Crystal Structure of the Human Histone Methyltransferase ASH1L Catalytic Domain and Its Implications for the Regulatory Mechanism (original) (raw)
Abstract
Absent, small, or homeotic disc1 (Ash1) is a trithorax group histone methyltransferase that is involved in gene activation. Although there are many known histone methyltransferases, their regulatory mechanisms are poorly understood. Here, we present the crystal structure of the human ASH1L catalytic domain, showing its substrate binding pocket blocked by a loop from the post-SET domain. In this configuration, the loop limits substrate access to the active site. Mutagenesis of the loop stimulates ASH1L histone methyltransferase activity, suggesting that ASH1L activity may be regulated through the loop from the post-SET domain. In addition, we show that human ASH1L specifically methylates histone H3 Lys-36. Our data implicate that there may be a regulatory mechanism of ASH1L histone methyltransferases.
Keywords: Chromatin Histone Modification, Crystal Structure, Gene Regulation, Histone Methylation, Protein Structure
Introduction
Nucleosomes, the fundamental unit of the highly ordered chromatin structure, are composed of DNA wrapped around histone octamers. Histones have N-terminal tails that are exposed on the outside of nucleosomes. These tails are subjected to several post-translational modifications, including acetylation, phosphorylation, ubiquitination, sumoylation, and methylation (1).
The site-specific methylation of histone lysine residues is important for the epigenetic control of gene expression. These marks serve to regulate epigenetically the organization of chromatin structure and to recruit other chromatin modifiers (2, 3). Methylation can occur at multiple lysine residues, including lysines 4, 9, 27, 36, and 79 of histone H3 and lysine 20 of histone H4.
Absent, small, or homeotic disc1 (Ash1) is a member of the trithorax group proteins, which are essential for epigenetic gene activation (4, 5). Previous studies have shown that Drosophila Ash1 activates homeotic gene ultrabithorax expression in imaginal discs of the third leg (6) and interacts with trithorax to regulate and maintain ultrabithorax expression (7). It has also been reported that the mammalian homolog of Ash1, ASH1L, is a histone methyltransferase (HMTase)2 that is associated with transcribed regions of active genes (8, 9). ASH1L has several domains, including an associated with SET domain (AWS), a SET domain, a post-SET domain, a bromodomain, a bromo-adjacent homology domain (BAH), and a plant homeodomain finger (SMART database (10)). The SET domain in HMTases is responsible for catalyzing the formation of monomethylated, dimethylated, and trimethylated lysine, establishing an additional complex system with respect to methylated lysine recognition in signaling (11). Several crystal structures of SET domain proteins have been solved, and they revealed that the SET domain forms a knot-like structure that constitutes the active site of HMTases (12). Notably, ASH1L contains a SET domain in the middle of the protein, whereas other proteins possess a SET domain at the C terminus. Drosophila Ash1 has been previously shown to affect H3K4 methylation levels genetically and to methylate H3K4, H3K9, and H4K20 in in vitro assays (8, 13). The Tanaka group (14) has found H3K36 to be the enzymatic target of the human ASH1L (hASH1L)-SET domain, whereas another group has demonstrated that hASHlL methylates H3K4 (9).
To elucidate the molecular function of mammalian ASH1L, we determined the crystal structure of the hASH1L catalytic domain, including AWS, SET, and post-SET domains, bound to _S_-adenosylmethionine (AdoMet). Surprisingly, the crystal structure shows that the substrate binding pocket is blocked by a loop in the post-SET domain. Further mutagenesis studies and biochemical analyses suggest that the loop regulates the HMTase activity of hASH1L.
EXPERIMENTAL PROCEDURES
Protein Purification
The catalytic domains of hASH1L (NM_018489.2) and its mutants (2069–2260 amino acids) were expressed in Escherichia coli BL21 (DE3) cells. Transformed cells were lysed in buffer A containing 50 mm Tris-HCl (pH 8.0), 250 mm NaCl, 5% glycerol, and 10 mm ZnCl2. Cell debris was cleared by centrifugation, and the nucleic acid was cleared with polyethylenimine precipitation (15). The cleared cell lysates were incubated with nickel-nitrilotriacetic acid beads (Qiagen) in the presence of 20 mm imidazole for 4 h. The nickel-nitrilotriacetic acid beads were collected and washed with buffer A. hASH1L-SET was eluted with buffer A containing 100 mm imidazole, and the N-terminal His tag of hASH1L-SET was removed with the tobacco etch virus protease. hASH1L-SET was further purified with HiTrap Q anion exchange and HiTrap S cation exchange (GE Healthcare) chromatography. The protein was finally purified with a Superdex 75 (GE Healthcare) size exclusion column and concentrated up to 14 mg/ml in a buffer of 50 mm Tris-HCl (pH 8.0) and 100 mm NaCl. Mutants were generated with the QuikChange site-directed mutagenesis kit (Qiagen) and purified in the same way as the wild-type protein.
