11, TIF1β and CECR2 | [22] |
The dominant BRD group has 24 members, including the BRDs of several double-BRD-containing proteins such as BRD2, BRD3, BRD4, the TFIID (transcription initiation factor) 210-kDa subunit, TAF1 (the TFIID 250-kDa subunit) and the testis-specific protein BRDT [Sanchez R: unpublished data]. The same group includes BRDs from single-BRD-containing proteins such as the histone acetyltransferases PCAF and GCN5, TIF1α, TIF1γ, tripartite motif-containing protein 66, protein kinase C-binding protein 1 (Rack7), BAZ2A (BRD adjacent to zinc finger domain protein 2A), BAZ2B, BRD PHD finger transcription factor isoform 2, and the nuclear proteins Sp100, Sp110 and Sp140 [Sanchez R: unpublished data]. The structures of several proteins in this group have been solved [9,11,12,16]. The RIKEN Structural Genomics/Proteomics Initiative (RSGI) [25,26] and the Structural Genomics Consortium (SGC) [26] have determined the structures of the BRDs of BRD2, BRD3, BRD4, BRDT, TIF1α and BAZ2B. Huang et al determined the solution-state structure of the second BRD of BRD2 and demonstrated that this BRD is monomeric in solution and interacts dynamically with the acetylated Lys12 residue of histone H4 [27].
The second BRD group includes the BRDs of SNF2L2 and Brg-1, and the second, fourth and fifth BRDs of the Polybromo protein. The RSGI and the SGC have determined structures for the SNF2L2 BRD and the fifth BRD of the Polybromo protein [26,28], and research groups at two laboratories recently determined the structure of the Brg-1 BRD [17,29]. Shen et al used a solution structure and data from NMR perturbation studies to demonstrate that the Brg-1 BRD interacts with an H3K14ac peptide [17]. A crystal structure of the same domain exhibited an unusual small β-sheet in the ZA loop [29].
The third group of human BRD-containing proteins includes BRD7 and BRD9, as well as three proteins that contain a PHD in addition to BRDs. Sun et al reported a solution structure for the BRD7 BRD, and used NMR and titration analysis with several acetylated histone peptides to demonstrate that this BRD lacks inherent histone binding specificity in vitro [30]. The RSGI determined the BRD structure of HOTTL (tubulin-tyrosine ligase-like protein 3), one of the BRD and PHD containing proteins [28].
The next two groups of human BRDs correspond to the N-terminal and C-terminal BRDs of three BRD- and tryptophan-aspartate (WD)-repeat-containing proteins, respectively [Sanchez R: unpublished data]. No experimentally determined structures were available for these six proteins.
The remaining four groups of human BRDs contain pairs of similar BRDs: the BRDs of ATAD2 (ATPase family AAA domain-containing protein 2; ANCCA) and ATAD2B, the structures of which were determined by the RSGI and the SGC [26,28]; the first and third BRDs of human Polybromo; the BRDs of CBP and p300 (solution [14] and crystallographic (SGC) [26] structures were available for the BRD of CBP); and the two BRDs of BRD8.
The eight outliers in this classification of human BRDs correspond to the sixth BRD of Polybromo, and the BRDs of the zinc finger proteins HRX/ALL-1, ASH1L (absent small and homeotic disks protein 1 homolog), BAZ1B, BAZ1A, MYND (myeloid translocation protein 8, Nervy, and DEAF-1) domain-containing protein 11, TIF1β and CECR2 (cat eye syndrome critical region protein 2) [Sanchez R: unpublished data]. Of these BRDs, an experimentally determined structure was available for TIF1β, which formed a structural unit with the adjacent PHD finger [22].
