Interferon-Lambda: A New Addition to an Old Family (original) (raw)
Abstract
The discovery and initial description of the interferon-λ (IFN-λ) family in early 2003 opened an exciting new chapter in the field of IFN research. There are 3 IFN-λ genes that encode 3 distinct but highly related proteins denoted IFN-λ1, -λ2, and -λ3. These proteins are also known as interleukin-29 (IL-29), IL-28A, and IL-28B, respectively. Collectively, these 3 cytokines comprise the type III subset of IFNs. They are distinct from both type I and type II IFNs for a number of reasons, including the fact that they signal through a heterodimeric receptor complex that is different from the receptors used by type I or type II IFNs. Although type I IFNs (IFN-α/β) and type III IFNs (IFN-λ) signal via distinct receptor complexes, they activate the same intracellular signaling pathway and many of the same biological activities, including antiviral activity, in a wide variety of target cells. Consistent with their antiviral activity, expression of the IFN-λ genes and their corresponding proteins is inducible by infection with many types of viruses. Therefore, expression of the type III IFNs (IFN-λs) and their primary biological activity are very similar to the type I IFNs. However, unlike IFN-α receptors which are broadly expressed on most cell types, including leukocytes, IFN-λ receptors are largely restricted to cells of epithelial origin. The potential clinical importance of IFN-λ as a novel antiviral therapeutic agent is already apparent. In addition, preclinical studies by several groups indicate that IFN-λ may also be useful as a potential therapeutic agent for other clinical indications, including certain types of cancer.
Introduction
In early 2003, 2 groups independently reported the discovery of a trio of novel interferon (IFN)-like cytokines that are referred to as either IFN-λ1, -λ2, and -λ3 or interleukin-29 (IL-29), IL-28A, and IL-28B, respectively (Kotenko and others 2003; Sheppard and others 2003). Both groups also identified and characterized the novel receptor, IFN-λR1 (also known as IL-28RA), through which these cytokines mediate their biological activities. Since their original description in 2003, much has been learned about this exciting new group of cytokines and their functions. In the fall of 2009, several of the leading scientists who are conducting IFN-λ-related research presented summaries of their studies in a special focus session on these cytokines during the annual meeting of the International Society for Interferon and Cytokine Research in Lisbon, Portugal. This special issue of Journal of Interferon & Cytokine Research includes a series of review articles by all of the speakers who presented in that special session as well as several additional invited experts. This collection of articles provides an excellent summary of the current state of knowledge regarding many aspects of IFN-λ biology.
IFNs are key cytokines in the establishment of a multifaceted antiviral response. Three distinct types of IFNs are now recognized (type I, II, and III) based on their structural features, receptor usage and biological activities. Although all IFNs are important mediators of antiviral protection, their roles in antiviral defense vary. Type I IFNs (IFN-α/β/ω/ɛ/κ in humans) possess strong intrinsic antiviral activity, and are able to induce a potent antiviral state in a wide variety of cells (Levy and Garcia-Sastre 2001; Samuel 2001). The essential role of the type I IFNs in the induction of antiviral resistance has been clearly demonstrated using type I IFN receptor knockout mice because such animals are highly susceptible to many viral infections (Müller and others 1994; Hwang and others 1995; Steinhoff and others 1995). In contrast, studies with IFN-γ and IFN-γ receptor knock-out mice (Dalton and others 1993; Huang and others 1993; Lu and others 1998) as well as analysis of humans who possess inherited genetic mutations of the IFN-γ receptor (Dorman and others 2004; Novelli and Casanova 2004) revealed that antiviral activity is not the primary biological function of IFN-γ.
IFN-γ is classified as a Th1-type cytokine that stimulates cell-mediated immune responses that are critical for the development of host protection against pathogenic intracellular microorganisms such as Mycobacterium tuberculosis (Bach and others 1997; Boehm and others 1997; Pestka and others 1997). IFN-γ also plays a central role in the development of antitumor immune responses, and it can amplify the induction of antiviral activity by IFN-α or -β. Therefore, type I and type II IFNs often work together to activate a variety of innate and adaptive immune responses that result in the induction of effective antitumor immunity and the elimination of viral infections (Biron 2001; Le Bon and Tough 2002; Pestka and others 2004b).
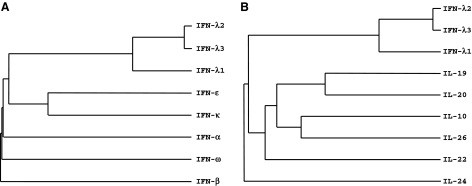
IFNs are part of the larger family of class II cytokines that also includes 6 IL-10-related cytokines: IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26 (Kotenko 2002; Renauld 2003; Pestka and others 2004a) as well as several viral IL-10-related cytokines (Kotenko and Langer 2004). IFNs and the IL-10-related cytokines can be grouped into the same family because they all signal via receptors that share common motifs in their extracellular domains. These receptors comprise the class II cytokine receptor family (CRF2). Consequently, IFNs and the IL-10-related cytokines are sometimes referred to as “CRF2 cytokines.” The type I IFN family in humans consists of 13 IFN-α species and a single species of IFN-β, IFN-κ, IFN-ω, and IFN-ɛ (LaFleur and others 2001; Hardy and others 2004; Langer and others 2004; Pestka and others 2004b). There is only one type II IFN known as IFN-γ. Although the tertiary structure of IFN-γ resembles that of IL-10, its primary structure has diverged the most from all of the CRF2 ligands. The most recent addition to the CRF2 family, the type III IFNs or IFN-λs, demonstrate structural features of the IL-10-related cytokines but also induce antiviral activity in a variety of target cells, which supports their functional classification as a new type of IFNs (Kotenko and others 2003; Sheppard and others 2003). In humans, 3 distinct but closely related IFN-λ proteins, IFN-λ1, -λ2, and -λ3 (also known as IL-29, IL-28A, and IL-28B, respectively) form the type III IFN family. Phylogenetically, the IFN-λ genes reside somewhere between the type I IFN and IL-10 gene families (Fig. 1). Amino acid sequence comparisons show that the type III IFNs exhibit about ∼5%–18% identity with either type I IFNs or the IL-10-related cytokines.
FIG. 1.
A phylogenetic alignment of the class II cytokine family genes. Alignment of the human interferon-λ (IFN-λ) genes with either (A) the human type I IFN genes or (B) the human interleukin-10 (IL-10)-related cytokines was used to generate a phylogenetic tree for the class II cytokine genes. Only one IFN-α was used in this alignment because the thirteen human IFN-α subtypes have nearly identical sequences. Because of the low sequence identity, these trees are subject to small changes, so these alignments are intended to be instructive, not definitive.
