Dynamin-like MxA GTPase: Structural Insights into Oligomerization and Implications for Antiviral Activity (original) (raw)
Abstract
The interferon-inducible MxA GTPase is a key mediator of cell-autonomous innate immunity against a broad range of viruses such as influenza and bunyaviruses. MxA shares a similar domain structure with the dynamin superfamily of mechanochemical enzymes, including an N-terminal GTPase domain, a central middle domain, and a C-terminal GTPase effector domain. Recently, crystal structures of a GTPase domain dimer of dynamin 1 and of the oligomerized stalk of MxA (built by the middle and GTPase effector domains) were determined. These data provide exciting insights into the architecture and antiviral function of the MxA oligomer. Moreover, the structural knowledge paves the way for the development of novel antiviral drugs against influenza and other highly pathogenic viruses.
Keywords: Antiviral Agents, Crystal Structure, Interferon, Protein Folding, X-ray Crystallography, Dynamin, Influenza, MxA GTPase, Protein Oligomerization, Viral Nucleocapsid
Introduction
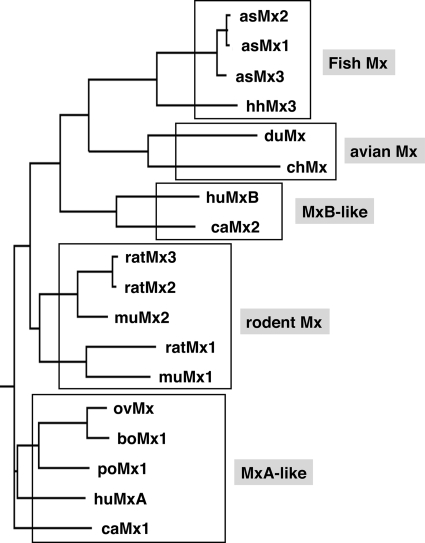
Mx proteins are key mediators of the interferon (IFN)2-induced antiviral response in vertebrates and hence of great biological interest and medical importance (1). Their discovery dates back to early studies on genetically determined inborn resistance of mice to influenza viruses (2, 3). The mouse Mx1 protein was originally found in influenza virus-resistant mice (4) and was shown to have intrinsic antiviral activity (5). It is encoded by the Mx1 gene (for myxovirus resistance gene 1), which is located on the distal part of chromosome 16 and is structurally altered in influenza virus-susceptible mice. Most inbred laboratory strains carry large deletions or nonsense mutations, indicating that they share parts of chromosome 16 with an ancestor founder mouse (6, 7). The mouse genome contains a second Mx gene, designated Mx2, which is closely linked to Mx1 on chromosome 16 and is also defective in inbred mouse strains (8). Both Mx genes are, however, intact in wild mice and in some laboratory strains derived from them (9, 10). The first evidence for Mx proteins in humans was obtained when a monoclonal antibody (2C12) against mouse Mx1 cross-reacted with an IFN-induced protein in human cells (11). Two proteins called MxA (myxovirus resistance protein 1) and MxB (myxovirus resistance protein 2) were found to be encoded by closely linked genes on the long arm of chromosome 21 (map position 21q22.3) that is syntenic with mouse chromosome 16 (12, 13). IFN-inducible Mx genes were subsequently identified in many vertebrates. They can be grouped into several subgroups according to their sequence similarities (Fig. 1). These comprise the fish, avian, and rodent subgroups as well as an MxA-like and MxB-like subgroup (for a review, see Ref. 1). Some polymorphisms have been reported, in particular for chicken and porcine Mx genes (14, 15), but their significance remains unclear (16).
FIGURE 1.
Phylogenetic tree of Mx proteins. The Mx proteins are grouped into five subgroups according to sequence similarities. Sequences were obtained from GenBankTM and aligned with ClustalW Version 1.81. The tree was constructed using the DrawGram function of Biology WorkBench 3.2. The following sequences were used for the alignment: human (hu) MxA and MxB; canine (ca) Mx1 and Mx2; porcine (po) Mx1; bovine (bo) Mx1; ovine (ov) Mx; murine (mu) Mx1 and Mx2; rat Mx1, Mx2, and Mx3; chicken (ch) Mx; duck (du) Mx; Atlantic salmon (as) Mx1, Mx2, and Mx3; and Atlantic halibut (hh) Mx. This figure was adapted from Ref. 1.