In total, the structures of 23 of the 56 human BRDs have been experimentally determined. Of the 33 BRDs without experimental structures, 21 share > 35% sequence identity with a protein of known structure, and it is therefore possible to construct reasonably accurate homology models for these BRDs [24]. Although these data represent a thorough coverage of the structures of the BRD family proteins, no structural information is available for 12 of the 56 human BRDs. The unique sequences of these 12 BRDs of unknown structure (which either have low overall sequence similarity or long insertions with respect to the sequences of proteins with known structures) may contain special structural or functional features that have not yet been observed in the better characterized BRDs. Additionally, although the structural description of the BRDs is almost complete, many more structures of BRD-ligand complexes are required to facilitate a detailed understanding of ligand-binding selectivity. The existence of tandem domains (BRD-BRD, PHD-BRD or BRD-PHD), which are tightly associated with each other in sequence and most likely also in the 3D structure, adds another level of complexity to the structural description of BRDs.
The association of bromodomains with other chromatin modules
BRDs are promiscuous domains in that they occur in a variety of proteins with different domain architectures, and can be considered functionally independent (ie, BRD-containing proteins do not all perform the same function) [31,32]. For example, the BRD-containing protein PCAF is a histone lysine acetyltransferase, whereas HRX/ALL-1 is a histone lysine _N_-methyltransferase and SNF2L2 is an ATP-dependent helicase. More than 15 different domain types have been identified to occur within the same proteins as BRDs, including the PHD, PWWP, B-box type zinc finger, ring finger, SAND, FY Rich, SET, TAZ zinc Finger, helicase, ATPase, BAH (bromo adjacent homolog) domain, WD40 repeat and MBD (methyl-CpG binding domain) [23].
The domain that is most frequently associated with the BRD is the PHD finger, which is a C4HC3 zinc-finger-like motif present in nuclear proteins. A PHD has been identified in 19 of the 42 human BRD-containing proteins. In 12 of these proteins the PHD and BRD are separated by a short amino acid sequence (< 30 residues) and may form structurally interdependent tandem PHD/BRD arrangements such as those observed in TIF1β [22]. The TIF1β structure contains a distinct scaffold that unifies the two protein modules, in which the Z helix of the BRD forms a hydrophobic core that anchors the other three helices of the BRD on one side and the PHD finger on the other. A comprehensive structure-function analysis correlating transcriptional repression, UBC9 (ubiquitin-conjugating enzyme 9) binding and SUMOylation demonstrated that the PHD finger and BRD of TIF1β cooperate as a functional unit to facilitate lysine SUMOylation, which is required for TIF1β corepressor activity in gene silencing [22,33]. These results identified a unified function for the tandem PHD/BRD as an intramolecular SUMO E3 ligase for transcriptional silencing. The ligase activity is a divergent function for the BRD, which does not bind to lysine-acetylated histones in this form. In contrast to TIF1β, the structure of BPTF, which also contains a PHD finger and a BRD separated by a short linker [16], did not demonstrate any significant structural interactions between the two domains. In BPTF, the PHD domain recognized the methylated Lys4 residue of histone H3 (H3K4me3) [16,34]. However, the histone binding specificity of the BPTF BRD was not established. These examples demonstrate that not only do BRD-containing proteins vary significantly in function, but that the BRD itself may have different binding activities as a consequence of other associated domains.
The second most common domain association for the BRD is an association with another BRD; of the 42 BRD-containing proteins, 12 contain 2 or more BRDs. With the exception of Polybromo, which contains six BRDs, all other proteins with multiple BRDs contain two BRDs. In the transcription initiation factor TAF1 and TFIID 210-kDa subunits, as well as in some of the Polybromo BRD pairs, the two BRDs are separated by short amino acid sequences (< 20 residues). The structure of the TAF1 BRDs suggests that they form a tandem arrangement that binds selectively to multiple acetylated histone H4 peptides [11]. Another tandem BRD arrangement was observed in the yeast Rsc4 protein, which is related to human Polybromo [35]. The yeast Rsc4 structure contains a compact BRD tandem that binds H3K14ac in the second BRD and the acetylated Lys25 residue of Rsc4 itself in the first BRD, suggesting an autoregulatory mechanism [35]. The arrangement of the tandem BRDs in TAF1 and yeast Rsc4 are different; therefore, whether the arrangement of tandem BRDs are protein-specific or are evolutionarily conserved is unclear. The structures of additional putative tandem BRDs (such as those from Polybromo) will be necessary to fully elucidate if BRD arrangements are evolutionarily conserved.
