Early identification of cardiovascular risk using genomics and proteomics (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 15.
Published in final edited form as: Nat Rev Cardiol. 2010 May 4;7(6):309–317. doi: 10.1038/nrcardio.2010.53
Abstract
Coronary heart disease (CHD) will soon become the leading cause of death and morbidity in the world. Early detection and treatment of CHD is thus imperative to improve global health. Atherosclerosis of the coronary arteries is a complex multifactorial disease process involving multiple pathways that can be influenced by both genetic and environmental factors. With the recent advances in genomics and proteomics, many new risk factors with small-to-moderate effects are likely to be identified. Additionally, individualized risk stratification and targeted therapy may become feasible; each individual could potentially be assessed with a panel of tests for genomic and proteomic markers and, on the basis of the individual’s composite risk profile, preventive and therapeutic steps could then be undertaken. With a multimarker approach, it may also be possible to identify alterations in pathways involved in atherogenesis, rather than focus on individual risk factors. In this article, we use the specific example of atherosclerosis to discuss the role of genomics and proteomics in cardiovascular risk assessment.
Introduction
Significant progress was made in reducing mortality from coronary heart disease (CHD) in the western world in the past century. These advances resulted from identification of risk factors for CHD, treatment of these risk factors, and improved care of patients with acute coronary syndromes. Despite these successes, CHD will soon be the leading cause of death and morbidity in the world.1 The first manifestation of CHD is often a devastating event, such as sudden death or myocardial infarction, and a substantial number of individuals develop acute coronary syndromes at a relatively young age or in the absence or paucity of conventional risk factors. Early-onset disease is particularly common in urban centers of developing countries, such as in Southeast Asia, where there is an ongoing epidemic of CHD.2 Refining methods for early detection and treatment of CHD is thus imperative to improve global health.
Prediction of cardiovascular events is based on conventional risk factors for atherosclerosis, such as age, male sex, hypertension, diabetes, dyslipidemia, and smoking.3,4 Because these risk factors are prevalent in much of the population, available risk-prediction algorithms have less than desired accuracy.5 For example, one study showed that the algorithms correctly predicted just ~11% of the CHD events occurring within 10 years.6 New tools to better predict the cardiovascular risk of an individual are clearly urgently needed.7 Whereas several drugs to treat risk factors such as dyslipidemia and hypertension have been developed over the past few decades, the identification of new biomarkers for early detection of atherosclerotic vascular disease has lagged behind.
In the past few years, substantial progress has been made in three areas—genomics, proteomics, and imaging (the latter of which is discussed by Fuster et al.8 in this issue of Nature Reviews Cardiology)—and is likely to lead to more-accurate cardiovascular risk prediction. Although outside the scope of this Review article (and reviewed in detail elsewhere9), metabolomics—the study of the total complement of small-molecule metabolites found in or produced by an organism—is another promising tool for early detection of disease and understanding of disease pathophysiology.
Knowledge of new plasma markers and genetic susceptibility variants may provide more-precise estimates of risk while also defining the pathways perturbed in individual patients, revealing new targets for intervention, and ultimately enabling an individualized approach to care.10–12 In this article, we discuss the current state of genomics and proteomics for cardiovascular risk assessment and how advances in these fields may promote a better understanding of the complexity of atherosclerotic vascular disease. These advances may refine cardiovascular risk assessment, allow early treatment and, therefore, improve global health.
Complexity of atherosclerosis
Our knowledge of the biology of atherosclerotic vascular disease has substantially increased over the past few decades. After an initial preoccupation with arterial lumen narrowing and ischemia, there has been gradual acceptance of the concept that atherosclerosis is a response to injury and that inflammation is involved in all stages of the disease. That acute coronary syndromes can occur in the absence of significant luminal stenosis is now well established. Alterations in several key pathways—including those involved in inflammation, thrombosis, lipoprotein metabolism, calcification, vascular remodeling, oxidative stress and cell death—are known to result in the development of atherosclerosis, as well as its progression and complications.13,14 Inclusion of potential markers of each of these pathways in CHD screening may lead to increased accuracy of CHD risk prediction. Furthermore, determination of how alterations in these pathways lead to atherosclerosis could allow implementation of therapies to restore perturbed pathways to their normal functions.
In addition to our increased knowledge of the cells and pathways involved in the pathogenesis of atherosclerosis, we are now also aware that the risk of acute coronary syndromes is dynamic and fluctuates over time. Although atherosclerosis generally progresses with age, there are periods of quiescence punctuated by episodic increases in plaque inflammation and growth. Most risk predictions are made on the basis of single cross-sectional profiles, and newer algorithms will need to incorporate the nonlinear behavior of atherosclerotic plaque growth and activity. One approach would be to have periodic risk assessments targeted to detect such changes in plaque behavior.
Our improved understanding of the biology of atherosclerosis has, therefore, highlighted the following two main paradigms important for CHD risk assessment: firstly, the complexity of CHD necessitates a multimodal and multimarker approach; and, secondly, there is need for periodic assessment, owing to the dynamic nature of plaque activity and CHD risk.