Crystallization and Structure Determination
Crystals of the hASH1L catalytic domain were obtained using the hanging drop vapor method at 18 °C in 70 mm Bis-Tris (pH 7.5), 30 mm citric acid, 20% polyethylene glycol 3350, and 10 mm spermine tetrahydrochloride. For cryoprotection, crystals were soaked for 1 min in a crystallization solution containing 45% PEG 3350. All data were collected under cryogenic conditions (105 K) at a beamline 4A-HFMX at the Pohang Accelerator Laboratory (PAL, Pohang, Korea). Data were processed with HKL2000. Phases were calculated from the single anomalous dispersion method at the zinc peak using the program SOLVE/RESOLVE (16). Models were built using the program O, and crystallographic refinement was performed with the program CNS.
Methyltransferase Assay
For the in vitro methyltransferase assay, hASH1L proteins (3.5 μm) were incubated in a reaction mixture containing mononucleosomes, a G5E4 nucleosomal array, or histone octamers (17) and 2 μm _S_-adenosyl-l-[_methyl_-3H]methionine (15 Ci/mmol, PerkinElmer Life Science NET155) for 30 min at 25 °C. Samples were then separated by 15% SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane. The membrane was applied to FLA-5000 (Fuji Film) for radioactive imaging. The membrane was stained with Ponceau S (Sigma) and scanned with LAS-3000 (Fuji Film) for protein quantification. The intensities of the detected bands were compared with Multi-Gauge (version 3.2). Histone purification and nucleosome reconstitution were carried out as described previously (18). H3K4A and H3K36A mutants were purified and reconstituted into nucleosomes in the same way as the wild-type histone H3. Antibodies against the H3K4me2, H3K36me2, or H3K36me3 (Upstate Biotech Millipore) were used to detect histone methylation. The intensities of the detected bands were scanned and compared using LAS-3000 (Fuji Film) with Multi-Gauge (version 3.2). Recombinant Xenopus histones or HeLa histones were used for the HMTase assay.
RESULTS
Crystal Structure of the hASH1L Catalytic Domain
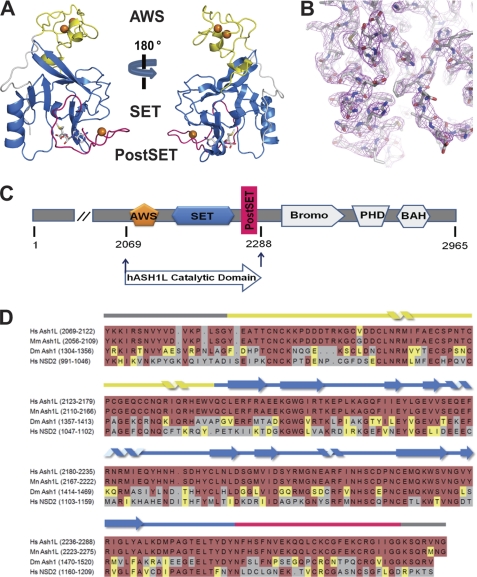
We determined a crystal structure of the hASH1L catalytic domain bound to AdoMet, including the AWS, SET, and post-SET domains (2069–2288 amino acids, Fig. 1, C and D) to 2.9 Å resolution using the zinc single anomalous dispersion method (Table 1, Fig. 1B). The structure of hASH1L shows that the N and C termini of the AWS domain are interwoven with the SET domain (Fig. 1A). The AWS domain is composed of mostly loops and two short α-helices. These loops are bound by two zinc atoms and coordinated by eight conserved cysteines in the AWS domain. The fact that the AWS domain in one of the two molecules of the asymmetric unit is partially disordered and that the temperature factor of the AWS domain is high suggests that the AWS domain is highly flexible. The structure of the hASH1L SET domain is similar to those of other known SET domain proteins.