The IFN-λ proteins bind and signal through a receptor complex composed of the unique IFN-λR1 chain (also known as IL-28RA) and the shared IL-10R2 chain which is also a part of the receptor complexes for IL-10, IL-22, and IL-26 (Kotenko and others 1997, 2001; Xie and others 2000; Donnelly and others 2004; Hör and others 2004; Sheikh and others 2004). In contrast, all type I IFNs exert their biological activities through a heterodimeric receptor complex composed of the IFN-αR1 (IFNAR1) and IFN-αR2 (IFNAR2) chains, and type II IFN (IFN-γ) engages the IFN-γR1 (IFNGR1) and IFN-γR2 (IFNGR2) chains to assemble its functional receptor complex. Although the IFN-λs do not use the IFN-α receptor complex for signaling, signaling through either IFN-λ or IFN-α receptor complexes results in the activation of the same Jak-STAT signal transduction cascade.
IFN-λ binds initially to the IFN-λR1 chain, and the binary complex formed by the association of IFN-λ with the IFN-λR1 chain causes a rapid conformational change that facilitates recruitment of the second receptor chain, IL-10R2, to the complex. Once assembly of the ternary complex is complete, the receptor-associated Janus tyrosine kinases, Jak1 and Tyk2, mediate _trans_-phosphorylation of the receptor chains which results in the formation of phosphotyrosine-containing peptide motifs on the intracellular domain (ICD) of the IFN-λR1 chain that provide transient docking sites for latent preformed cytosolic STAT proteins, including STAT1 and STAT2. Signaling through type I (IFN-α/β) or type III (IFN-λ) IFN receptor complexes results in the formation of a transcription factor complex known as IFN-stimulated gene factor 3 (ISGF3). This complex consists of 3 proteins, STAT1, STAT2, and IFN regulatory factor-9 (IRF-9) (also known as ISGF3γ or p48). Once assembled, ISGF3 then translocates to the nucleus where it binds to IFN-stimulated response elements in the promoters of various ISGs. Consequently, the biological activities induced by either type I or type III IFNs are very similar, including induction of antiviral activity and up-regulation of major histocompatibility complex (MHC) class I antigen expression on many cell types.
Organization of the IFN-λ and IFN-λ Receptor Genes
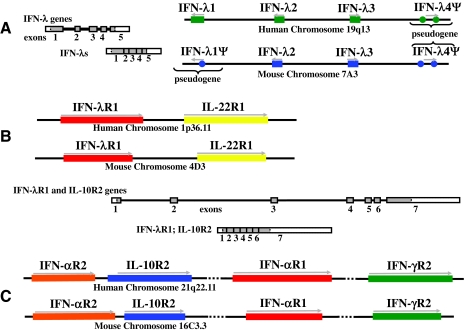
As shown in Fig. 2A, the IFN-λ genes are clustered together on human chromosome 19 (19q13.13 region) or murine chromosome 7 (7A3 region). The IFN-λ3 gene (IL28B) is transcribed in the opposite direction of the IFN-λ1 (IL29) and IFN-λ2 (IL28A) genes. The coding region for each of these genes is divided into 5 exons. The intron-exon organization of the genes encoding the IFN-λs correlates well with the common conserved architecture of the genes encoding the IL-10-related cytokines (Kotenko 2002; Kotenko and Donnelly 2006). Although the intron sizes vary significantly, the exon sizes and positions and frames of the intron/exon junctions are highly conserved within the genes for the type III IFNs and the IL-10-related cytokines. In contrast, the type I IFN genes lack introns.
FIG. 2.
Organization of the genes encoding the IFN-λs and their receptors. Schematic representations of the chromosomal regions of the human and mouse genomes that encode the IFN-λ (A) and IFN-λ receptor (B and C) genes. The genes are transcribed in the direction indicated by the arrows. Although the human and murine IFN-λ and IFN-λ receptor loci are colinear, the human genome encodes 3 functional IFN-λ genes and 1 pseudogene (denoted IFN-λ4Ψ), whereas there are only 2 functional IFN-λ coding genes in the murine genome: Il28a (IFN-λ2) and Il28b (IFN-λ3). mIFN-λ1Ψ and mIFN-λ4Ψ genes are pseudogenes. Unspliced transcripts (right panel for the IFN-λ genes, and bottom panel for the IFN-λ receptor genes) are schematically shown as strings of filled or open boxes (exons) joined by intervening lines (introns). Spliced transcripts are also shown as shaded/open boxes with vertical lines indicating the relative positions of former introns. The coding regions of exons are shaded and the segments corresponding to the 5' and 3' untranslated regions are open (not shaded).
As already mentioned, there are 3 functional IFN-λ genes in the human genome. It appears that, after the divergence of the IFN-λ1 and IFN-λ2 genes, a more recent duplication event occurred in which a fragment containing the IFN-λ1 and IFN-λ2 genes was copied and integrated back into the genome in a “head-to-head” orientation with the IFN-λ1- IFN-λ2 genomic segment. Divergence within this region created the IFN-λ3 gene, which is almost identical to the IFN-λ2 gene not only in the coding region but also in the upstream and downstream flanking sequences. However, in the duplicated fragment, the segment that contained the IFN-λ1 gene was extensively mutated so that only separate pieces that do not encode a functional gene (denoted IFN-λ4Ψ in Fig. 2A) can be found in this region.
Analysis of the murine genome showed that the region that is colinear with the human IFN-λ (hIFN-λ) gene cluster is located on chromosome 7A3, and it has a similar organization to the hIFN-λ locus (Fig. 2A). Two full-length genes colinear with the hIFN-λ2 and hIFN-λ3 genes were predicted to encode functional proteins, and were designated murine IFN-λ2 (mIFN-λ2) and mIFN-λ3 in accordance with the corresponding human genes (Lasfar and others 2006). The corresponding mIFN-λ2 and IFN-λ3 proteins demonstrate higher amino acid sequence homology to hIFN-λ2 and IFN-λ3 than to hIFN-λ1. In contrast to the IFN-λ2 and IFN-λ3 genes that are conserved in both the murine and human genomes, the IFN-λ1 gene equivalent in mice lacks the entire exon 2 and contains a stop codon within exon 1. Despite these aberrations, exons 3, 4, and 5 are intact. Sequence analysis of the IFN-λ1 genomic fragment from several mouse strains (CD1, FVB, C57BL/6, 129/Sv and wild-type feral mice) revealed significant sequence variations in the region between exons 1 and 3 (Lasfar and others 2006). Nevertheless, the stop codon within exon 1 was present in all of the strains, and exon 2 could not be predicted in any of the strains. Studies conducted in several labs have shown that this mutated mIFN-λ1 gene does not encode a functional IFN-λ1 protein. Therefore, the mouse IFN-λ1 gene is a pseudogene. It is noteworthy that, in the case of the hIFN-λ genes, IFN-λ1 is less homologous to IFN-λ2 or IFN-λ3 than IFN-λ2 and IFN-λ3 are to one another. It is also interesting that among the 3 hIFN-λ proteins, only IFN-λ1 is N-linked glycosylated, whereas both of the mIFN-λ proteins (IFN-λ2 and -λ3) are glycosylated (Kotenko and others 2003; Bartlett and others 2005; Lasfar and others 2006).