The critical role of mouse and human Mx proteins in mediating the antiviral activity of IFNs against specific viruses became evident early on. IFN had almost no protective effect against influenza viruses in mice and cell cultures lacking Mx1 (17, 18). However, constitutive expression of recombinant mouse Mx1 or human MxA protein protected _Mx1_-deficient cells against infection with Mx-sensitive viruses (5, 19). Also, when monoclonal antibody 2C12 was microinjected into Mx1-carrying cells, IFN lost its protective activity against influenza virus but could still protect the injected cells against unrelated viruses (20). Likewise, microinjection of this antibody neutralized the antiviral activity of human MxA against Thogoto virus (THOV), an exquisitely MxA-sensitive influenza-like orthomyxovirus (21). The power of human MxA as an efficient in vivo host defense mechanism was illustrated in experiments with MxA transgenic mice. These animals were produced in such a way that they constitutively expressed the human MxA protein in all tissues but were unable to express functional IFN type I (α/β) receptors for genetic reasons. They survived an otherwise lethal infection with MxA-sensitive viruses despite their inability to mount a type I IFN response (22). These early experiments clearly demonstrated that the mouse and human Mx proteins have intrinsic antiviral activity and act in an autonomous way without the need for other type I IFN-induced factors. The nature of Mx proteins remained unclear until sequence analyses revealed that Mx proteins contain putative guanine nucleotide-binding sites (23), and an Mx-related protein, Vps1p (vacuolar protein sorting 1 protein), was discovered in yeast and shown to perform an essential function in vacuolar protein sorting (24). Also, molecular cloning of rat dynamin revealed extensive homology to Vps1p and Mx1, establishing a new family of GTP-binding proteins (25).
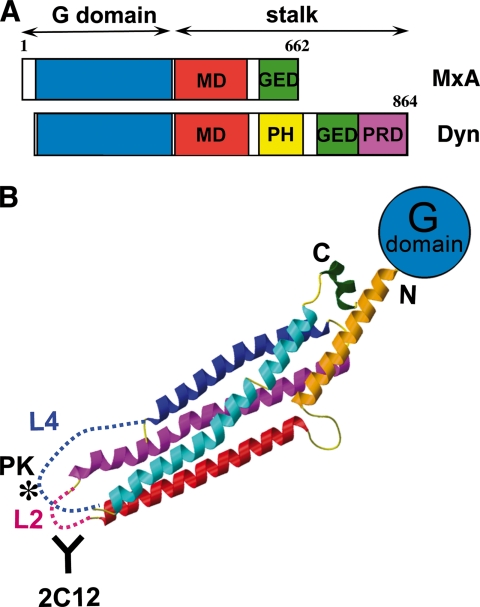
It is now clear that Mx proteins form a distinct subclass within the dynamin superfamily of GTPases. Mx GTPases have a molecular mass of ∼75,000 Da and, like dynamins, display a relatively low affinity for GTP and a high intrinsic rate of GTP hydrolysis. They consist of three domains, namely an N-terminal GTPase domain (G domain) that binds and hydrolyzes GTP, a middle domain (MD) that mediates self-assembly and oligomerization, and a C-terminal GTPase effector domain (GED) that is involved in viral target recognition (26, 27) and also self-assembly (Fig. 2A) (28). Unlike dynamin, Mx proteins do not have a pleckstrin homology (PH) domain, which is implicated in membrane binding of classical dynamin. They also lack a proline/arginine-rich domain, which is involved in dynamin-protein interactions, but feature short N-terminal extensions of 25–90 residues of unknown function. Of note, the C terminus of mouse Mx1 but not human MxA has a stretch of basic amino acids that constitutes a nuclear localization signal and mediates nuclear accumulation (5, 29). The G domains of MxA and dynamin share 40% overall sequence identity, whereas the MDs and GEDs are 20% identical, suggesting a similar fold and basic catalytic machinery of these types of GTPases. A common feature of dynamin-like GTPases is their ability to self-assemble into highly ordered oligomers and to show cooperativity in GTP hydrolysis. Biochemical analysis suggested that the GED of Mx GTPases folds back to the G domain and regulates their GTP-hydrolyzing activity (30). In dynamin, the structural details of this intramolecular interaction have now been elucidated. An α-helix at the very C terminus of the GED binds into a hydrophobic groove formed by two α-helices preceding and following the G domain. Together, they form a three-helix bundle called the “bundle signaling element,” which was suggested to transmit signals between the G domain and the MD/GED (31, 32). Purified Mx proteins form high molecular mass homo-oligomers and self-assemble into helical arrays at high protein and low salt concentrations (Fig. 3, A and B) (33–35). MxA binds to negatively charged membranes and oligomerizes in ring-like structures around liposomes much in the same way as dynamin, inducing liposome tubulation (Fig. 3C) (35, 36). In IFN-treated cells, mouse Mx1 accumulates in distinct nuclear dots in close proximity to promyelocytic leukemia nuclear bodies (37), whereas human MxA forms punctate granula in the cytoplasm, which partially co-localizes with COPI-positive membranes of the smooth endoplasmic reticulum-Golgi intermediate compartment (38). These cellular Mx assemblies appear to provide an intracellular storage form from where Mx proteins can be recruited (39). Both human and mouse Mx proteins have a half-life of >24 h, suggesting that oligomerization most likely prevents their rapid degradation (40, 41).