Use of genomics for risk assessment
Family history has been described as a “…free, well-proven, personalized genomic tool that captures many of the genes and environmental interactions and can serve as the cornerstone for individualized disease prevention”.15 CHD has a significant heritable component, as exemplified by twin and family studies,16,17 and high-risk families make up a considerable proportion of early CHD cases in the general population.18 A history of early CHD in a first-degree relative approximately doubles the risk of CHD, although the reported relative risk ranges from 1.3 to 11.3.17 The familial clustering of CHD can be partly explained by heritable variation in known CHD risk factors, which can thus be quantified by using currently available risk-prediction algorithms; however, existing evidence suggests that family history contributes to an increased risk of CHD independently of the known risk factors.19 Multiple, as yet undiscovered, genetic susceptibility variants that mediate the familial clustering of CHD must, therefore, exist. Both candidate-gene and agnostic genomic approaches have been used to identify genetic variants that influence susceptibility to CHD.
Candidate-gene approach
The candidate-gene approach involves genotyping variants in genes with important roles in etiologic pathways of atherosclerosis, such as lipoprotein metabolism, to determine whether the variants—typically single nucleotide polymorphisms (SNPs)—are associated with adverse cardiovascular events or quantitative measures of subclinical atherosclerotic vascular disease. Candidate-gene studies of lipoprotein-related genes, which include APOE, LPA and PCSK9, have reinforced the importance of lipoprotein metabolism in atherogenesis. Presence of one or more APOE ε4 alleles is associated with dyslipidemia and increased risk of CHD.20 Similarly, variants in LPA that are associated with higher levels of lipoprotein(a) are in turn associated with an increased risk of CHD.21 Contrarily, mutations in PCSK9 that lead to lower levels of LDL cholesterol are associated with lower lifetime risk of CHD.22
By contrast to the candidate-gene studies of the lipoprotein metabolism pathway, findings from the majority of candidate-gene studies have not been replicable. Typically, between one and several SNPs in a candidate gene were genotyped in cases and controls and statistical significance was set at P <0.05. The newer generation of candidate-gene studies attempts a more-comprehensive coverage of the genetic variation at the candidate gene locus, on the basis of knowledge of neighboring SNPs being strongly correlated with each other (that is, in linkage disequilibrium), and replicates the findings either within a study sample or in an independent cohort.23 There is also growing interest in sequencing the ‘exome’—the gene-coding parts of the human genome—to identify rare variants that are associated with human diseases.24
Agnostic genomic approach
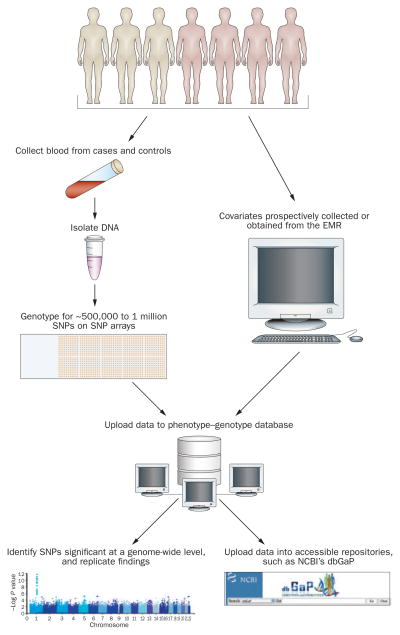
Genome-wide association studies (GWAS; Figure 1) have become feasible owing to our knowledge of patterns of linkage disequilibrium across the genome and availability of high-throughput genotyping platforms.25 By contrast to family-based linkage studies, GWAS are easier to initiate and simply require an adequate number of cases and controls. DNA is typically extracted from white blood cells and then subjected to genotyping of anywhere from 500,000 to 1 million SNPs across the genome. Although at least 7 million common SNPs (minor allele frequency >5%) exist in the human genome, neighboring SNPs are often in linkage disequilibrium. GWAS take advantage of patterns of linkage disequilibrium, such that genotyping approximately 500,000 SNPs will cover more than 80% of the common SNPs in the genomes of nonblack populations, whereas 1 million SNPs are needed to obtain similar coverage in black populations.25
Figure 1.
Flow diagram illustrating the use of GWAS to identify new genetic markers. Covariates might include age, sex, body-mass index, smoking history, levels of total and HDL cholesterol, diabetes, blood pressure, and statin use. Abbreviations: dbGaP, database for genotypes and phenotypes;71 EMR, electronic medical record; GWAS, genome-wide association studies; NCBI, National Center for Biotechnology Information; SNP, single-nucleotide polymorphism.
SNPs that differ in frequency between cases and controls are then said to be ‘associated’ with the disease of interest. To account for multiple testing, the P value for statistical significance is usually set at 5 × 10−8. Of note, a statistically significant SNP is not necessarily causal, but may be in linkage disequilibrium with the causal SNP. Establishing causality typically requires bioinformatics study of the locus of interest, additional genotyping, and functional assays in vitro or in experimental animal models.26
Over the past few years, GWAS have generated hundreds of replicable SNP associations for complex traits and diseases, including at least 35 loci influencing various atherosclerotic vascular diseases and related intermediate traits.25 Loci implicated in susceptibility to CHD and replicated in different populations are listed in Table 1. The susceptibility variants that have been uncovered increase the risk modestly, typically by 10–40% per risk allele. Notably, however, many of the alleles associated with increased risk are frequently found in the population, so the population-attributable risk is often substantial.27 Many more such loci associated with weak effects on risk will probably be identified by GWAS of large sample sizes and also as a result of meta-analyses that combine results of several studies. The discovery of new, robust and replicable CHD-associated SNPs in GWAS might confer clinical value by facilitating multiplex testing of these SNPs in an individual, thereby yielding predictive information that is incremental to assessment of conventional risk factors.
Table 1.