FIGURE 1.
Crystal structure of the human histone methyltransferase ASH1L catalytic domain. A, overall structure of the Ash1L-SET catalytic domain. The protein is colored according to panel C. The three zinc ions are shown in orange. The AWS domain shown in yellow is composed of mostly loops, with two short helices interwoven into the SET domain. A loop (shown in magenta) from the post-SET domain is blocking the substrate binding pocket. B, 2_Fo_ − Fc electron density map at 1 σ around the auto-inhibitory loop is shown in magenta. C, schematic representation of the full-length ASH1L and the construct containing the catalytic domain used in this current study (residues 2069–2288). The AWS domain is in yellow, the SET domain is in marine blue, and the post-SET domain is in magenta. BAH, bromo-adjacent homology domain. D, sequence alignment of the catalytic domains of human ASH1L (2069–2288), mouse ASH1L (2056–2275), fly Ash1 (1304–1520), and human NSD2 (991–1209). Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster.
TABLE 1.
Data collection and refinement
| Zn2+ SADa | |
|---|---|
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | |
| α, β, γ (°) | a = b = 90, γ = 120 |
| Resolution (Å)b | 50-2.9 (2.95-2.9) |
| R_sym (%)b,c_ | 9.7 (62) |
| I/σ(I)b | 46.2 (4.7) |
| Completeness (%)b | 99 (100) |
| Redundancyb | 14.9 (14.4) |
| Refinement | |
| Reflections used (all data) | 50.0-2.90 Å |
| Number of atoms | |
| Protein | 3306 |
| AdoMet | 54 |
| Zn2+ | 6 |
| Water | 27 |
| Total | 3393 |
| _R_workd (reflections) | 0.2348 (18,803) |
| _R_freed (reflections) | 0.2994 (963) |
| Ramachandran statistics | 69.5 (30.5) |
| r.m.s.e deviations | |
| Bond length (Å) | 0.009299 |
| Bond angle (°) | 1.73975 |
The structure of the hASH1L catalytic domain, however, is striking because of its substrate binding pocket. It was previously determined that the SET-I subdomain serves as a substrate binding platform with the post-SET domain (12). It was also determined that an AdoMet methyl donor is positioned at the inner pocket between the SET-I subdomain and the post-SET domain. In our structure, however, a loop from the post-SET domain is found in between the SET-I subdomain and the post-SET domain, blocking the substrate binding pocket (Figs. 1A and 2A). We refer to the loop occupying the substrate binding pocket as the “auto-inhibitory loop.” The auto-inhibitory loop is highly flexible, as indicated by its high temperature factor, suggesting that the loop can be repositioned upon substrate binding. In this configuration, the methyl donor AdoMet is totally buried inside the protein, and the auto-inhibitory loop must be repositioned before the substrates can bind. The two molecules in the asymmetric unit have basically identical conformations of the auto-inhibitory loop, indicating that the loop blocking conformation is not due to crystal packing.
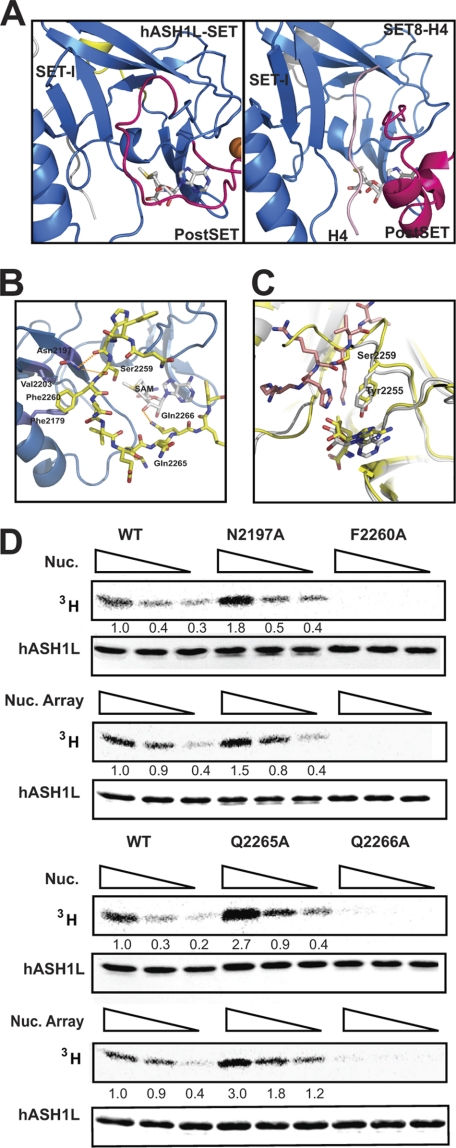
FIGURE 2.