FIGURE 2.
Structure and domain composition of human MxA. A, schematic diagram of human MxA and human dynamin 1 (Dyn). MxA consists of three domains: the G domain (blue), the MD (red), and the GED (green). The MD and GED form the MxA stalk. Dynamin features two additional domains, namely a PH domain (yellow) and a proline/arginine-rich domain (PRD; magenta). B, structure of the MxA stalk composed of the MD and GED (amino acids 366–633) combined with the N-terminal globular G domain (blue). The unstructured loops L2 and L4 are indicated by dashed lines. The positions of the antibody 2C12-binding site (positions 432–471) in loop L2 (47) and the proteinase K cleavage site at position 564 (PK, asterisk) in loop L4 (30) are indicated. This figure was adapted from Ref. 46.
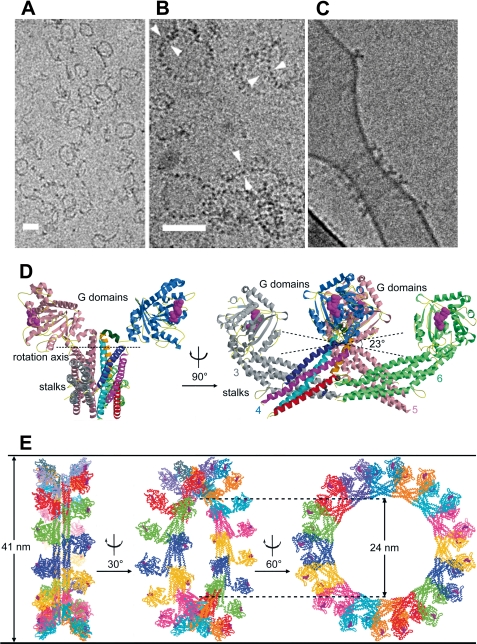
FIGURE 3.
Oligomerization and liposome binding of human MxA. A–C, cryo-transmission EM images of purified MxA dialyzed against low salt buffer in the presence of 1 mm GMP-PCP. Self-assemblies show rings and open arcs (A); higher magnification of the rings reveals a structure of two parallel sets of electron-dense globular domains (B, double arrowheads); and incubation with phosphatidylserine-containing liposomes in the presence of GTP shows MxA ring formation and tubulation (C) (36). Scale bars = 50 nm. D, shown is a model of oligomeric MxA composed of the MxA stalks connected to the G domains of dynamin. Side and front views are shown. E, a complete MxA ring composed of 16 MxA dimers was designed according to cryo-EM reconstructions of dynamin as described previously (46) but with an angle of only 23° between the associating stalks to accomplish formation of a 41-nm oligomeric ring.
Although self-assembly of Mx proteins was found to be critical for GTPase activity and presumably protein stability, it was not known whether oligomerization is also crucial for recognition of viral target structures and antiviral activity. Despite tremendous efforts, previous attempts to obtain structural information for Mx GTPases by x-ray crystallography were unsuccessful. However, the crystal structure of GBP1 (guanylate-binding protein 1) represented the first structure of a large GTPase within the dynamin superfamily (42). Later, structures of monomeric G domains of rat dynamin and Dictyostelium discoideum dynamin A were determined (43, 44). In contrast, the organization and architecture of the MD and GED of dynamin and MxA remained elusive. Low resolution cryo-electron microscopy (EM) structures suggested that the MD and GED of dynamin-like GTPases constitute a stalk that mediates oligomerization and transmits conformational changes from the G domain to the target structure (45).