Replicated single-nucleotide polymorphisms associated with CHD that have been identified in genome-wide association studies
| SNP | Chr | Gene(s) in that location | Function of gene product(s) | RAF | OR | Initial report | Replication studies |
|---|---|---|---|---|---|---|---|
| rs10757278 | 9p21.3 | Cyclin-dependent kinase inhibitor 2A/2B (CDKN2A,2B) | Regulation of cell cycle | 0.45 | 1.28 | Helgadottir, A. et al. (2007)72 | Samani, N. J. et al. (2007);27 Wellcome Trust Case Control Consortium (2007);73 Coronary Artery Disease Consortium (2008);74 Kathiresan, S. et al. (2009)75 |
| rs599839 | 1p13.3 | Cadherin, EGF LAG seven-pass G-type receptor 2 (CELSR2) | Involved in contact-mediated communication | 0.78 | 1.20 | Willer, C. J. et al. (2008)76 | Samani, N. J. et al. (2007);27 Wellcome Trust Case Control Consortium (2007);73 Coronary Artery Disease Consortium (2008);74 Kathiresan, S. et al. (2009)75 |
| Proline/serine-rich coiled-coil 1 (PSRC1) | May participate in p53-mediated growth suppression | ||||||
| Sortilin 1 (SORT1) | An essential player in adipocyte and muscle glucose metabolism | ||||||
| rs3008621 | 1q41 | Melanoma inhibitory activity family, member 3 (MIA3) | Regulates cell adhesion and migration | 0.26 | 1.10 | Samani, N. J. et al. (2007)27 | Coronary Artery Disease Consortium (2008);74 Kathiresan, S. et al. (2009)75 |
| rs501120 | 10q11.2 | Unknown | Unknown | 0.16 | 1.33 | Samani, N. J. et al. (2007)27 | Coronary Artery Disease Consortium (2008)74 |
| rs9818870 | 3q22.3 | Muscle RAS oncogene homolog (MRAS) | Responsible for a variety of normal cellular functions | 0.15 | 1.15 | Erdmann, J. et al. (2009)77 | Erdmann, J. et al. (2009)77 |
| rs3184504 | 12q24 | SH2B adaptor protein 3 (SH2B3) | Mediates the interaction between the extracellular receptors and intracellular signaling pathway | 0.39 | 1.13 | Gudbjartsson, D. F. et al. (2009)78 | Gudbjartsson, D. F. et al. (2009)78 |
| rs9982601 | 21q22 | Solute carrier family 5, member 3 (SLC5A3) | Permits sodium ion transport | 0.13 | 1.20 | Kathiresan, S. et al. (2009)75 | Kathiresan, S. et al. (2009)75 |
| Mitochondrial ribosomal protein S6 (MRPS6) | Involved in protein synthesis within the mitochondrion | ||||||
| Potassium voltage- gated channel, Isk-related family, member 2 (KCNE2) | Regulates neurotransmitter release, heart rate, insulin secretion, neuronal excitability, epithelial electrolyte transport, smooth muscle contraction, and cell volume | ||||||
| rs12526453 | 6p24 | Phosphatase and actin regulator 1 (PHACTR1) | Inhibits phosphoprotein phosphatase inhibitor | 0.65 | 1.12 | Kathiresan, S. et al. (2009)75 | Kathiresan, S. et al. (2009)75 |
| rs6725887 | 2q33 | WD repeat domain 12 (WDR12) | Affects maturation of the large ribosomal subunit | 0.14 | 1.17 | Kathiresan, S. et al. (2009)75 | Kathiresan, S. et al. (2009)75 |
| rs1122608 | 19p13 | Low-density lipoprotein receptor (LDLR) | Involved in receptor-mediated endocytosis of specific ligands | 0.75 | 1.15 | Kathiresan, S. et al. (2009)75 | Kathiresan, S. et al. (2009)75 |
| rs11206510 | 1p32 | Proprotein convertase subtilisin/kexin type 9 (PCSK9) | Involved in cholesterol homeostasis | 0.81 | 1.15 | Kathiresan, S. et al. (2009)75 | Kathiresan, S. et al. (2009)75 |
| rs1746048 | 10q11 | Chemokine (C-X-C motif) ligand 12 (CXCL12) | Activates leukocytes | 0.84 | 1.17 | Samani, N. J. et al. (2007)27 | Kathiresan, S. et al. (2009)75 |
Technology for SNP genotyping
Over the past several years, substantial advances have taken place in genotyping technology, including SNP ‘arrays’ that can allow assays of up to 1 million SNPs. Detection of SNPs is by means of specific oligonucleotide probes (25–50 base pairs in length), which are placed either on beads 2 μm in diameter or ligated directly on to glass slides (SNP array technology is reviewed in detail elsewhere28). In addition to rapid technological advances, the cost per genotype has gone down dramatically. These developments have enabled the widespread use of GWAS and the identification of many new genetic variants associated with common diseases. Custom-designed SNP assays can be utilized for follow-up replication and validation studies.
In addition to sequence variation, structural variation contributes substantially to genetic variation in humans. Copy-number variants—defined as fragments of the genome that are larger than 1 kb and vary in copy number between individuals—are being investigated for their contribution to common diseases including cardiovascular diseases.29 To date, few copy-number variants implicated in susceptibility to atherosclerosis have been identified; however, one example is variation in the region on chromosome 6 that harbors the LPA gene.30 The current SNP genotyping arrays incorporate assays for copy-number variants, and new analysis algorithms are being tested to infer copy-number variants from SNP genotyping data.