In human proteins, the association of BRDs with domains other than PHDs or BRDs occurs infrequently. For example, BRDs can associate with either PWWP or B-box zinc finger domains, but in each case these domains are present in only four human BRD-containing proteins. Additionally, of the non-BRD domains that associate with BRDs, none occur in as close proximity to a BRD within the protein sequence as either the PHD or BRDs that form the tandem motifs [23]. The association between PHDs and BRDs often observed in human proteins is absent in yeast, although yeast does express proteins that contain both BRDs and PHDs [23].
Functions of human bromodomain proteins
The complexity and variability of the domain composition of human BRD-containing proteins, and the influence of neighboring domains (such as the PHD) on the function of the BRD itself, make it difficult to predict the function of BRD-containing proteins based on sequence similarity alone. Many of the human BRD-containing proteins do not have well-characterized functions, although some have been implicated in disease processes. The most recent data on human BRD-containing protein function and disease involvement are reviewed in this section. However, the specific function of any of the BRDs remains to be elucidated.
BRD4 plays an important role in various biological processes by means of its two BRDs. This protein functions in the inflammatory response as a co-activator for the transcriptional activation of NFκB, via the binding of the BRDs to the acetylated-Lys310 residue on the RelA subunit of NFκB [36]. BRD4 also plays a cellular role by stimulating G1 gene transcription and promoting cell-cycle progression to the S phase [37]. Additionally, BRD4 can control the transcription of viral genes. For example, this protein regulates HIV transcription by inducing the phosphorylation of CDK9 (cyclin-dependent kinase 9) at the Thr29 residue in the HIV transcription initiation complex, thereby inhibiting CDK9 kinase activity and leading to the inhibition of HIV transcription [38]. BRD4 is also involved in the inhibition of the proteasomal degradation of the papillomavirus E2 protein [39]. Furthermore, BRD4 associates with Kaposi's sarcoma-associated herpesvirus-encoded LANA-1 (latency-associated nuclear antigen) through molecular interactions involving the C-terminal region [40] and extraterminal domain [41] of BRD4. Additionally, both BRD4 and BRD2 interact with the murine γ-herpesvirus 68 protein or f73, which is required to establish viral latency in vivo [42]. Finally, BRD4 activation may also predict the survival of patients with breast cancer [43]. Crawford et al proposed that the activation of BRD4 manipulates the response of the tumor to its microenvironment in vivo, resulting in a reduction of tumor growth and pulmonary metastasis in mice [44]. Microarray analysis of multiple human mammary tumor cell lines demonstrated that the activation of BRD4 was predictive of progression and/or survival. These results suggest that the dysregulation of BRD4-associated pathways may play an important role in breast cancer progression.
The coupling of histone acetylation to transcription in vivo by BRD2 and BRD3 was demonstrated; in human 293 cells, these proteins preferentially associated with specific H4 modifications along the entire lengths of genes, and allowed RNA polymerase II to transcribe through the nucleosomes [43]. BRD2 also exhibited histone chaperone activity [43]. In mice, BRD2 is essential for embryonic development [45], and an association between BRD2 and juvenile myoclonic epilepsy in humans has been reported [46].
In mice, the BRD and WD-repeat-containing protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition [47]. A mutation in BRWD1 leads to phenotypically normal, but infertile mice.
The BRD of transcriptional co-activator p300 was suggested to play a role in the IL-6 signaling pathway, by mediating the interaction of the STAT3 amide-terminal domain with p300, thereby stabilizing enhanceosome assembly [48].
ATAD2 is an estrogen-regulated ATPase co-activator with a BRD that functions in both estrogen receptor α and androgen receptor signaling. This protein is required for the formation of transcriptional coregulator complexes at chromatin and the modification of chromatin [49]. Chen and colleagues suggested that ATAD2 plays an important role in prostate cancer by mediating specific androgen receptor functions involved in cancer cell survival and proliferation [50].