Structure of the MxA Stalk
Early modeling of MxA protein folding was based on biochemical data and the known structure of human GBP1 (39). All available evidence indicated that the Mx GTPases had an N-terminal large globular G domain followed by a bundle of elongated antiparallel α-helices built from the MD and GED. The molecular architecture of the MD and GED has now been solved (46). The resulting x-ray structure encompasses nearly the complete MD and the N-terminal part of the GED, which together form an antiparallel four-helix bundle, the stalk of MxA (Fig. 2B). Two major loops, L2 and L4, at the opposite position of the G domain were not resolved in the crystal structure. Loop L2 (amino acids 438–447) is the target of monoclonal antibody 2C12, which neutralizes the antiviral activity of human and mouse Mx proteins when injected into living cells (20, 21, 47). The 41-amino acid long loop L4 (residues 532–572) is at the equivalent sequence position as the PH domain of dynamin and may serve a similar function in lipid/target binding. Loop L4 is predicted to be unstructured and is known to harbor a proteinase K cleavage site (Fig. 2B) (30). The stalk of MxA structurally differs from the corresponding domains of other dynamin superfamily members such as human GBP1 (42), BDLP (bacterial dynamin-like protein) (48), and EHD2 (Esp15 homology domain-containing protein 2) (49), but its structure is consistent with predictions for the stalk of dynamin (46). From an overall perspective, the MxA stalk connects the globular G domain (the “head”) to the two intrinsically disordered loops L2 and L4 (the “legs”), which reach out of the four-helix bundle at the opposite side (Fig. 2B) and might constitute target interaction sites, as briefly discussed below.
MxA Self-assembles in a Crisscross Pattern into Highly Ordered Oligomers
Initially, the self-assembly properties of purified MxA protein were assessed using EM techniques developed to study the oligomerization of dynamin by the group of J. E. Hinshaw (50, 51). When incubated with GDP or GDPβS, MxA formed rod- and ring-like structures resembling the structures formed by dynamin (Fig. 3, A and B) (34, 35). Addition of GTPγS changed the oligomerization state of MxA, leading to the appearance of spirals and stacks of rings (34, 35) as originally also reported for mouse Mx1 (52). It was therefore of great interest to elucidate the molecular basis of MxA oligomerization to understand the structural changes induced by nucleotide binding and hydrolysis. Interestingly, each MxA stalk in the obtained crystals was assembled in a crisscross pattern, resulting in a linear oligomer (Fig. 3D). This arrangement involves three distinct interfaces and one loop in each stalk and agrees well with the proposed crisscross arrangement of the dynamin stalks deduced by EM reconstructions (45, 53). Furthermore, this stalk arrangement pattern allows the G domains to be located on one side of the MxA oligomer, whereas the putative membrane- or virus-binding sites in L2 and L4 are located on the opposite side (Fig. 3, D and E).
Analytical ultracentrifugation experiments demonstrated that wild-type MxA is a stable tetramer in solution (46), similar to dynamin (54). Each of the three interfaces contributes to the native assembly of full-length MxA, as demonstrated by mutational analyses and protein-protein interaction experiments. Strikingly, individual mutations at two positions in interface 2 (M527D in the MD and F602D in the GED) led to a complete disruption of the tetramer, resulting in a predominantly monomeric form. Consequently, these mutants eluted as monomers in analytical gel filtration experiments and did not interact with wild-type MxA in co-immunoprecipitation assays or in an in vivo co-translocation assay (46). It was reported previously that substitution of lysine for leucine at position 612 (L612K) resulted in monomeric MxA (28) and that the mutant was still antivirally active (41). The crystal structure now reveals that Leu-612 is located between interfaces 1 and 2 and points toward the hydrophobic core of the stalk but does not directly participate in intermolecular interactions. However, a mutation to lysine is likely to destabilize the stalk, leading to disruption of interface 2. Other mutations in interfaces 1 and 3 and the two loops also led to the disruption of the tetramer but resulted predominantly in a dimeric form of the protein. Interestingly, the corresponding mutations in yeast dynamin DNM1 (G385D) and in dynamin 1 (e.g. R361S and R399A) also generated stable dimers (54, 55). These results demonstrated that mutations involving interfaces 1 and 3 and L4 disrupt tetramerization and induce a stable dimeric form of MxA, whereas mutations in interface 2 are completely disruptive, leading to monomeric MxA. They further suggest that equivalent interfaces exist in dynamin and additional dynamin-like proteins.
Previous EM studies of full-length MxA and dynamin revealed the formation of ring-like oligomers of various diameters (Fig. 3, A–C) (34–36). Importantly, the linear oligomeric stalk model of MxA could be fitted into an EM reconstruction map of oligomerized dynamin (45) by introducing a simple rotation in interface 1 (Fig. 3, D and E). This finding supported the notion that oligomers of MxA and dynamin have a similar three-dimensional architecture. The oligomeric model accounts for the “T-bar” shape seen in side views of oligomerized dynamin (45) and MxA (Fig. 3, C and D) (36). Furthermore, it illustrates the connectivity of the G domain with the PH domain in oligomerized dynamin and is in agreement with the formation of a bundle signaling element involving the C-terminal part of the GED as mentioned above (32).