The high degree of homology between the IFN-λ genes suggests that these genes evolved from a common predecessor relatively recently. Particularly, the IFN-λ3 gene (IL28B) is almost identical to the IFN-λ2 (IL28A) gene not only in the coding region but also in the upstream and downstream flanking sequences. Thus, the promoters of the IFN-λ2 and IFN-λ3 genes are very similar and share several common elements with the IFN-λ1 promoter, suggesting that all 3 genes are likely to be regulated in a similar manner (Onoguchi and others 2007; Osterlund and others 2007; Thomson and others 2009). Computer analysis predicts the existence of potential binding sites for a variety of transcription factors, including AP1 (dimeric transcription factor containing members of the Jun, Fos, ATF, and Maf protein families [reviewed in Eferl and Wagner (2003)] and nuclear factor-κB (Silverman and Maniatis 2001; Li and Verma 2002), as well as multiple virus response elements that provide the binding sites for various IRF proteins. These same transcription factors have been shown to be involved in the regulation of transcription of the type I IFN genes (Wathelet and others 1998). Indeed, studies have shown that the type III IFN genes are expressed in response to many of the same stimuli that activate expression of the type I IFN genes (Coccia and others 2004; Ank and others 2006). These stimuli include many types of viruses and a variety of _Toll_-like receptor (TLR) agonists.
As shown in Fig. 2B, the genes encoding the IFN-λ receptor subunits, IFN-λR1 (also known as IL-28RA or CRF2-12) and IL-10R2 are located on human chromosome 1 (1p36.11 region) and chromosome 21 (21q22.11 region), or murine chromosome 4 (4D3 region) and chromosome 16 (16C3.3 region), respectively (Kotenko and others 2003; Sheppard and others 2003; Lasfar and others 2006). It is noteworthy that the IFN-λR1 gene is located in very close proximity to the gene for another important class II cytokine receptor, IL-22R1 (IL22RA1). Both the IFN-λR1 gene and the adjacent IL-22R1 gene are transcribed in the same direction. Interestingly, the IL-10R2 gene, IL10RB, is clustered together with 3 other genes that encode members of the CRF2 family: IFN-αR1 (IFNAR1), IFN-αR2 (IFNAR2), and IFN-γR2 (IFNGR2). All 4 of these genes are transcribed in the same direction. The IFN-λR1 and IL-10R2 genes have a similar intron/exon structure that is also shared by other genes encoding class II cytokine receptor proteins (Kotenko 2002). The coding regions of the receptor genes are composed of 7 exons (Fig. 2B). Exon 1 encodes the 5'-UTR and the signal peptide, the extracellular domain is encoded by exons 2, 3, 4, 5, and part of exon 6. Exon 6 also encodes the transmembrane domain and the beginning of the ICD. Exon 7 encodes the rest of the ICD and the 3'-UTR.
The primary IFN-λR1 mRNA transcript in human cells is ∼5 kb long. However, several variant IFN-λR1 transcripts of different sizes have been detected. Differential mRNA splicing of the hIFN-λR1 gene generates at least 3 splice variants. When all 7 exons of the IFN-λR1 gene are present in the transcript, a full-length, membrane-associated, signaling-competent IFN-λR1 protein is generated. When exon 6 is spliced out of the transcript, a secreted soluble IFN-λR1 protein is generated. Another splice variant, generated by a distinct splicing event, encodes a membrane-bound receptor with a shorter intracytoplasmic domain than that found in the full-length IFN-λR1 protein. This particular IFN-λR1 splice variant is likely to be signaling-incompetent. All 3 of these variant forms of IFN-λR1 share the same extracellular domain, and therefore have similar affinity for binding of IFN-λs. It remains to be seen whether alternative splicing of the mouse IFN-λR1 gene also occurs. It is possible that these IFN-λ receptor variants evolved to mediate unique biological functions. For example, the soluble IFN-λR1 protein may function as a naturally occurring receptor antagonist to limit signaling through the membrane-bound form of this receptor (Witte and others 2009).
Although IFN-λR1 is constitutively expressed by a broad range of cell lines and tissues, there are many cell types that do not express IFN-λ receptors, and, as a consequence, cannot respond to treatment with this cytokine. For example, only type I IFNs but not type III IFNs can induce STAT activation in primary fibroblasts, human umbilical vein endothelial cells, and murine splenocytes (Lasfar and others 2006; Sommereyns and others 2008). Peripheral blood mononuclear cells and bone marrow cells demonstrate only very weak activation in response to IFN-λ treatment (Lasfar and others 2006), and it appears that most leukocytes do not respond to IFN-λ even at high concentrations. Cells of epithelial origin appear to be the primary targets for IFN-λ because, unlike leukocytes, they express significant levels of IFN-λR1 (Lasfar and others 2006; Sommereyns and others 2008; Witte and others 2009). The membrane expression pattern of IFN-λR1 appears to be very similar to IL-22R1, and like the IL-22R1 chain, IFN-λR1 is not expressed on leukocytes (Witte and others 2009). Because the IFN-λR1 and IL-22R1 genes are clustered together on chromosome 1 (Fig. 2B), it is tempting to speculate that expression of these genes is coregulated under certain conditions.
The IFN-λ Receptor Complex and Signaling Pathway
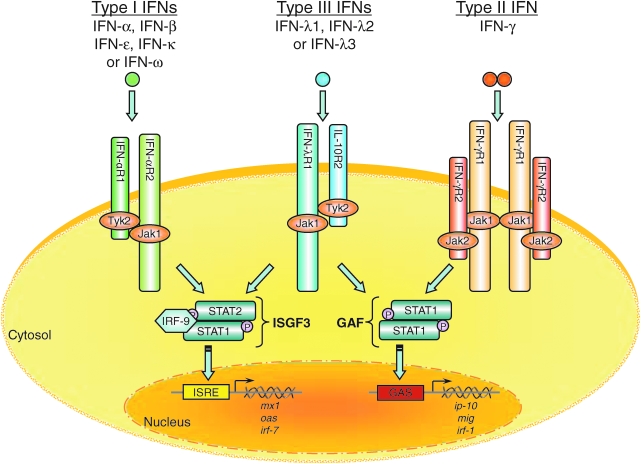
IFN-λs exert their biological activities by signaling through a heterodimeric receptor complex composed of IFN-λR1 and IL-10R2 (Kotenko and others 2003; Sheppard and others 2003). The IL-10R2 chain is also an essential component of the receptor complexes for IL-10, IL-22, and IL-26 (Donnelly and others 2004). In contrast, the IFN-λR1 chain is a private receptor that is used for signaling only by the IFN-λ proteins. IFN-λs appear to be monomers as demonstrated by cross-linking experiments with radioactively labeled IFN-λ1 and by gel filtration chromatography. As depicted in Fig. 3, the IFN-λ proteins require both receptor chains (IFN-λR1 and IL-10R2) to achieve high affinity binding, and one molecule of IFN-λ is likely to physically interact with a single molecule each of the IFN-λR1 and IL-10R2 subunits.