Conclusion
The role of the BRD as the sole protein domain known to recognize acetyl-lysine residues on proteins is more complex than initially thought [9,11,51]. Studies of individual BRDs, which have focused on the structural characterization of the domains and their interactions with ligands, identified varied ligand-binding specificities that were dependent not only on the characteristics of the BRD itself, but also on the other domains (BRD and non-BRD) present in the same protein. Studies of BRD-containing proteins have highlighted the role of these domains in many important biological processes and their association with disease. The characterization of the multiplicity of molecular interactions mediated by BRDs is therefore essential for deciphering the role of individual domains and proteins. This challenging task may be facilitated by the high structural coverage of the human BRD family, which presents a unique opportunity for the rational design of selective small molecules that could serve as tools to modulate and control gene expression in the human biology.
Acknowledgements
The authors were supported by the grants GM081713 (Roberto Sanchez), MCB0517352 (Ming-Ming Zhou/R Sanchez), CA87658 and HG004508 (MM Zhou).
Abbreviations
ATAD
ATPase family AAA domain-containing protein
BPTF
BRD and PHD finger-containing transcription factor
BRD
bromodomain
CBP
CREB binding protein
H
histone
PCAF
p300/CREB-binding protein-associated factor
PHD
plant homeodomain
SUMO
small ubiquitin-like modifier
TAF1
transcription initiation factor TFIID subunit 1
TIF
transcription intermediate factor
TFIID
transcription initiation factor
WD
tryptophan-aspartate
ZA
αZ-αA
References
•• of outstanding interest
• of special interest
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Turner BM. Cellular memory and the histone code. Cell. 2002;111(3):285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 3.Nightingale KP, O'Neill LP, Turner B. Histone modifications: Signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16(2):125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51(5):786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54(1):21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 7.Neely K, Workman J. Histone acetylation and chromatin remodeling: Which comes first? Mol Genet Metab. 2002;76(1):1–5. doi: 10.1016/s1096-7192(02)00014-8. [DOI] [PubMed] [Google Scholar]
- 8.Zeng L, Zhou MM. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513(1):124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 9.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 10.Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9(3):575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson R, Ladurner AG, King DS, Tijan R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288(5470):1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 12.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the Gcn5 bromodomain. J Mol Biol. 2000;304(3):355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 13.Owen D, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19(22):6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13(2):251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, Uda-Tochio H, Hamana H, Terada T, Adachi N, Matsumoto T, et al. Crystal structure of the human BRD2 bromodomain: Insights into dimerization and recognition of acetylated histone H4. J Biol Chem. 2007;282(6):4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46(8):2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen P, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416(6876):103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295(5562):2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 20.Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22(5):151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16(4):643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ, 3rd, Zhou MM. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15(6):626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The structure of a tandem PHD finger-BRD reveals a tight structural and functional association between the two domains and a new role for a BRD. The tandem facilitates the lysine SUMOylation required for gene silencing.