As in the case of dynamin and other dynamin-related GTPases, GTP hydrolysis of MxA is regulated by oligomerization and exhibits cooperative stimulation with increasing protein concentrations (46, 56). The structure of the G domain dimer of dynamin demonstrated that GTP hydrolysis of dynamin-like proteins is activated by direct contacts between the G domains (32). In the oligomeric MxA model, the G domains of one MxA ring are not in contact with other G domains of the same ring but are pointing away from the central stalk of the ring and facilitate inter-ring contacts (Fig. 3, D and E). Consequently, we propose that MxA oligomerization initially proceeds via association of the stalks, until a complete ring is formed. Only then can G domains from neighboring rings approach each other, and nucleotide hydrolysis is stimulated in a coordinated fashion.
MxA Oligomerization and Antiviral Activity
The molecular basis of the antiviral activity of Mx GTPases is still poorly understood. MxA inhibits a diverse range of viruses that represent distinct RNA and DNA virus families (for a review, see Ref. 1). Among them are influenza A, B, and C viruses; influenza-like THOV; LaCrosse virus (LACV); and also African swine fever virus (57). Most studies were performed with influenza A virus. Early work indicated that overexpression of the influenza polymerase subunit PB2 abolishes the antiviral effect of mouse Mx1, suggesting that PB2 might be a putative Mx1 target (58, 59). However, such a role for PB2 could not be substantiated in subsequent work based on reverse genetics and minireplicon systems. These new approaches demonstrated that the nucleoprotein (NP) rather than PB2 was the Mx target. NP is associated with the viral genome and, together with the viral polymerase complex, forms the nucleocapsid structure of influenza and influenza-like viruses. Influenza virus strains were shown to differ in their sensitivity to the antiviral effect of Mx proteins in cultured cells and minireplicon assays (60). Artificial reassortments between relatively susceptible and relatively resistant strains demonstrated that Mx sensitivity clearly segregated with the viral NP and not PB2 or any other viral component (60). Importantly, a physical interaction of MxA with the viral NP was demonstrated for influenza A virus (61) and THOV (62). Furthermore, nucleocapsid recognition was demonstrated in co-sedimentation assays using glycerol gradient centrifugation: when MxA-containing cell lysates were mixed with THOV nucleocapsids, MxA co-sedimented with the nucleocapsids into fractions of higher density, suggesting a tight association of MxA with these viral structures (62). Recognition of viral nucleocapsids also occurs in intact living cells. MxA blocked the movement of viral nucleocapsids into the nucleus, where they normally accumulate after microinjection into the cytoplasm (21). This transport block could be released by anti-Mx monoclonal antibody 2C12 when microinjected along with the nucleocapsids (21).
The capacity of MxA to relocate viral components has been extensively studied also in bunyavirus-infected cells (63) as well as in cells infected with African swine fever virus (57). MxA was demonstrated to bind to the nucleocapsid protein N of LACV and to form intracellular complexes with N (63). The interaction of MxA with viral N occurred on membranes of the smooth endoplasmic reticulum-Golgi intermediate compartment and led to a depletion of N protein from the viral replication sites (64). EM revealed that stacks of filamentous bundles were accumulating near the nuclear membrane. Labeling with specific antibodies demonstrated that these filaments were composed of both MxA and viral N protein (63). It is conceivable that intracellular membranes serve as a scaffold to facilitate the interaction between MxA and viral target structures, but the role of lipid binding for antiviral activity of MxA is not clear at present.
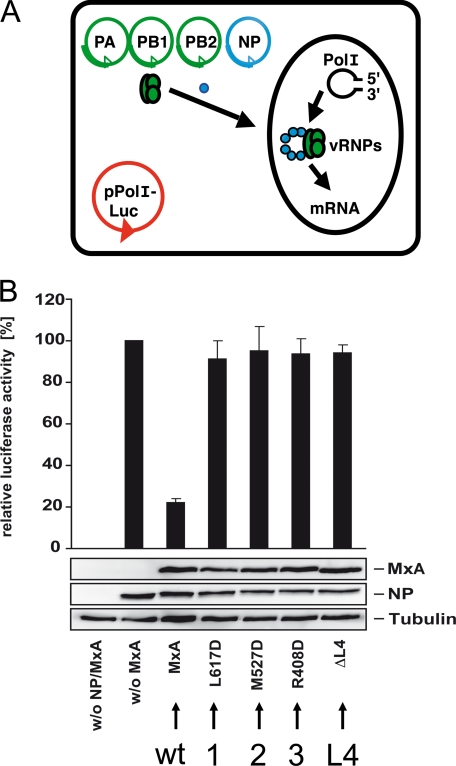
Mutational analyses revealed that the stalk region of MxA is involved in target interaction (26), but the structural requirements for antiviral activity were unknown. Therefore, wild-type and assembly-deficient mutant forms of MxA were assessed for their inhibitory activity against influenza virus in a minireplicon reporter assay (Fig. 4A). The minireplicon system mimics infection and is amenable to experimental modifications: coexpression of a luciferase reporter minigenome together with the three subunits of the viral polymerase and the viral NP generates nucleocapsids that transcribe and replicate the minigenome. In this reporter assay, luciferase production correlates with the transcriptional and replicative activity of the viral polymerase and is sensitive to inhibition by the MxA GTPase. Although wild-type MxA inhibited viral polymerase activity by 80%, mutations in each of the three interfaces identified by the structural analysis or deletions in loop L4 abolished antiviral activity almost completely (Fig. 4B). Likewise, assembly-deficient MxA proteins lost their antiviral activity against LACV and failed to form complexes with the viral N protein (46).