FIG. 3.
A model of the IFN-λ receptor signaling pathway. The type I, type II, and type III IFNs bind to distinct receptor complexes on the cell membrane. Signal transduction activated by the binding of IFNs to their cognate receptors induces expression of many IFN-stimulated genes (ISGs). The proteins encoded by these genes in turn mediate the antiviral activity of the IFNs, particularly the type I and III IFNs. The functional IFN-λ receptor complex consists of 2 distinct receptor chains: the ligand-specific IFN-λR1 chain (also known as IL-28RA) and the IL-10R2 chain. The binding of IFN-λ to its receptor induces a signaling cascade that results in the activation of STAT1 and STAT2 which together with IRF-9 (p48) form ISGF3 transcription factor complexes. The newly formed ISGF3 complexes then translocate from the cytosol to the nucleus where they bind to IFN-stimulated response elements (ISRE) in the promoters of ISGs such as IRF7, MX1, and OAS1.
Binding of IFN-λ1, -λ2, or -λ3 to the membrane-associated IFN-λ receptor complex leads to the activation of the Janus kinases, Jak1 and Tyk2, subsequent tyrosine phosphorylation of the IFN-λR1 ICD, and activation of the latent transcription factors, STAT1 and STAT2 (Kotenko and others 2003; Dumoutier and others 2004). It can also induce activation of STAT3, STAT4, and STAT5 in some cell types (Dumoutier and others 2003). The activated STATs are tyrosine-phosphorylated and form homo- and heterodimers via reciprocal interaction of their SH2 domains with phosphorylated tyrosine residues on other STAT molecules. The STAT homo- and heterodimers translocate to the nucleus where they bind to specific DNA elements such as IFN-γ activated sequence (GAS) elements in the promoters of IFN-responsive genes and modulate their transcription. Activated STAT1 and STAT2 molecules form heterodimers and recruit IRF-9 to form a trimeric transcription factor complex known as ISGF3. ISGF3 regulates gene transcription by binding to IFN-stimulated response elements (ISRE) in the promoters of other ISGs. These include a number of genes that are classically associated with the antiviral phenotype, including OAS1, MX1, EIF2AK2 (double-stranded RNA-activated protein kinase), and IRF7. Recent comparative cDNA microarray analyses by several groups have shown that the repertoire of genes that are induced by type III IFNs (IFN-λ) is essentially the same as that which are induced by type I IFNs (IFN-α/β) (Doyle and others 2006; Marcello and others 2006).
STAT recruitment occurs mainly through the phosphotyrosine-containing peptide motifs on the IFN-λR1 ICD. The murine and hIFN-λR1 polypeptide chains share ∼67% overall homology. There are 2 conserved tyrosine residues on the ICD of mouse and hIFN-λR1. In the case of the hIFN-λR1 chain, these 2 tyrosine residues, Tyr343 and Tyr517, can independently mediate STAT2 activation by IFN-λs (Dumoutier and others 2004). Interestingly, the Tyr341-based motif of mIFN-λR1 (YLERP) shows similarities with that surrounding Tyr343 of hIFN-λR1 (YIEPP). In addition, the C-terminal amino acid sequence of mIFN-λR1 containing Tyr533 (YLVRstop) is very similar to the C-terminal amino acid sequence of hIFN-λR1 containing Tyr517 (YMARstop). Therefore, both the mouse and hIFN-λR1 chains contain similar docking sites for STAT2 recruitment and activation, YΦEXP and YΦXRstop (where Φ is hydrophobic). Thus, the Tyr341- and Tyr533-based motifs on mIFN-λR1 are also likely to mediate STAT2 recruitment and activation of ISGF3 that are responsible for most of the IFN-λ-induced biological activities. Although activation of STAT2 requires the presence of either Tyr343 or Tyr517 on the hIFN-λR1 chain, activation (phosphorylation) of STAT4, and to a lesser degree, STAT1 and STAT3, occurs independently of the tyrosine residues on the ICD of IFN-λR1 (Dumoutier and others 2004).
Biological Activities
As discussed above, the IFN-λ signal transduction cascade is very similar to that induced by type I IFNs (IFN-α or -β). Therefore, it is not surprising that type I and type III IFNs induce similar biological activities. Both types of IFN possess the intrinsic ability to induce antiviral activity in cells. IFN-λ-induced antiviral activity has been demonstrated against many different viruses, including encephalomyocarditis virus and vesicular stomatitis virus in several different cell types (Kotenko and others 2003; Sheppard and others 2003; Ank and others 2006). IFN-λ has been shown to inhibit hepatitis B virus (HBV) replication in a differentiated murine hepatocyte cell line (Robek and others 2005). IFN-λ also inhibits replication of subgenomic and full-length HCV replicons in the human hepatoma cell line, Huh7 (Robek and others 2005; Doyle and others 2006; Marcello and others 2006). Consistent with its antiviral activity, IFN-λ induces expression of several classical biomarkers of the antiviral response, including double-stranded RNA-activated protein kinase, 2',5'-oligoadenylate synthetase, and the Mx proteins (Kotenko and others 2003; Brand and others 2005a, 2005b). Interestingly, one group showed that suppressor of cytokine signaling (SOCS)-1 inhibited induction of the OAS1 and MX1 genes (Brand and other 2005b). IFN-λ can also significantly up-regulate expression of the inhibitory gene, SOCS3, suggesting a possible mechanism for feedback inhibition of IFN-λ signaling (Brand and others 2005a). MHC class I antigen expression is also up-regulated on the surface of cells following exposure to either type I or type III IFNs (Kotenko and others 2003). Up-regulation of MHC class I antigen expression may enhance the ability of the immune system to recognize and destroy virus-infected cells.
The importance of type III IFNs in antiviral defense was highlighted by the recent discovery of a virus defense mechanism that directly targets type III IFNs (Huang and others 2007). A secreted glycoprotein, known as Y136, produced by the Yaba-like disease virus binds and inhibits signaling by both type I and type III IFNs. Y136 can also block IFN-mediated biological activities, including up-regulation of MHC class I antigen expression and induction of antiviral activity. The ability of Y136 to bind both type I and type III IFNs distinguishes this viral protein from the B18R protein which only blocks binding of type I IFNs.