Sequencing
In the near future, high-density genotyping and candidate-gene multiplex-SNP testing might both be replaced by sequencing of entire genomes or ‘exomes’. The milestone of the $1,000 genome, once thought improbable, now seems to be likely in the not-too-distant future. Sequencing of candidate genes, exons, or whole genomes will allow identification of ‘rare’ susceptibility variants that may have stronger effects on disease susceptibility.
Predictive utility of genetic markers
Because of small relative risks and odds ratios, most genetic susceptibility variants have low discriminative accuracy and contribute only marginally to the c-statistic, compared with existing risk factors, such as presence of diabetes or history of smoking.31–33 The predictive value of genetic profiling for CHD and related traits has been investigated in several studies. Talmud et al. found that a chromosome 9p21.3 SNP (rs10757274) did not add substantially to the Framingham risk score for predicting CHD events in 2,742 men, but led to reclassification of risk in 22%, of whom 63% moved into more-accurate categories (defined by the observed risk corresponding better to the predicted risk in the new category).34 Two studies published in 2009 also investigated whether the addition of the chromosome 9p21 allele to conventional risk factors improved CHD risk prediction; these studies yielded conflicting results.35,36
The effect of susceptibility genetic variants may vary in different environmental backgrounds. The study of gene–environment interactions in the setting of GWAS is important to improve understanding of disease pathways, facilitate individualized medicine to maximize response or minimize adverse effects on the basis of genetic susceptibility, and improve prediction of an individual’s risk of disease or prognosis, and of potential changes in risk in relation to modifiable environmental factors.37 Improved methods to quantify environmental exposures and large sample sizes are needed to accelerate progress in this area.
Clinical utility of genetic markers
Several web-based companies have already started to market genotyping of disease-susceptibility variants to the public.38 Estimation of CHD risk is a prominent feature of the reports given to the consumers after genotyping. Given the proliferation of direct-to-consumer genetic testing, there is a need to study the integration of predictive genetic risk assessment in clinical practice.39,40 Furthermore, although genetic testing may improve accuracy of risk profiles and thereby aid in early detection of CHD, whether genetic testing will improve outcomes remains to be established. Box 1 details important points to consider when evaluating the utility of genetic testing, as proposed by Haddow and Palomaki in 2004.41 Several aspects require further investigation, including whether genetic testing could lead to potential harm (for example, by causing undue anxiety), how physicians should deal with predictive genetic information, how to communicate risk to patients such that they can make sense of it, and the effect of communication of genetic risk on patient behavior and lifestyle.
Box 1. The ACCE41 criteria for genetic testing.
Analytical validity
Ability to measure the genotype of interest both accurately and reliably.
Clinical validity
Ability of a genetic test to detect or predict the presence or absence of the phenotype or clinical disease.
Clinical utility
The likelihood that the genetic test will lead to an improved outcome, including test and disease characteristics.
Ethical, legal, and social implications
Impact of genetic test results on insurance and employment, privacy and confidentiality, equity of access, and stigmatization.
Use of proteomics for risk assessment
As for genetic variation, the expectation that a single protein marker will provide significant incremental information that will be useful for early detection of cardiovascular disease is unrealistic.13,42 Given the complexity of atherosclerotic vascular disease, a multimarker approach is likely to be more informative than use of a single marker because it might help detect perturbations in one or more of the various etiologic pathways of atherosclerosis.13,43 One caveat to the multimarker approach is that if the candidate biomarkers derive from pathways that are correlated with clinical characteristics that are already being measured (for example, lipid levels or inflammatory markers), their incremental predictive value is limited. The failure of correlated biomarkers to provide incremental predictive value was illustrated by Pepe and colleagues.44
Proteomic technology
To identify new biomarkers that are not correlated with the known candidate markers and are associated with risk of CHD, an unbiased approach is needed. The utility of an agnostic approach—which assumes no prior knowledge of disease pathophysiology—has been highlighted by the often unexpected genetic susceptibility variants identified by GWAS. The results of these studies also highlight that our knowledge of disease pathophysiology is far from complete.