FIGURE 4.
Antiviral activity of MxA requires oligomerization. A, influenza A virus minireplicon system. Coexpression of a luciferase reporter minigenome (pPolI-luc) together with the three subunits of the viral polymerase (PA, PB1, and PB2) and the viral NP generates nucleocapsids that transcribe and replicate the minigenome. Luciferase production correlates with the transcriptional and replicative activity of the viral polymerase. vRNPs, viral ribonucleoproteins. B, wild-type (wt) MxA inhibits the polymerase activity of the avian H5N1 influenza A/Vietnam/1203/04 virus in a minireplicon system. Expression plasmids for the indicated MxA constructs were used, and the relative luciferase activity was determined. Assembly-defective MxA variants with mutations either in one of the three interfaces (1, 2, or 3) or in L4 (Δ_L4_) have lost antiviral activity. Protein expression was analyzed by Western blotting using specific antibodies. This figure was adapted from Ref. 46.
In summary, these findings demonstrate that proper assembly of the MxA stalk is essential for the antiviral function. Whereas tetramers appear to constitute the cytoplasmic inactive form of MxA, oligomerization of tetramers around the target structure induces the formation of functional ring-like MxA units. A straightforward scenario postulates that MxA forms oligomeric rings around the tubular structure of viral nucleocapsids, thereby blocking their function. As a consequence, MxA may immobilize nucleocapsids or direct them to special sites in the cytoplasm where they will eventually be degraded.
Future Prospects
The structural characterization of the MxA stalk provides new insights into the formation of higher order MxA assemblies and demonstrates that multimerization is required for antiviral activity, as discussed here. Together with recent progress in dynamin, it allows the conclusion that these GTPases are built of three structural entities that do not strictly reflect the predicted domain organization, namely the G domain, the bundle signaling element (consisting of the C-terminal portion of the GED and two helices connected with the G domain), and the stalk (composed of the MD and the N-terminal part of the GED). Further structural studies should help to clarify the basic setup of the antiviral machinery, which is presently not understood. It will be important to characterize the viral target recognition site of MxA. The present model features two unstructured loops that might represent ideal target-binding sites due to their position in the oligomeric MxA rings. Detailed molecular knowledge of the interaction between MxA and viral target structures should provide a starting point to develop low molecular mass compounds with antiviral activity as a new strategy to combat influenza and other highly pathogenic viruses.
Supplementary Material
Author profile
Acknowledgments
We thank our colleagues working on Mx and other large GTPases for helpful discussions.
*
This work was supported by German-Israeli Foundation for Scientific Research and Development Grant GIF-841/04 (to O. H. and G. K.), Deutsche Forschungsgemeinschaft Grants Ko1579/1-8 and SFB 740 (From Molecules to Modules), and a career development fellowship from the International Human Frontier Science Program Organization (to O. D.). This is the third article in the Thematic Minireview Series on Influenza Virus. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
2
The abbreviations used are:
IFN
interferon
THOV
Thogoto virus
G domain
GTPase domain
MD
middle domain
GED
GTPase effector domain
PH
pleckstrin homology
EM
electron microscopy
GDPβS
guanosine 5′-(β-thio)diphosphate
GTPγS
guanosine 5′-(γ-thio)triphosphate
LACV
LaCrosse virus
NP
nucleoprotein
N
nucleocapsid protein
GMP-PCP
guanylyl β,γ-methylenediphosphonate.