Overall, the pattern of gene expression induced by either type I IFN (IFN-α) or type III IFN (IFN-λ) is very similar. However, the relative magnitude of gene expression induced by IFN-α is often greater than that induced by IFN-λ in many cell types. This may reflect a difference in the relative strength of signaling through type I IFN receptors versus type III IFN receptors. Alternatively, this difference may simply reflect a significant difference in the relative levels of expression of these receptors on the cell membrane.
The IFN-λ antiviral system is highly conserved throughout evolution, and can be traced back at least to birds (class: Aves). The chicken genome encodes at least one IFN-λ gene (Karpala and others 2008). Although there are only 2 functional IFN-λ genes encoded in the mouse genome, the mIFN-λ system appears to be fully capable of mediating effective antiviral responses both in vitro and in vivo (Bartlett and others 2005; Lasfar and others 2006). Forced expression of mIFN-λ in a vaccinia virus construct strongly attenuated replication of this virus in several infection models in mice (Bartlett and others 2005). Mice infected intranasally with recombinant vaccinia virus engineered to produce IFN-λ (vIFN-λ) did not show any signs of illness or weight loss. In addition, vIFN-λ-expressing vaccinia virus was cleared more rapidly from infected lungs, and, in contrast to the control virus, did not disseminate to the brain. Attenuation of vIFN-λ2 was associated with increases in both lymphocytes in bronchial alveolar lavages and CD4+ T cells in total lung lymphocyte isolates.
Consistent with their antiviral activity, the IFN-λs are usually coexpressed together with type I IFNs by virus-infected cells (Kotenko and others 2003; Sheppard and others 2003). Virtually any cell type can express IFN-λ following viral infection, and presumably infection by most viruses induces IFN-λ expression. For example, we showed originally that 4 distinct viruses (Sindbis virus, Dengue virus, vesicular stomatitis virus, and encephalomyocarditis virus) induce coexpression of IFN-λ1, -λ2, and -λ3 together with IFN-α and -β in several different human cell lines, including HeLa (cervical epithelial carcinoma), HT-29 (colorectal carcinoma), and Huh7 (hepatoma) (Kotenko and others 2003). It has also been shown that infection of human epithelial cells by respiratory syncytial virus induces coexpression of type I and type III IFNs (Spann and others 2004). In mice, infection with murine cytomegalovirus up-regulates IFN-λ mRNA expression in vivo (Brand and others 2005a). Other viruses, such as influenza virus and Sendai virus have also been shown to induce expression of IFN-λ in human monocyte-derived dendritic cells (mDC) (Coccia and others 2004; Osterlund and others 2005; Sirén and others 2005). Furthermore, it was shown that IFN-α amplifies induction of IFN-λ by influenza or Sendai virus (Osterlund and others 2005; Sirén and others 2005).
The ability of IFN-α to up-regulate induction of IFN-λ may be due to the ability IFN-α to up-regulate expression of the TLR and IRF7 genes (Osterlund and others 2005; Sirén and others 2005; Tissari and others 2005). Viral infection or treatment with diverse TLR agonists induces differential expression of the IFN-α, -β, and -λ genes in plasmacytoid dendritic cell (pDC) and mDC (Coccia and others 2004; Gautier and others 2005). pDCs are a highly specialized and relatively rare population of cells that can produce large amounts of IFN-α and -λ in response to viral infection. Influenza virus infection of pDCs or mDCs induces coexpression of all of the IFN-α subtypes, IFN-β and IFN-λ. Certain TLR agonists such as CpG DNA which signals via TLR9 in pDCs also induces coexpression of IFN-α, -β, and -λ (Coccia and others 2004). In contrast, other TLR agonists such as lipopolysaccharide and poly I:C which signal via TLR4 and TLR3, respectively, induce expression of IFN-β and IFN-λ, but do not induce expression of IFN-α in monocyte-derived DCs (Coccia and others 2004). Therefore, infection by live viruses induces coexpression of IFN-α, -β, and -λ, whereas other microbial agents or their structural components such as bacterial DNA, endotoxin and double-stranded RNA induce a more selective expression of the IFN subtypes. The coexpression of type I and type III IFNs following viral infection is consistent with the existence of common upstream regulatory elements in the IFN-α and IFN-λ genes (Onoguchi and others 2007; Osterlund and others 2007; Thomson and others 2009).
Based on our current knowledge, it appears that both the IFN-α/β and IFN-λ ligand-receptor systems are activated independently in response to viral infections. Inhibition of signaling through type I IFN receptors with neutralizing anti-IFNAR antibodies does not block signaling through type III IFN receptors. Similarly, inhibition of signaling through type III IFN receptors using neutralizing anti-IL-10R2 antibodies does not block signaling through type I IFN receptors. Signaling through IFN-α/β receptors or IFN-λ receptors leads to the establishment of an antiviral state through the shared use of a common downstream signaling pathway and a common set of ISGs. The evolution and conservation of a second antiviral ligand-receptor system (i.e., the IFN-λ receptor pathway) that promotes induction of antiviral activity underscores the critical importance of IFNs in host defense against pathogenic viruses.
Although type I IFNs are able to induce antiproliferative responses in many cell types, the antiproliferative activity of IFN-λ appears to be more limited (Maher and others 2008). The antiproliferative activity of the IFN-λs has been demonstrated using several target cell types, including intestinal epithelial cells (Brand and others 2005a) and the human glioblastoma cell line, LN319 (Meager and others 2005). The ability of IFN-λs to induce antiproliferative activity in target cells may depend on the relative levels of IFN-λR1 expression because IFN-λs can effectively inhibit proliferation of cells engineered to express high levels of IFN-λR1 by forced expression of an IFN-λR1 expression plasmid (Dumoutier and others 2004) or a chimeric receptor that recapitulates IFN-λ signaling in cells (Li and others 2008).
Type I IFNs are recognized not only for their antiviral activities, but also for their antitumor activity. IFN-α is used clinically as a treatment for various cancers, including Hairy cell and myelogenous leukemias, multiple myeloma, non-Hodgkins lymphoma, renal cell carcinoma, Kaposi' sarcoma and metastatic melanoma (De Maeyer and De Maeyer-Guignard 1998; Belardelli and others 2002; Pestka and others 2004b). Because the signaling pathway and biological activities of the IFN-λs are essentially the same as those of the type I IFNs, the potential antitumor activity of IFN-λ was examined by several groups (Lasfar and others 2006; Sato and others 2006; Numasaki and others 2007; Abushahba and others 2010). For example, Lasfar and others (2006) used a gene therapy approach to examine the possible antitumor activity of IFN-λ in the murine B16 melanoma model. B16 cells constitutively expressing mIFN-λ2 (B16.IFN-λ2 cells) were generated and evaluated for their tumorigenicity in syngeneic C57BL/6 mice. Although constitutive expression of mIFN-λ2 in melanoma cells did not affect their proliferation in vitro, the growth of B16.IFN-λ2 cells, when injected subcutaneously into mice, was either retarded or completely prevented. Rejection of the modified tumor cells correlated with their level of IFN-λ2 expression. We then developed IFN-λ-resistant B16.IFN-λ2 cells (B16.IFN-λ2Res cells), and demonstrated that their tumorigenicity was also highly impaired or completely abolished similar to the B16.IFN-λ2 cells, indicating that IFN-λ can activate host antitumor mechanisms that inhibit the growth of certain tumors. These in vivo experiments demonstrate the inherent antitumor activity of IFN-λ and suggest an important additional clinical indication for type III IFNs.