The auto-inhibitory loop of hASH1L. A, comparison of the hASH1L-SET substrate binding pocket with that of the SET8-histone H4 complex. The auto-inhibitory loop in hASH1L occupies the substrate binding pocket where the histone substrate (colored in pink) binds in the SET8-histone complex structure. B, the auto-inhibitory loop interacts with the SET-I subdomain in the hASH1L SET domain. The carbonyl oxygens of His-2258 and Ser-2259 make hydrogen bonds with Asn-2197. Phe-2260 in the loop interacts with two hydrophobic residues, Val-2203 and Phe-2179. The auto-inhibitory loop also forms a hydrogen bond with AdoMet via Gln-2266. C, the hASH1L SET domain (shown in yellow) is superimposed on the SET8-histone H4 complex structure (SET8 shown in gray and H4 shown in pink). Ser-2259 of hASH1L occupies the substrate Lys position. D, disrupting the interactions between the auto-inhibitory loop and the SET-I subdomain or AdoMet affects the activity of hASH1L. Mutating Asn-2197 or Gln-2265 to alanine (N2197A, Q2265A) increases HMTase activity, whereas F2260A or Q2266A mutations abolish the activity. The HMTase activity was performed with tritium-labeled AdoMet with mononucleosome (Nuc.) or G5E4 nucleosomal array (Nuc. Array) as substrates.
Configuration of the Auto-inhibitory Loop of hASH1L
To better understand the configuration of the auto-inhibitory loop of hASH1L, we compared our structure of the hASH1L catalytic domain with that of SET8 bound to the histone H4 peptide (Fig. 2A). In the structure of the SET8-histone H4 substrate complex, the histone H4 substrate forms a parallel β-sheet with one of the two hairpin β-strands in the SET-I subdomain via hydrogen bonding between the backbones (19). Although the auto-inhibitory loop in hASH1L also forms hydrogen bonds with the β-strand in the SET-I subdomain in an anti-parallel manner, the major interactions between the loop and the SET-I subdomain are contributed by the following (Fig. 2, A and B). First, the highly flexible auto-inhibitory loop is held by an interaction between the side chain of Asn-2197 from the SET-I subdomain and the carbonyl oxygens of His-2258 and Ser-2259 at the start of the loop. This interaction is particularly interesting because the auto-inhibitory loop makes a sharp turn between His-2258 and Ser-2259, suggesting that the turning confirmation of the auto-inhibitory loop is assisted by the interaction between Asn-2197, His-2258, and Ser-2259. Second, the auto-inhibitory loop is held by the hydrophobic interaction between Phe-2260 in the middle of the loop and the Phe-2179 and Val-2203 residues in the SET-I subdomain. Last, the auto-inhibitory loop interacts with the AdoMet molecule. The Gln-2266 side chain in the loop forms a hydrogen bond with the 3′-hydroxyl group of the ribose ring of AdoMet at the final position of the loop. This 3′-hydroxyl group of AdoMet has been shown to interact with the histone substrates in the SET8-H4 complex (19). In this configuration, the auto-inhibitory loop is hinged by Ser-2259 and Gln-2265, and the middle of the loop is held by Phe-2260 (Fig. 2B).
It has been previously identified that a Phe/Tyr in the SET domain guides the side chain of the Lys substrate (12). When we superimposed the hASH1L and SET8-H4 structures, the Tyr-2255 in hASH1L can be identified as the guide Phe/Tyr, maintaining an identical position to other known guide phenylalanines (Fig. 2C). The Ser-2259 in the auto-inhibitory loop occupies the position of the target Lys at the substrate binding pocket. These data also suggest that the auto-inhibitory loop must be reconfigured for the substrate to bind to hASH1L.