Proteomic technologies for biomarker discovery
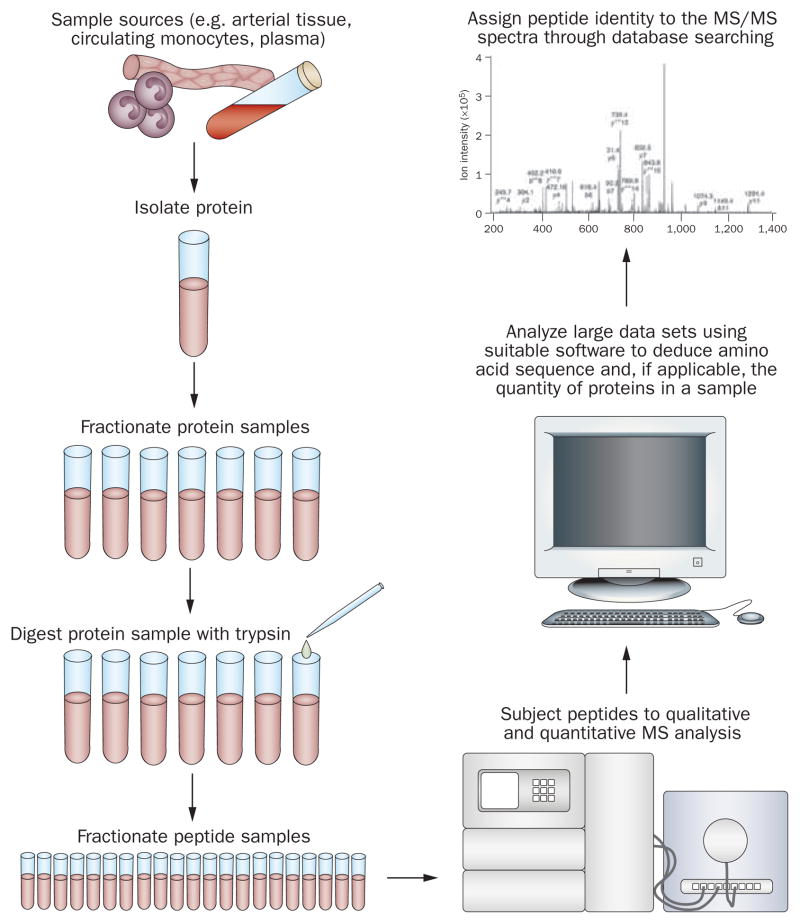
The principal technology of proteomic discovery is mass spectrometry (MS) (Figure 2). In what is often termed ‘shotgun proteomics’, proteins in a particular sample are initially digested by trypsin into their constituent peptides and then separated by high-performance liquid chromatography. The sequences of these peptides are then established by tandem MS (MS/MS) experiments in which parent ions are fragmented by collision with nitrogen or helium gas to produce fragment ions. The resulting product ion mass spectra plot the mass-to-charge ratio of the ions observed versus detected ion abundance.45 Software tools can then be used to deduce the amino-acid sequence and, if applicable, the quantity of the proteins in a sample and the peptide identity to the MS/MS spectra, through database searching.46 MS can also be useful in the triage and validation of novel proteins found in discovery efforts. Sensitivity can be increased if MS is coupled with immunogenic extraction and enrichment methods to increase low concentration target selection.47,48 To date, no specific markers for clinical use in CHD risk stratification have emerged from MS studies.
Figure 2.
Flow diagram illustrating the use of MS to identify new protein biomarkers. Abbreviation: MS, mass spectrometry. Tandem MS image reproduced with permission from Nature Publishing Group © Hodges, A. et al. Nat. Immunol. 8, 569–577 (2007).
Additional technology platforms that incorporate MS for proteomic biomarker discovery include pattern-based methods that produce MS-derived protein patterns via surface-enhanced laser desorption–ionization (SELDI) or matrix-assisted laser desorption–ionization (MALDI). Further details of these technologies are beyond the scope of the present article and are provided in reviews published elsewhere.45,46,48 A substantial disadvantage of these approaches is that once a differential pattern is detected, identification of the peaks in the pattern is often challenging.49 Without knowing the identity of the candidate peaks, it is difficult to gain insight into the disease pathophysiology and to move to higher-throughput, widely disseminated, clinical assay platforms, which are typically immunoassays (see the section on assay standardization below). Furthermore, pattern-only approaches face considerable challenges when used as diagnostic tests in the clinical setting.10
Pattern-based and identity-based methods can be combined, as in MS/MS analysis of selected spots from differential protein displays, such as two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). The limitations of this approach include limited sensitivity, reproducibility and throughput; these limitations are ameliorated to a certain degree by the method of upfront liquid-chromatography followed by MS/MS.
Challenges for discovery proteomics
Although proteomic technologies have the potential to identify novel proteins in the plasma that can improve accuracy of cardiovascular risk prediction, many challenges exist. Some of the main technical challenges for discovery proteomics are associated with depleting abundant proteins in the plasma to obtain the less-abundant proteins (the ‘deep proteome’). Another issue is that, by contrast to only ~25,000 genes in the human genome, hundreds of thousands of unique proteins circulate in the blood, owing to alternative splicing of genes leading to different transcripts from the same gene, as well as to post-translational modification of proteins. The latter includes glycosylation, which is particularly relevant to atherosclerosis given the role of advanced glycation end-products in increasing oxidative stress in the vascular wall.50
Unlike the GWAS agnostic approach, which is a productive method for identification of genetic variants that influence susceptibility to common chronic diseases, an agnostic proteomics approach has not yet had much success. Advances in proteomic technology, standardizing the collection and storage of specimens, preventing degradation during storage, and using well-defined cases and controls will increase the likelihood of success in identifying novel markers that may help in disease prediction and prognostication.46,51
Assay standardization for routine clinical use
Considerable work is required for biomarkers to be successfully transitioned from proteomic discovery to clinical use for risk prediction.52 Most research-based protein marker assays are not validated according to Clinical Laboratory Improvement Amendments (CLIA) standards. Before clinical use in the USA, these assays must meet CLIA requirements53 in terms of accuracy, precision, analytical sensitivity, analytical specificity, reportable range of test results, and reference intervals. Furthermore, these procedures must be clinically validated using samples from patients with well-characterized clinical profiles. This translation process demands an established infrastructure and multidisciplinary expertise, particularly for assays that provide multiple, parallel protein measurements on the same specimen (‘multiplex assays’).49,54–56 Extensive validation is required for multiplex protein test panels intended for use in clinical trials or diagnostic laboratories.57 To date, FDA-cleared protein multiplex assays consist primarily of the lateral-flow immunoassays used for point-of-care evaluation.58 No multiplex assays are currently available for CHD risk prediction in asymptomatic individuals.