Future Directions
The collection of review articles in this special issue of Journal of Interferon & Cytokine Research provides a very informative overview of our current understanding of IFN-λ structure and function. The article by Gad and others (2010) reviews what is known so far regarding the physical structure of the IFN-λ proteins. Based on structural modeling conducted by Rune Hartmann's group in Denmark, it is now known that the structure of IFN-λ is more closely related to members of the IL-10 family such as IL-22 than to type I IFNs such as IFN-α or -β. This is perhaps not entirely surprising because the IFN-λ receptor, IFN-λR1 (or IL-28RA), heterodimerizes with the second chain of the IL-10 receptor complex, IL-10R2, to generate functional receptor complexes for IFN-λ. IL-22 signals through receptor complexes composed of the ligand-specific IL-22R1 chain and the shared IL-10R2 chain (Kotenko and others 2001; Donnelly and others 2004).
The article by Iversen and Paludan (2010) in this issue reviews the molecular basis for induction of IFN-λ gene expression by viral infection. They discuss the mechanisms by which viruses induce coexpression of type I and type III IFNs. They also summarize what is known currently regarding the transcription factors that orchestrate the induction of IFN-λ gene expression. Particular attention is given to the roles of nuclear factor-κB, IRF-3 and IRF-7 in the induction of IFN-λ gene expression. The article by Mordstein and others (2010) reviews what is known regarding the role of the IFN-λ receptor in innate immune responses to different types of viral infections in mice. By comparing the responses to several different types of viruses in mice with genetic deletions of the IFN-α receptor, IFN-λR1 (Il28ra) or both, these authors have shown that deletion of both type I and type III receptor genes is necessary to markedly inhibit antiviral responses to many viruses. These authors have found that an intact IFN-λ receptor signaling system is essential for effective immune responsiveness to viral infections that preferentially infect epithelial cells in the lung and/or gastrointestinal tract.
The review by Pagliaccetti and Robek (2010) discusses the important emerging role for IFN-λ as a potential new therapeutic agent for the treatment of HBV and HCV infections. The authors summarize several key preclinical studies that demonstrated that recombinant hIFN-λ can inhibit replication of HCV replicons in the Huh-7 human hepatoma cell line model (Robek and others 2005; Doyle and others 2006; Marcello and other 2006). They also discuss the differential expression of IFN-λR1 and IL-10R2 by various cell types, and provide data that shows that primary human hepatocytes express significant levels of the IFN-λR1 and IL-10R2 genes. These findings compliment the review by Eleanor Ramos (2010) in which she discusses the preclinical rationale for the use of pegylated-recombinant hIFN-λ1 as a novel therapeutic agent for the treatment of chronic HCV infection. She also discusses the results of the initial Phase-1 clinical studies of IFN-λ1 in patients with chronic HCV infection.
Type I IFNs are used clinically for the treatment of viral infections, various cancers, and multiple sclerosis (IFN-β). However, severe adverse events associated with type I IFN therapy, including inhibition of hematopoiesis, neuropsychiatric effects and influenza-like symptoms, underscore the need for more effective and less toxic therapeutic agents. Although IFN-λs signal via a unique receptor complex, they resemble type I IFNs in terms of their pattern of expression, primary signaling pathway and biological activities. However, the more limited tissue expression of IFN-λ receptors suggests that type III IFNs do not simply recapitulate the type I IFN antiviral system. The fact that most if not all hematopoietic cell types do not express IFN-λ receptors predicts that, unlike IFN-α, IFN-λ may exhibit less hematopoietic toxicity than IFN-α when administered clinically as a therapeutic antiviral agent. Indeed, the results from early clinical trials of recombinant pegylated-IFN-λ1 in patients with chronic HCV infection support the prediction that IFN-λ will be less toxic than IFN-α as a therapeutic agent (Miller and others 2009; Muir and others 2010).
Although recombinant IFN-λ is being evaluated initially as a potential therapeutic alternative to IFN-α for the treatment of HCV infection, it is possible that this cytokine may also be useful as a therapeutic agent for the treatment of other types of viral infections, including viral infections of the upper respiratory tract. IFN-λ might also be useful for treating certain types of cancer. The article by Steen and Gamero (2010) in this special issue summarizes the information now available regarding the antiproliferative and antitumor activities of IFN-λ. The results from a number of published studies using murine tumor models support the hypothesis that IFN-λ may have potential as a novel antitumor agent for treatment of at least some types of cancer. Finally, the article by Wolk and others (2010) summarizes studies to date that have explored the role of IFN-λ in regulating the functions of skin epithelial cells. As mentioned several times throughout this overview article, epithelial cells such as keratinocytes express IFN-λ receptors, and treatment with IFN-λ induces activation of ISGF3 and subsequent expression of many ISGs in keratinocytes. The ability of type I and type III IFNs to rapidly activate the antiviral state in skin epithelial cells may provide critical first-line defense against environmental challenge by many types of pathogenic viruses.
Although a complete understanding of the role of the IFN-λs in antiviral and antitumor defense has not yet been reached, the discovery and characterization of this novel group of cytokines has greatly advanced our understanding of the innate response to viral infections. It has also paved the way to the development and clinical application of an exciting new group of therapeutic proteins.
Acknowledgments
We wish to thank all of the authors who contributed to this special issue of JICR.
This work was supported in part by U.S. Public Health Service Grants AI057468 and AI076937 from the National Institutes of Health (NIH) and a TIL Grant from the Alliance for Lupus Research to S.V.K.
Author Disclosure Statement
No competing financial interests exist.