Mutations on the Auto-inhibitory Loop Stimulate hASH1L HMTase Activity
Previous work has suggested that the post-SET domain may be involved in regulating HMTase activity (20). To investigate the effect of the auto-inhibitory loop of hASH1L on enzyme activity, we generated several mutations in the loop that would be predicted to affect the interaction between the loop and the other parts of hASH1L. We then measured the HMTase activity using mononucleosomes or nucleosomal arrays as substrates.
In the first set of mutants, which were designed to make the auto-inhibitory loop more flexible, we mutated Asn-2197 to Ala to disrupt the interaction between the loop and the SET-I subdomain at the beginning of the loop. We also mutated Gln-2265 to Ala at the final position of the loop to make the hinge region of the loop more flexible. In these mutants, the auto-inhibitory loop would be expected to become more flexible due to loss of the interaction between the loop and the SET-I subdomain. We crystallized the Q2265A mutant protein, and preliminary structural analysis of the mutant protein shows that the auto-inhibitory loop is highly disordered. With these mutants, we observed an increased level of HMTase activity for hASH1L, with higher levels of activity observed for Q2265A than for N2197A (Fig. 2D). This observation suggests that mutating Asn-2197 or Gln-2265 to alanine makes it easier for the substrate to access the active site by making the auto-inhibitory loop more flexible. In the second mutant, we generated the F2260A mutant to disrupt the hydrophobic interaction between the loop and the SET-I subdomain and measured the HMTase activity of the mutant. Unexpectedly, the F2260A mutant showed no activity toward either the nucleosomal array or the mononucleosome substrates. Phe-2260 is highly conserved in the hASH1L subfamily; the result that the mutation of Phe-2260 abolishes HMTase activity indicates that this residue may also have other critical roles for regulating HMTase activity. In a third mutant, we mutated the Gln-2266 residue to Ala to perturb the interaction of the protein with AdoMet. In this mutant, we were not able to see any HMTase activity, suggesting that the Gln-2266 residue is critical for activity. In a fourth mutant, Glu-2263 was mutated to Ala. This mutant showed neither increased nor decreased HMTase activity (supplemental Fig. S1).
To examine whether the increase of activities of two mutants results from the substrate binding, we performed kinetic analysis with wild-type, Q2265A, and N2197A mutant proteins (supplemental Fig. S2). Our kinetic analysis shows roughly similar _V_max values for all proteins. However, there is significant decrease in Km values of the mutant proteins when compared with that of wild-type protein, indicating that the mutants have higher substrate binding affinity.
It should also be noted that all mutant proteins behaved identically to the wild-type protein in gel filtration chromatography, indicating that the mutant proteins maintain overall structures that are similar to that of the wild type. Our mutagenesis study on the auto-inhibitory loop indicates that the interaction between the loop and the SET-I domain may be involved in regulating hASH1L HMTase activity. These results also suggest that there may be a regulatory mechanism to reconfigure and open up the auto-inhibitory loop.
hASH1L Catalytic Domain Methylates Histone H3 Lys-36
Drosophila Ash1 was previously shown to affect the levels of histone H3 Lys-4 (H3K4) methylation genetically and to methylate H3K4, H3K9, and H4K20. Recently, however, the Tanaka group (14) showed using nucleosomal substrates that the hASH1L catalytic domain methylates H3K36 but not H3K4. The other group used free histone as the substrate to demonstrate that hASH1L methylates H3K4 (9).