- 23.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28(1):231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez R, Pieper U, Melo F, Eswar N, Martí-Renom MA, Madhusudhan MS, Mirković N, Sali A. Protein structure modeling for structural genomics. Nat Struct Biol. 2000;7(Suppl):986–990. doi: 10.1038/80776. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama S, Terwilliger TC, Kuramitsu S, Moras D, Sussman JL. RIKEN aids international structural genomics efforts. Nature. 2007;445(7123):21. doi: 10.1038/445021a. [DOI] [PubMed] [Google Scholar]
- 26.The Structural Genomics Consortium . University of Oxford; University of Toronto; Karolinska Institutet; Oxford, UK: Toronto, ON, Canada: Stockholm, Sweden: 2009. www.thesgc.org/index.php. [Google Scholar]
- 27.Huang H, Zhang J, Shen W, Wang X, Wu J, Wu J, Shi Y. Solution structure of the second bromodomain of BRD2 and its specific interaction with acetylated histone tails. BMC Struct Biol. 2007;7:57. doi: 10.1186/1472-6807-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.RIKEN Structural Genomics/Proteomics Initiative . RIKEN Genomic Sciences Center/RIKEN SPring-8 Center; Yokohama, Japan: 2009. www.rsgi.riken.go.jp/rsgi_e/index.html. [Google Scholar]
- 29.Singh M, Popowicz GM, Krajewski M, Holak TA. Structural ramification for acetyl-lysine recognition by the bromodomain of human Brg1 protein, a central ATPase of the SWI/SNF remodeling complex. Chembiochem. 2007;8(11):1308–1316. doi: 10.1002/cbic.200600562. [DOI] [PubMed] [Google Scholar]
- 30.Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, Dai H, Wu J, Shi Y. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun. 2007;358(2):435–441. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- 31.Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18(3):449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu MK, Poliakov E, Rogozin IB. Domain mobility in proteins: Functional and evolutionary implications. Brief Bioinform. 2009;10(3):205–216. doi: 10.1093/bib/bbn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, Sadofsky MJ, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28(5):823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 35.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the Rsc4 tandem bromodomain by Gcn5 acetylation. Mol Cell. 2007;27(5):817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. BRD4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• BRDs of BRD4 bind to the acetylated RelA subunit of NFκB, enhancing its transcriptional activation, and play a role in inflammatory response.
- 37.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein BRD4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem. 2008;283(14):9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, Huang K, Jung KJ, Cho WK, Klase Z, Kashanchi F, Pise-Masison CA, Brady JN. Bromodomain protein BRD4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J Virol. 2009;83(2):1036–1044. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagnon D, Joubert S, Sénéchal H, Fradet-Turcotte A, Torre S, Archambault J. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of bromodomain-containing protein 4. J Virol. 2009;83(9):4127–4139. doi: 10.1128/JVI.02468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the role ofBRD4 in increasing levels of the E2 protein from human papillomavirus.
- 40.Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol. 2006;80(21):10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Güntert P. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17(12):2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ottinger M, Pliquet D, Christalla T, Frank R, Stewart JP, Schulz TF. The interaction of the γ-herpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters. J Virol. 2009;83(9):4423–4434. doi: 10.1128/JVI.02274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins BRD2 and BRD3 couple histone acetylation to transcription. Mol Cell. 2008;30(1):51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes how BRD2 and BRD3 bind to hyperacetylated chromatin of transcribed genes and allow RNA polymerase II to transcribe through nucleosomes.
- 44.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, Lee MP, Ozato K, Hunter KW. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci USA. 2008;105(17):6380–6385. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The BRD4 activation signature, derived from microarray analyses, is a good predictor of human breast cancer progression and patient survival.
- 45.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene BRD2 is essential for embryonic development in mouse. Dev Dyn. 2009;238(4):908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pal DK, Evgrafov OV, Tabares P, Zhang F, Durner M, Greenberg DA. BRD2 (RING3) is a probable major susceptibility gene for common juvenile myoclonic epilepsy. Am J Hum Genet. 2003;73(2):261–270. doi: 10.1086/377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, Handel M, Eppig JJ, Schimenti JC. The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol. 2008;317(1):72–82. doi: 10.1016/j.ydbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou T, Ray S, Lee C, Brasier AR. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem. 2008;283(45):30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The N-terminal domain of STAT3 interacts with the p300 BRD in an acetylation-dependent manner and stabilizes enhanceosome assembly.
- 49.Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERα, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci USA. 2007;104(46):18067–18072. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The ATAD2 BRD-containing protein interacts with the androgen receptor and enhances its transcriptional activity, thus playing a role in prostate cancer. Suggests the ATPase and BRD of ATAD2 represent potential new targets for prostate cancer therapeutics.
- 50.Zou JX, Guo L, Revenko AS, Tepper CG, Gemo AT, Kung HJ, Chen HW. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69(8):3339–3346. doi: 10.1158/0008-5472.CAN-08-3440. [DOI] [PubMed] [Google Scholar]
- 51.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26(37):5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]