REFERENCES
- 1.Haller O., Stertz S., Kochs G. (2007) Microbes Infect. 9, 1636–1643 [DOI] [PubMed] [Google Scholar]
- 2.Lindenmann J. (1962) Virology 16, 203–204 [DOI] [PubMed] [Google Scholar]
- 3.Haller O., Lindenmann J. (1974) Nature 250, 679–680 [DOI] [PubMed] [Google Scholar]
- 4.Horisberger M. A., Staeheli P., Haller O. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1910–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staeheli P., Haller O., Boll W., Lindenmann J., Weissmann C. (1986) Cell 44, 147–158 [DOI] [PubMed] [Google Scholar]
- 6.Reeves R. H., O'Hara B. F., Pavan W. J., Gearhart J. D., Haller O. (1988) J. Virol. 62, 4372–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staeheli P., Grob R., Meier E., Sutcliffe J. G., Haller O. (1988) Mol. Cell. Biol. 8, 4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staeheli P., Sutcliffe J. G. (1988) Mol. Cell. Biol. 8, 4524–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haller O., Acklin M., Staeheli P. (1987) J. Interferon Cytokine Res. 7, 647–656 [DOI] [PubMed] [Google Scholar]
- 10.Jin H. K., Yamashita T., Ochiai K., Haller O., Watanabe T. (1998) Biochem. Genet. 36, 311–322 [DOI] [PubMed] [Google Scholar]
- 11.Staeheli P., Haller O. (1985) Mol. Cell. Biol. 5, 2150–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aebi M., Fäh J., Hurt N., Samuel C. E., Thomis D., Bazzigher L., Pavlovic J., Haller O., Staeheli P. (1989) Mol. Cell. Biol. 9, 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horisberger M. A., Wathelet M., Szpirer J., Szpirer C., Islam Q., Levan G., Huez G., Content J. (1988) Somatic Cell Mol. Genet. 14, 123–131 [DOI] [PubMed] [Google Scholar]
- 14.Seyama T., Ko J. H., Ohe M., Sasaoka N., Okada A., Gomi H., Yoneda A., Ueda J., Nishibori M., Okamoto S., Maeda Y., Watanabe T. (2006) Biochem. Genet. 44, 437–448 [DOI] [PubMed] [Google Scholar]
- 15.Palm M., Leroy M., Thomas A., Linden A., Desmecht D. (2007) J. Interferon Cytokine Res. 27, 147–155 [DOI] [PubMed] [Google Scholar]
- 16.Haller O., Staeheli P., Kochs G. (2009) Rev. Sci. Tech. Off. Int. Epizoot. 28, 219–231 [DOI] [PubMed] [Google Scholar]
- 17.Haller O., Arnheiter H., Lindenmann J., Gresser I. (1980) Nature 283, 660–662 [DOI] [PubMed] [Google Scholar]
- 18.Haller O., Arnheiter H., Gresser I., Lindenmann J. (1979) J. Exp. Med. 149, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlovic J., Zürcher T., Haller O., Staeheli P. (1990) J. Virol. 64, 3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnheiter H., Haller O. (1988) EMBO J. 7, 1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochs G., Haller O. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2082–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hefti H. P., Frese M., Landis H., Di Paolo C., Aguzzi A., Haller O., Pavlovic J. (1999) J. Virol. 73, 6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horisberger M. A., McMaster G. K., Zeller H., Wathelet M. G., Dellis J., Content J. (1990) J. Virol. 64, 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothman J. H., Raymond C. K., Gilbert T., O'Hara P. J., Stevens T. H. (1990) Cell 61, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 25.Obar R. A., Collins C. A., Hammarback J. A., Shpetner H. S., Vallee R. B. (1990) Nature 347, 256–261 [DOI] [PubMed] [Google Scholar]
- 26.Zürcher T., Pavlovic J., Staeheli P. (1992) EMBO J. 11, 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannes L., Kambadur R., Lee-Hellmich H., Hodgkinson C. A., Arnheiter H., Meier E. (1997) J. Virol. 71, 9792–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumacher B., Staeheli P. (1998) J. Biol. Chem. 273, 28365–28370 [DOI] [PubMed] [Google Scholar]
- 29.Zürcher T., Pavlovic J., Staeheli P. (1992) J. Virol. 66, 5059–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwemmle M., Richter M. F., Herrmann C., Nassar N., Staeheli P. (1995) J. Biol. Chem. 270, 13518–13523 [PubMed] [Google Scholar]
- 31.Chappie J. S., Acharya S., Liu Y. W., Leonard M., Pucadyil T. J., Schmid S. L. (2009) Mol. Biol. Cell 20, 3561–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chappie J. S., Acharya S., Leonard M., Schmid S. L., Dyda F. (2010) Nature 465, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melén K., Ronni T., Broni B., Krug R. M., von Bonsdorff C. H., Julkunen I. (1992) J. Biol. Chem. 267, 25898–25907 [PubMed] [Google Scholar]