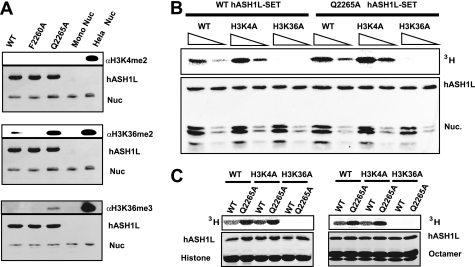
To investigate the substrate specificity of hASH1L, we performed an HMTase assay with the wild type as well as two mutant hASH1L proteins (F2260A, Q2265A) using mononucleosomes as the substrate. The methylation state was examined with H3K4me2-, H3K36me2-, and H3K36me3-specific antibodies (Fig. 3A). Fig. 3A clearly shows that the methylated H3K36-specific antibodies, but not the methylated H3K4-specific antibodies, recognize the product of hASH1L. These data indicate that hASH1L methylates histone H3 Lys-36 in vitro under our conditions. Moreover, with the hyperactive mutant (Q2265A), the signal specific for H3K36me2 greatly increased, whereas that specific for the H3K4 remained undetectable. We were also able to detect trimethylation of histone H3 Lys-36 with the hyperactive hASH1L mutant. This observation is also consistent with the notion that hASH1L specifically methylates H3K36. To rule out the possibility that this result was caused by cross-reactivity of the antibody to other methylated histones, we assembled nucleosomes with mutant histones that have Lys-4 mutated to Ala or Lys-36 mutated to Ala. HMT assays were performed with the wild type and hyperactive hASH1L mutants (Fig. 3B). hASH1L was not able to methylate nucleosomes containing histones with H3 Lys-36 mutated to Ala, whereas it was found to have full activity toward the H3 K4A mutant nucleosomes, as well as wild-type nucleosomes. Similar results were obtained when the hyperactive mutant was used in the HMT assay. In addition, when we used histone H3 or the histone octamer, we also observed that hASH1L specifically methylates histone H3 Lys-36 (Fig. 3C). Taken together, the hASH1L catalytic domain specifically methylates histone H3 Lys-36 in vitro.
FIGURE 3.
The hASH1L catalytic domain methylates histone H3 Lys-36. A, HMTase assay of the hASH1L catalytic domain was performed with wild-type hASH1L (WT), F2260A, and the hyperactive mutant (Q2265A) (lanes 1, 2, and 3) using recombinant mononucleosomes. Recombinant mononucleosomes (Nuc) and HeLa nucleosomes (Hela Nuc) were used as controls for the antibodies (lanes 4 and 5). The methylation product of the hASH1L catalytic domain is specifically recognized by the anti methyl-H3K36 antibody but not by the methyl-H3K4 antibody. The hyperactive mutant (Q2265A) is able to also produce H3K36 trimethylation. B and C, the wild-type hASH1L SET catalytic domain and the hyperactive mutant methylate the wild-type or H3K4A mutant histone, octamer, and nucleosome (Nuc.), but not the H3K36A mutant histone, octamer, and nucleosome.
DISCUSSION
The results presented here reveal that the hASH1L substrate binding pocket is blocked by an auto-inhibitory loop. Our data suggest that this loop regulates hASH1L activity. At this moment, however, it is not clear what the physiological role of the auto-inhibitory loop is.
We were able to measure the HMTase activity with the hASH1L catalytic domain alone; thus, the auto-inhibitory loop may be reconfigured upon substrate binding. Alternatively, there may be other regulatory mechanisms that function to open the auto-inhibitory loop. This mechanism needs to be further studied.
Considering that an auto-inhibitory mechanism has also been observed in other enzymes (21–23) and that histone methyltransferases need to be tightly regulated to coordinate gene expression, it is likely that the auto-inhibitory loop plays an important role in coordinating histone methylation. Based on sequence alignment (Fig. 1D) and the accompanying study (25), other SET domain proteins containing a SET domain in the middle, such as SET2 and nuclear receptor SET domain-containing protein (NSD1), may adopt a similar loop configuration as hASH1L in the post-SET domain.
The auto-inhibitory conformation of hASH1L catalytic domain is unlikely due to an artifact of using a fragment of the catalytic domain. The accompanying study (25) on the structure of NSD1 catalytic domain shows a similar auto-inhibitory conformation, suggesting that the auto-inhibitory conformation may be a common mechanism of the methyltransferases having a SET domain in the middle. In addition, the auto-inhibitory loop is well ordered in the crystal structure, and the interaction between the auto-inhibitory loop and the SET domain is specific. Also, the disruption of the specific interaction increases the activity. These observations suggest that the auto-inhibitory conformation is physiological relevant, although it is possible that the auto-inhibitory conformation can be transient. Consistent with our data, it has been suggested that the post-SET domain may be involved in regulating the activity in other methyltransferases (mixed-lineage leukemia) (20). However, the exact mechanism how the auto-inhibitory loop functions needs to be studied in the full-length protein. It would be interesting to examine the roles of plant homeodomains, bromo domains, and BAH domains in regulating HMTase activity.