Predictive utility of biomarkers
Several studies have tested whether multiple markers increase the ability to predict adverse cardiovascular outcomes. Wang and colleagues evaluated 10 biomarkers of cardiovascular disease in over 3,000 study participants who were followed for development of cardiovascular disease for a median follow-up of 7.4 years.59 Of the 10 biomarkers measured, many were significant predictors of cardiovascular events (B-type natriuretic peptide and urinary albumin excretion) and mortality (C-reactive protein, B-type natriuretic peptide, urinary albumin excretion, renin, and homocysteine). Risk prediction was improved by combining biomarkers into a multimarker score; however, combining the biomarkers yielded only a modest increment in the c-statistic. By contrast, a study of elderly men in Sweden revealed that four biomarkers—N-terminal pro-B-type natriuretic peptide (NT-proBNP), C-reactive protein, cystatin, and troponin—led to significant improvement in the c-statistic for predicting myocardial infarction and death during a median follow-up of 10 years.60
Evaluation of diagnostic or predictive tests uses the c-statistic as a measure of the test’s ability to discriminate individuals with disease from those without disease.61 Distributions of biomarkers in individuals with and without cardiovascular disease typically overlap a great deal, which provides one explanation for the modest increase in the c-statistic seen in most clinical bio-marker studies of cardiovascular risk.62 Because it is difficult to demonstrate improvements in the c-statistic, some investigators have advocated use of other metrics (described below) to evaluate the predictive utility of new biomarkers.63,64
The net reclassification index refers to the proportion of persons who change risk categories when prediction models incorporate new biomarkers.63 Reclassification refers to the ability of new biomarkers to move people between discrete risk categories, so that some low-risk individuals may be reclassified as high-risk and vice versa. For CHD, reclassification is meaningful and clinically relevant, given the widespread use of low-risk, intermediate-risk, and high-risk categories of 10-year CHD risk. Movement between risk categories might not be clinically relevant if there is no management strategy explicitly linked to the categorization, or if most of the movements involve small shifts in absolute risk from just below to just above the cutoff point.65 However, if risk categories are defined according to cutoff points used to indicate type or intensity of interventions, as is true for CHD, reclassification can impact on clinical management.
Integrated discrimination improvement is another method of assessing the discriminative value of a bio-marker beyond known risk factors,64 and has been used in the setting of CHD.37 This method measures the difference in discriminative ability between two models according to their predicted survival probabilities. Comparing one model to another, an increased probability of an outcome among individuals who eventually did have the outcome, and a decreased probability of an outcome among subjects who did not have the outcome implies better predictive ability, whereas the opposite implies worse predictive ability. These two changes are summed to give the integrated discrimination improvement (with improvement always being considered positive).
Clinical utility of biomarkers
Clinical utility refers to the likelihood that measurement of a biomarker will lead to an improved outcome.41 To determine clinical utility of a new biomarker, several features in addition to the ability of the marker to discriminate between individuals who will or will not develop cardiovascular disease need to be evaluated. These considerations include the potential added benefits (including prevention of adverse cardiovascular outcomes and reduction of health-related costs) and harms (for example, anxiety, excessive downstream testing and unwarranted pharmacotherapy) of using the new biomarker. Although this concept remains to be proven, it is hoped that measurement of new biomarkers will promote cardiovascular health by enabling early disease detection and management.66 The availability of clinically useful biomarkers for the evaluation of cardiovascular risk might enable the health-care system to become more proactive, moving the focus away from treatment of end-stage disease and towards early detection of disease risk and prevention of adverse outcomes.
Conclusions
Atherosclerosis is a complex multifactorial disease process involving multiple pathways that are influenced by genetic and environmental factors.14 At the population level, efforts to reduce the prevalence of conventional risk factors, and to treat these factors when present, are enormously important for reduction of the burden of cardiovascular disease. However, to refine cardiovascular risk assessment in an individual, and thereby promote cardiovascular health, further work is needed to elucidate the role of the newer genomic and proteomic markers. Ideally, a new marker should add to the accuracy of risk prediction beyond the traditional risk factors, have a standardized and reproducible assay with established cutoff points to guide interpretation of the results, and have a therapeutic intervention available that leads to a reduction in the incidence of cardiovascular events.67–69
An important component of individualized medicine—a concept currently surrounded by a great deal of interest—is knowledge of which risk factors are increased in the individual. With advances in genomics and proteomics, robust, individualized risk stratification and targeted therapy may become feasible in the near future. Many risk factors with small-to-moderate effects are likely to be identified as a result of advances in the genomic and proteomic sciences,70 and each individual could potentially be assessed with a panel of tests for genomic and proteomic markers. With a multimarker approach, it may also be possible to identify alterations in specific pathways of atherogenesis, rather than focus on individual risk factors; this approach might aid in provision of the best preventative and therapeutic interventions for each individual, which would restore the specific perturbed pathways to their normal function.
Key points.
- Conventional risk factors for atherosclerosis are prevalent in much of the general population and, therefore, available risk-prediction algorithms based on such risk factors have less than desired accuracy
- New genomic and proteomic markers are likely to have an important role in refining cardiovascular risk assessment, thereby permitting individualized risk stratification and targeted therapy
- Genome-wide association studies have identified variants that have modest effects on disease propensity; sequencing of entire genomes, or parts thereof, might allow identification of rare variants with stronger effects
- Proteomic discovery technology is improving and holds promise for identifying new biomarkers that will refine cardiovascular risk stratification
- In the future, individuals could potentially be assessed with a panel of tests for the many new genomic and proteomic markers that each have small-to-moderate effects
- A multimarker approach might also enable us to identify alterations in specific pathways of atherogenesis as a means to assess cardiovascular risk, rather than assessing risk by focusing on individual risk factors
Review criteria.