References
- Abushahba W. Balan M. Castaneda I. Yuan Y. Reuhl K. Raveche E. de la A. Lasfar A. Kotenko SV. Antitumor activity of type I and type III interferons in BNL hepatoma model. Cancer Immunol Immunother. 2010;59(7):1059–1071. doi: 10.1007/s00262-010-0831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N. West H. Bartholdy C. Eriksson K. Thomsen AR. Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80(9):4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach EA. Aguet M. Schreiber RD. The IFN-gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- Bartlett NW. Buttigieg K. Kotenko SV. Smith GL. Murine interferon lambdas (type-III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J Gen Virol. 2005;86(Pt 6):1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- Belardelli F. Ferrantini M. Proietti E. Kirkwood JM. Interferon-alpha in tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002;13(2):119–134. doi: 10.1016/s1359-6101(01)00022-3. [DOI] [PubMed] [Google Scholar]
- Biron CA. Interferons alpha and beta as immune regulators—a new look. Immunity. 2001;14(6):661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- Boehm U. Klamp T. Groot M. Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Brand S. Beigel F. Olszak T. Zitzmann K. Eichhorst ST. Otte JM. Diebold J. Diepolder H. Adler B. Auernhammer CJ. Göke B. Dambacher J. IL-28A and IL-29 mediate anti-proliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol. 2005a;289(5):G960–G968. doi: 10.1152/ajpgi.00126.2005. [DOI] [PubMed] [Google Scholar]
- Brand S. Zitzmann K. Dambacher J. Beigel F. Olszak T. Vlotides G. Eichhorst ST. Göke B. Diepolder H. Auernhammer CJ. SOCS-1 inhibits expression of the antiviral proteins, 2′,5′-OAS and MxA, induced by the novel interferon-lambdas, IL-28A and IL-29. Biochem Biophys Res Commun. 2005b;331(2):543–548. doi: 10.1016/j.bbrc.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Coccia EM. Severa M. Giacomini E. Monneron D. Remoli ME. Julkunen I. Cella M. Lande R. Uzé G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34(3):796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- Dalton DK. Pitts-Meek S. Keshav S. Figari IS. Bradley A. Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259(5102):1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- De Maeyer E. De Maeyer-Guignard J. Type I interferons. Int Rev Immunol. 1998;17(1–4):53–73. doi: 10.3109/08830189809084487. [DOI] [PubMed] [Google Scholar]
- Donnelly RP. Sheikh F. Kotenko SV. Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004;76(2):314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- Dorman SE. Picard C. Lammas D. Heyne K. van Dissel JT. Baretto R. Rosenzweig SD. Newport M. Levin M. Roesler J. Kumararatne D. Casanova JL. Holland SM. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364(9451):2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- Doyle SE. Schreckhise H. Khuu-Duong K. Henderson K. Rosler R. Storey H. Yao L. Liu H. Barahmand-Pour F. Sivakumar P. Chan C. Birks C. Foster D. Clegg CH. Wietzke-Braun P. Mihm S. Klucher KM. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44(4):896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- Dumoutier L. Lejeune D. Hor S. Fickenscher H. Renauld JC. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem J. 2003;370(Pt 2):391–396. doi: 10.1042/BJ20021935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoutier L. Tounsi A. Michiels T. Sommereyns C. Kotenko SV. Renauld JC. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and anti-proliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem. 2004;279(31):32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- Eferl R. Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Gad HH. Hamming OJ. Hartmann R. The structure of human interferon-lambda and what it has taught us. J Interferon Cytokine Res. 2010;30(8):565–571. doi: 10.1089/jir.2010.0062. [DOI] [PubMed] [Google Scholar]
- Gautier G. Humbert M. Deauvieau F. Scuiller M. Hiscott J. Bates EE. Trinchieri G. Caux C. Garrone P. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12 p70 secretion by dendritic cells. J Exp Med. 2005;201(9):1435–1446. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy MP. Owczarek CM. Jermiin LS. Ejdebäck M. Hertzog PJ. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84(2):331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Hör S. Pirzer H. Dumoutier L. Bauer F. Wittmann S. Sticht H. Renauld JC. de Waal Malefyt R. Fickenscher H. The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004;279(32):33343–33351. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- Huang J. Smirnov SV. Lewis-Antes A. Balan M. Li W. Tang S. Silke GV. Pütz MM. Smith GL. Kotenko SV. Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc Natl Acad Sci U S A. 2007;104(23):9822–9827. doi: 10.1073/pnas.0610352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Hendriks W. Althage A. Hemmi S. Bluethmann H. Kamijo R. Vilcek J. Zinkernagel RM. Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259(5102):1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- Hwang SY. Hertzog PJ. Holland KA. Sumarsono SH. Tymms MJ. Hamilton JA. Whitty G. Bertoncello I. Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci U S A. 1995;92(24):11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen MB. Paludan SR. Mechanisms of type III interferon expression. J Interferon Cytokine Res. 2010;30(8):573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- Karpala AJ. Morris KR. Broadway MM. McWaters PG. O'Neil TE. Goossens KE. Lowenthal JW. Bean AG. Molecular cloning, expression, and characterization of chicken IFN-lambda. J Interferon Cytokine Res. 2008;28(6):341–350. doi: 10.1089/jir.2007.0117. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13(3):223–240. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. Donnelly RP. Type-III interferons: the interferon-λ family. In: Meager A, editor. The interferons: Characterization and application. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co; 2006. pp. 141–163. [Google Scholar]
- Kotenko SV. Gallagher G. Baurin VV. Lewis-Antes A. Shen M. Shah NK. Langer JA. Sheikh F. Dickensheets H. Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. Izotova LS. Mirochnitchenko OV. Esterova E. Dickensheets H. Donnelly RP. Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10R-beta) is a common chain of both the IL-10 and IL-22 receptor complexes. J Biol Chem. 2001;276(4):2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. Krause CD. Izotova LS. Pollack BP. Wu W. Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16(19):5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV. Langer JA. Full house: 12 receptors for 27 cytokines. Int Immunopharmacol. 2004;4(5):593–608. doi: 10.1016/j.intimp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- LaFleur DW. Nardelli B. Tsareva T. Mather D. Feng P. Semenuk M. Taylor K. Buergin M. Chinchilla D. Roshke V. Chen G. Ruben SM. Pitha PM. Coleman TA. Moore PA. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J Biol Chem. 2001;276(43):39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- Langer JA. Cutrone EC. Kotenko S. The class II cytokine receptor (CRF2) family: overview and patterns of receptor-ligand interactions. Cytokine Growth Factor Rev. 2004;15(1):33–48. doi: 10.1016/j.cytogfr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Lasfar A. Lewis-Antes A. Smirnov SV. Anantha S. Abushahba W. Tian B. Reuhl K. Dickensheets H. Sheikh F. Donnelly RP. Raveche E. Kotenko SV. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66(8):4468–4677. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- Le Bon A. Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14(4):432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- Levy DE. Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12(2–3):143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Li Q. Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Li W. Lewis-Antes A. Huang J. Balan M. Kotenko SV. Regulation of apoptosis by type III interferons. Cell Prolif. 2008;41(6):960–979. doi: 10.1111/j.1365-2184.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. Ebensperger C. Dembic Z. Wang Y. Kvatyuk M. Lu T. Coffman RL. Pestka S. Rothman PB. Targeted disruption of the interferon-gamma receptor 2 gene results in severe immune defects in mice. Proc Natl Acad Sci U S A. 1998;95(14):8233–8238. doi: 10.1073/pnas.95.14.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher SG. Sheikh F. Scarzello AJ. Romero-Weaver AL. Baker DP. Donnelly RP. Gamero AM. IFN-alpha and IFN-lambda differ in their anti-proliferative effects and duration of JAK/STAT signaling. Cancer Biol Ther. 2008;7(7):1109–1115. doi: 10.4161/cbt.7.7.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T. Grakoui A. Barba-Spaeth G. Machlin ES. Kotenko SV. MacDonald MR. Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- Meager A. Visvalingam K. Dilger P. Bryan D. Wadhwa M. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine. 2005;31(2):109–118. doi: 10.1016/j.cyto.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Miller DM. Klucher KM. Freeman JA. Hausman DF. Fontana D. Williams DE. Interferon-lambda as a potential new therapeutic for hepatitis C. Ann N Y Acad Sci. 2009;1182:80–87. doi: 10.1111/j.1749-6632.2009.05241.x. [DOI] [PubMed] [Google Scholar]
- Mordstein M. Michiels T. Staeheli P. What have we learned from the IL-28 receptor knockout mouse? J Interferon Cytokine Res. 2010;30(8):579–584. doi: 10.1089/jir.2010.0061. [DOI] [PubMed] [Google Scholar]
- Muir AJ. Shiffman ML. Zaman A. Yoffe B. de la Torre A. Flamm S. Gordon SC. Marotta P. Vierling JM. Carlos Lopez-Talavera J. Byrnes-Blake K. Fontana D. Freeman J. Gray T. Hausman D. Hunder NN. Lawitz E. Phase 1b study of pegylated interferon-lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010 May 14; doi: 10.1002/hep.23743. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Müller U. Steinhoff U. Reis LF. Hemmi S. Pavlovic J. Zinkernagel RM. Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Novelli F. Casanova JL. The role of IL-12, IL-23 and IFN-gamma in immunity to viruses. Cytokine Growth Factor Rev. 2004;15(5):367–377. doi: 10.1016/j.cytogfr.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M. Tagawa M. Iwata F. Suzuki T. Nakamura A. Okada M. Iwakura Y. Aiba S. Yamaya M. IL-28 elicits antitumor responses against murine fibrosarcoma. J Immunol. 2007;178(8):5086–5098. doi: 10.4049/jimmunol.178.8.5086. [DOI] [PubMed] [Google Scholar]
- Onoguchi K. Yoneyama M. Takemura A. Akira S. Taniguchi T. Namiki H. Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282(10):7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- Osterlund P. Veckman V. Sirén J. Klucher KM. Hiscott J. Matikainen S. Julkunen I. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J Virol. 2005;79(15):9608–9617. doi: 10.1128/JVI.79.15.9608-9617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund PI. Pietilä TE. Veckman V. Kotenko SV. Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179(6):3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- Pagliaccetti NE. Robek MD. Interferon-lambda in the immune response to hepatitis B virus and hepatitis C virus. J Interferon Cytokine Res. 2010;30(8):585–590. doi: 10.1089/jir.2010.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Kotenko SV. Muthukumaran G. Izotova LS. Cook JR. Garotta G. The interferon gamma (IFN-gamma) receptor: a paradigm for the multi-chain cytokine receptor. Cytokine Growth Factor Rev. 1997;8(3):189–206. doi: 10.1016/s1359-6101(97)00009-9. [DOI] [PubMed] [Google Scholar]
- Pestka S. Krause CD. Sarkar D. Walter MR. Shi Y. Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004a;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- Pestka S. Krause CD. Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004b;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Ramos EL. Preclinical and clinical development of pegylated interferon-lambda 1 in chronic hepatitis C. J Interferon Cytokine Res. 2010;30(8):591–595. doi: 10.1089/jir.2010.0066. [DOI] [PubMed] [Google Scholar]
- Renauld JC. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol. 2003;3(8):667–676. doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- Robek MD. Boyd BS. Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79(6):3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A. Ohtsuki M. Hata M. Kobayashi E. Murakami T. Antitumor activity of IFN-λ in murine tumor models. J Immunol. 2006;176(12):7686–7694. doi: 10.4049/jimmunol.176.12.7686. [DOI] [PubMed] [Google Scholar]
- Sheikh F. Baurin VV. Lewis-Antes A. Shah NK. Smirnov SV. Anantha S. Dickensheets H. Dumoutier L. Renauld JC. Zdanov A. Donnelly RP. Kotenko SV. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J Immunol. 2004;172(4):2006–2010. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- Sheppard P. Kindsvogel W. Xu W. Henderson K. Schlutsmeyer S. Whitmore TE. Kuestner R. Garrigues U. Birks C. Roraback J. Ostrander C. Dong D. Shin J. Presnell S. Fox B. Haldeman B. Cooper E. Taft D. Gilbert T. Grant FJ. Tackett M. Krivan W. McKnight G. Clegg C. Foster D. Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Silverman N. Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15(18):2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- Sirén J. Pirhonen J. Julkunen I. Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J Immunol. 2005;174(4):1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- Sommereyns C. Paul S. Staeheli P. Michiels T. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4(3):e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM. Tran KC. Chi B. Rabin RL. Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J Virol. 2004;78(8):4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen HC. Gamero AM. Interferon-lambda as a potential therapeutic agent in cancer treatment. J Interferon Cytokine Res. 2010;30(8):597–602. doi: 10.1089/jir.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff U. Müller U. Schertler A. Hengartner H. Aguet M. Zinkernagel RM. Antiviral protection by vesicular stomatitis virus-specific antibodies in alpha/beta interferon receptor-deficient mice. J Virol. 1995;69(4):2153–2158. doi: 10.1128/jvi.69.4.2153-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SJ. Goh FG. Banks H. Krausgruber T. Kotenko SV. Foxwell BM. Udalova IA. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissari J. Sirén J. Meri S. Julkunen I. Matikainen S. IFN-alpha enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J Immunol. 2005;174(7):4289–4294. doi: 10.4049/jimmunol.174.7.4289. [DOI] [PubMed] [Google Scholar]
- Wathelet MG. Lin CH. Parekh BS. Ronco LV. Howley PM. Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1(4):507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Witte K. Gruetz G. Volk HD. Looman AC. Asadullah K. Sterry W. Sabat R. Wolk K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type-III interferons. Genes Immun. 2009;10(8):702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- Wolk K. Witte K. Sabat R. Interleukin-28 and interleukin-29: novel regulators of skin biology. J Interferon Cytokine Res. 2010;30(8):617–628. doi: 10.1089/jir.2010.0064. [DOI] [PubMed] [Google Scholar]
- Xie MH. Aggarwal S. Ho WH. Foster J. Zhang Z. Stinson J. Wood WI. Goddard AD. Gurney AL. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275(40):31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]