- 34.Kochs G., Haener M., Aebi U., Haller O. (2002) J. Biol. Chem. 277, 14172–14176 [DOI] [PubMed] [Google Scholar]
- 35.Accola M. A., Huang B., Al Masri A., McNiven M. A. (2002) J. Biol. Chem. 277, 21829–21835 [DOI] [PubMed] [Google Scholar]
- 36.Kochs G., Reichelt M., Danino D., Hinshaw J. E., Haller O. (2005) Methods Enzymol. 404, 632–643 [DOI] [PubMed] [Google Scholar]
- 37.Engelhardt O. G., Ullrich E., Kochs G., Haller O. (2001) Exp. Cell Res. 271, 286–295 [DOI] [PubMed] [Google Scholar]
- 38.Stertz S., Reichelt M., Krijnse-Locker J., Mackenzie J., Simpson J. C., Haller O., Kochs G. (2006) J. Interferon Cytokine Res. 26, 650–660 [DOI] [PubMed] [Google Scholar]
- 39.Haller O., Kochs G. (2002) Traffic 3, 710–717 [DOI] [PubMed] [Google Scholar]
- 40.Horisberger M. A., De Staritzky K. (1989) J. Interferon Res. 9, 583–590 [DOI] [PubMed] [Google Scholar]
- 41.Janzen C., Kochs G., Haller O. (2000) J. Virol. 74, 8202–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash B., Praefcke G. J., Renault L., Wittinghofer A., Herrmann C. (2000) Nature 403, 567–571 [DOI] [PubMed] [Google Scholar]
- 43.Niemann H. H., Knetsch M. L., Scherer A., Manstein D. J., Kull F. J. (2001) EMBO J. 20, 5813–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reubold T. F., Eschenburg S., Becker A., Leonard M., Schmid S. L., Vallee R. B., Kull F. J., Manstein D. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mears J. A., Ray P., Hinshaw J. E. (2007) Structure 15, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao S., von der Malsburg A., Paeschke S., Behlke J., Haller O., Kochs G., Daumke O. (2010) Nature 465, 502–506 [DOI] [PubMed] [Google Scholar]
- 47.Flohr F., Schneider-Schaulies S., Haller O., Kochs G. (1999) FEBS Lett. 463, 24–28 [DOI] [PubMed] [Google Scholar]
- 48.Low H. H., Löwe J. (2006) Nature 444, 766–769 [DOI] [PubMed] [Google Scholar]
- 49.Daumke O., Lundmark R., Vallis Y., Martens S., Butler P. J., McMahon H. T. (2007) Nature 449, 923–927 [DOI] [PubMed] [Google Scholar]
- 50.Hinshaw J. E., Schmid S. L. (1995) Nature 374, 190–192 [DOI] [PubMed] [Google Scholar]
- 51.Sweitzer S. M., Hinshaw J. E. (1998) Cell 93, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 52.Nakayama M., Yazaki K., Kusano A., Nagata K., Hanai N., Ishihama A. (1993) J. Biol. Chem. 268, 15033–15038 [PubMed] [Google Scholar]
- 53.Klockow B., Tichelaar W., Madden D. R., Niemann H. H., Akiba T., Hirose K., Manstein D. J. (2002) EMBO J. 21, 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramachandran R., Surka M., Chappie J. S., Fowler D. M., Foss T. R., Song B. D., Schmid S. L. (2007) EMBO J. 26, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. (2005) J. Cell Biol. 170, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richter M. F., Schwemmle M., Herrmann C., Wittinghofer A., Staeheli P. (1995) J. Biol. Chem. 270, 13512–13517 [PubMed] [Google Scholar]
- 57.Netherton C. L., Simpson J., Haller O., Wileman T. E., Takamatsu H. H., Monaghan P., Taylor G. (2009) J. Virol. 83, 2310–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang T., Pavlovic J., Staeheli P., Krystal M. (1992) J. Virol. 66, 4154–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stranden A. M., Staeheli P., Pavlovic J. (1993) Virology 197, 642–651 [DOI] [PubMed] [Google Scholar]
- 60.Dittmann J., Stertz S., Grimm D., Steel J., García-Sastre A., Haller O., Kochs G. (2008) J. Virol. 82, 3624–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turan K., Mibayashi M., Sugiyama K., Saito S., Numajiri A., Nagata K. (2004) Nucleic Acids Res. 32, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kochs G., Haller O. (1999) J. Biol. Chem. 274, 4370–4376 [DOI] [PubMed] [Google Scholar]
- 63.Kochs G., Janzen C., Hohenberg H., Haller O. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reichelt M., Stertz S., Krijnse-Locker J., Haller O., Kochs G. (2004) Traffic 5, 772–784 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Author profile