Database searched included PubMed and MEDLINE, for articles published from 1995 to present, using the search terms “genomics”, “proteomics”, “genome-wide association studies”, “biomarker discovery”, and “mass spectrometry”. Only articles published in English were considered.
Acknowledgments
This work was supported in part by grants HL81331 and HG04599 from the NIH, USA, and a generous gift from the Marriot family.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Murray CJL, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: global burden of disease study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 2.Yusuf S, Reddy S, Ounpuu S, Anand S. Global burden of cardiovascular diseases: Part II: variations in cardiovascular disease by specific ethnic groups and geographic regions and prevention strategies. Circulation. 2001;104:2855–2864. doi: 10.1161/hc4701.099488. [DOI] [PubMed] [Google Scholar]
- 3.Executive Summary of the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 4.Wilson PW, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 5.Kullo IJ, Ballantyne CM. Conditional risk factors for atherosclerosis. Mayo Clin Proc. 2005;80:219–230. doi: 10.4065/80.2.219. [DOI] [PubMed] [Google Scholar]
- 6.Cooper JA, Miller GJ, Humphries SE. A comparison of the PROCAM and Framingham point-scoring systems for estimation of individual risk of coronary heart disease in the Second Northwick Park Heart Study. Atherosclerosis. 2005;181:93–100. doi: 10.1016/j.atherosclerosis.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Wald NJ, Hackshaw AK, Frost CD. When can a risk factor be used as a worthwhile screening test? BMJ. 1999;319:1562–1565. doi: 10.1136/bmj.319.7224.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuster V, Lois F, Franco M. Early identification of atherosclerotic disease by noninvasive imaging. Nat Rev Cardiol. doi: 10.1038/nrcardio.2010.54. [DOI] [PubMed] [Google Scholar]
- 9.Mayr M. Metabolomics: ready for the prime time? Circ Cardiovasc Genet. 2008;1:58–65. doi: 10.1161/CIRCGENETICS.108.808329. [DOI] [PubMed] [Google Scholar]
- 10.Morrison AC, et al. Prediction of coronary heart disease risk using a genetic risk score: The Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 11.Khoury MJ, Jones K, Grosse SD. Quantifying the health benefits of genetic tests: the importance of a population perspective. Genet Med. 2006;8:191–195. doi: 10.1097/01.gim.0000206278.37405.25. [DOI] [PubMed] [Google Scholar]
- 12.Cortese DA. A vision of individualized medicine in the context of global health. Clin Pharmacol Ther. 2007;82:491–493. doi: 10.1038/sj.clpt.6100390. [DOI] [PubMed] [Google Scholar]
- 13.Kim CX, et al. Sex and ethnic differences in 47 candidate proteomic markers of cardiovascular disease: The Mayo Clinic Proteomic Markers of Arteriosclerosis Study. PLoS ONE. 2010;5:e9065. doi: 10.1371/journal.pone.0009065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttmacher AE, Collins FS, Carmona RH. The family history—more important than ever. N Engl J Med. 2004;351:2333–2336. doi: 10.1056/NEJMsb042979. [DOI] [PubMed] [Google Scholar]
- 16.Scheuner MT. Family history: where to go from here. Genet Med. 2003;5:66–68. doi: 10.1097/01.GIM.0000056980.14050.88. [DOI] [PubMed] [Google Scholar]
- 17.Kullo IJ, Ding K. Mechanisms of disease: The genetic basis of coronary heart disease. Nat Clin Pract Cardiovasc Med. 2007;4:558–569. doi: 10.1038/ncpcardio0982. [DOI] [PubMed] [Google Scholar]
- 18.Williams RR, et al. Usefulness of cardiovascular family history data for population-based preventive medicine and medical research (the Health Family Tree Study and the NHLBI Family Heart Study) Am J Cardiol. 2001;87:129–135. doi: 10.1016/s0002-9149(00)01303-5. [DOI] [PubMed] [Google Scholar]
- 19.Murabito JM, et al. Sibling cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults. JAMA. 2005;294:3117–3123. doi: 10.1001/jama.294.24.3117. [DOI] [PubMed] [Google Scholar]
- 20.Utermann G, et al. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604–607. doi: 10.1038/269604a0. [DOI] [PubMed] [Google Scholar]
- 21.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 22.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in pcsk9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 23.Kullo IJ, et al. Association of polymorphisms in NOS3 with the ankle-brachial index in hypertensive adults. Atherosclerosis. 2008;196:905–912. doi: 10.1016/j.atherosclerosis.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding K, Kullo IJ. Genome-wide association studies for atherosclerotic vascular disease and its risk factors. Circ Cardiovasc Genet. 2009;2:63–72. doi: 10.1161/CIRCGENETICS.108.816751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visel A, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragoussis J. Genotyping technologies for genetic research. Annu Rev Genomics Hum Genet. 2009;10:117–133. doi: 10.1146/annurev-genom-082908-150116. [DOI] [PubMed] [Google Scholar]
- 29.Pollex RL, Hegele RA. Copy number variation in the human genome and its implications for cardiovascular disease. Circulation. 2007;115:3130–3138. doi: 10.1161/CIRCULATIONAHA.106.677591. [DOI] [PubMed] [Google Scholar]
- 30.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Agostino RB, Sr, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 32.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100:1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssens AC, van Duijn CM. Genome-based prediction of common diseases: advances and prospects. Hum Mol Genet. 2008;17:R166–R173. doi: 10.1093/hmg/ddn250. [DOI] [PubMed] [Google Scholar]
- 34.Talmud PJ, et al. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467–474. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 35.Brautbar A, et al. Impact of adding a single allele in the 9p21 locus to traditional risk factors on risk classification for coronary heart disease and implications for lipid-modifying therapy in the white population of the Atherosclerosis Risk in Communities (ARIC) study. Circ Cardiovasc Genet. 2009;2:279–285. doi: 10.1161/CIRCGENETICS.108.817338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paynter NP, et al. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle—will we get our wish? N Engl J Med. 2008;358:105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 39.Khoury MJ, et al. The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009;11:559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weston AD, Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 42.Haddow J, Palomaki G. ACCE: A model process for evaluating data on emerging genetic tests. Oxford University Press; New York: 2004. [Google Scholar]
- 43.Khawaja FJ, et al. Association of novel risk factors with the ankle brachial index in African American and non-Hispanic white populations. Mayo Clin Proc. 2007;82:709–716. doi: 10.4065/82.6.709. [DOI] [PubMed] [Google Scholar]
- 44.Pepe MS. An interpretation for the ROC curve and inference using GLM procedures. Biometrics. 2000;56:352–359. doi: 10.1111/j.0006-341x.2000.00352.x. [DOI] [PubMed] [Google Scholar]
- 45.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 46.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 47.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti- Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 48.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling MM, Ricks C, Lea P. Multiplexing molecular diagnostics and immunoassays using emerging microarray technologies. Expert Rev Mol Diagn. 2007;7:87–98. doi: 10.1586/14737159.7.1.87. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt AM, Yan SD, Wautier JL, Stern D. Activation of receptor for advanced glycation end products: a mechanism for chronic vascular dysfunction in diabetic vasculopathy and atherosclerosis. Circ Res. 1999;84:489–497. doi: 10.1161/01.res.84.5.489. [DOI] [PubMed] [Google Scholar]
- 51.Cooper LT, Jr, et al. Genomic and proteomic analysis of myocarditis and dilated cardiomyopathy. Heart Fail Clin. 2010;6:75–85. doi: 10.1016/j.hfc.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 52.Granger CB, Van Eyk JE, Mockrin SC, Anderson NL. National Heart, Lung, and Blood Institute Clinical Proteomics Working Group Report. Circulation. 2004;109:1697–1703. doi: 10.1161/01.CIR.0000121563.47232.2A. [DOI] [PubMed] [Google Scholar]
- 53.Department of Health and Human Services, Centers for Disease Control and Prevention. Current CLIA Regulations (including all changes through 01/24/2004) 2004 [online], http://wwwn.cdc.gov/clia/regs/toc.aspx.
- 54.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2009;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Measurement and quality control issues in multiplex protein assays: a case study. Clin Chem. 2009;55:1092–1099. doi: 10.1373/clinchem.2008.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.US Food and Drugs Administration. Draft guidance for industry and FDA staff—pharmacogenetic tests and genetic tests for heritable markers. 2006 [online], http://www.fda.gov/cdrh/oivd/guidance/1549.html.
- 58.Rosen S. In: Lateral Flow Immunoassay. Wong RC, Tse HY, editors. Humana Press; New York: 2009. pp. 35–49. [Google Scholar]
- 59.Wang TJ, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- 60.Zethelius B, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 61.Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem. 2008;54:17–23. doi: 10.1373/clinchem.2007.096529. [DOI] [PubMed] [Google Scholar]
- 62.Ware JH. The limitations of risk factors as prognostic tools. N Engl J Med. 2006;355:2615–2617. doi: 10.1056/NEJMp068249. [DOI] [PubMed] [Google Scholar]
- 63.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 64.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 65.Wang TJ. New cardiovascular risk factors exist, but are they clinically useful? Eur Heart J. 2008;29:441–444. doi: 10.1093/eurheartj/ehm644. [DOI] [PubMed] [Google Scholar]
- 66.Prainsack B, et al. Personal genomes: Misdirected precaution. Nature. 2008;456:34–35. doi: 10.1038/456034a. [DOI] [PubMed] [Google Scholar]
- 67.Mosca L. C-reactive protein—to screen or not to screen? N Engl J Med. 2002;347:1615–1617. doi: 10.1056/NEJMe020127. [DOI] [PubMed] [Google Scholar]
- 68.Hackam DG, Anand SS. Emerging risk factors for atherosclerotic vascular disease: a critical review of the evidence. JAMA. 2003;290:932–940. doi: 10.1001/jama.290.7.932. [DOI] [PubMed] [Google Scholar]
- 69.Manolio T. Novel risk markers and clinical practice. N Engl J Med. 2003;349:1587–1589. doi: 10.1056/NEJMp038136. [DOI] [PubMed] [Google Scholar]
- 70.Braunwald E. The Simon Dack lecture. Cardiology: the past, the present, and the future. J Am Coll Cardiol. 2003;42:2031–2041. doi: 10.1016/j.jacc.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 71.National Center for Biotechnology Information. The database of Genotypes and Phenotypes (dbGaP) 2010 [online], http://www.ncbi.nlm.nih.gov/gap.
- 72.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 73.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coronary Artery Disease Consortium et al. Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscler Thromb Vasc Biol. 2009;29:774–780. doi: 10.1161/ATVBAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erdmann J, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]