Recent work on the nuclear receptor NSD1 protein shows that substrate specificity depends on the nature of the substrate (24). It would be interesting to investigate whether the regulation of the specificity is also through an auto-inhibitory loop in the post-SET domain of the NSD protein.
Although the Drosophila Ash1 mutant has a defect in histone H3 Lys-4 methylation, the substrate specificity of mammalian ASH1L is controversial (9, 14). However, in our in vitro assay, hASH1L clearly has histone methyltransferase activity that is specific for H3 Lys-36. This result is consistent with those obtained by the Tanaka group (14), whereas the Blobel group (9) observed specificity for H3 Lys-4. This discrepancy may be attributed to the different substrates that were used. We and the Tanaka group used nucleosomes, whereas the Blobel group used a histone peptide as the substrate and used a long incubation time. We also detected activity for histone H3 Lys-36 when octamers or histone H3s were used as substrates. Furthermore, the hyperactive mutant of hASH1L (Q2265A) is able to trimethylate histone H3 Lys-36, suggesting that hASH1L is a histone H3 Lys-36-specific methyltransferase. We cannot totally rule out the possibility that hASH1L may have dual specificity toward histone H3K4 as well as H3K36 or that it actually prefers H3K36 over H3K4. It has been reported recently that the substrate specificity of NSD1 methyltransferase depends on the nature of the substrate (24). The substrate specificity of hASH1L may be similarly regulated, although we have not observed any activity other than the H3 Lys-36-specific activity when nucleosomes, nucleosomal arrays, histone octamers, or even histone H3 molecules are used as substrates. To unequivocally address the substrate specificity of hASH1L, further structural studies on hASH1L in complex with its substrate will be required. Taken together, our data demonstrate that the hASH1L SET domain adopts a novel configuration of the substrate binding pocket that is limited by an auto-inhibitory loop from the post-SET domain. This result suggests that there is a mechanism to regulate hASHL1 HMTase activity.
Supplementary Material
Supplemental Data
Acknowledgments
We thank the members of the Song laboratory, David Helfman, and Jun Kyu Rhee for critical reading of the manuscript and Dr. Yujiro Tanaka for providing human ASH1L cDNA. We also thank Bob Kingston and Karim Bouazoune for providing the HeLa histone octamer. We also thank Dr. Yeongil Kim for support with data collection. The data for this study were collected at the beamline 4A HFMX4 of the Pohang Accelerator Laboratory.
*
This work was supported by the Brain Korea 21 (BK21) program from the Korea Ministry of Education, Science and Technology (to S. A.). This work was partially supported by the World Class University (WCU) program Grant R31-2008-000-10071-0 (to J.-J. S.), a grant from the Korea Healthcare Technology R&D project of the Ministry of Health, Welfare and Family Affairs (Grant A100525) and Grants KRF-2010-007869 and KRF-2010-0016263 (to J.-J. S.), and a grant from the High Field NMR Research Program (to Y. H. J.) awarded through the National Research Foundation of Korea and funded by the Ministry of Education, Science and Technology.
The atomic coordinates and structure factors (code 3OPE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
2
The abbreviations used are:
HMTase
histone methyltransferase
NSD
nuclear receptor SET domain-containing protein
AdoMet
_S_-adenosylmethionine
Bis-Tris
2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
h
human.
REFERENCES
- 1.Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 2.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 4.Gould A. (1997) Curr. Opin. Genet. Dev. 7, 488–494 [DOI] [PubMed] [Google Scholar]
- 5.Tripoulas N., LaJeunesse D., Gildea J., Shearn A. (1996) Genetics 143, 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaJeunesse D., Shearn A. (1995) Mech. Dev. 53, 123–139 [DOI] [PubMed] [Google Scholar]
- 7.Rozovskaia T., Tillib S., Smith S., Sedkov Y., Rozenblatt-Rosen O., Petruk S., Yano T., Nakamura T., Ben-Simchon L., Gildea J., Croce C. M., Shearn A., Canaani E., Mazo A. (1999) Mol. Cell Biol. 19, 6441–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beisel C., Imhof A., Greene J., Kremmer E., Sauer F. (2002) Nature 419, 857–862 [DOI] [PubMed] [Google Scholar]
- 9.Gregory G. D., Vakoc C. R., Rozovskaia T., Zheng X., Patel S., Nakamura T., Canaani E., Blobel G. A. (2007) Mol. Cell Biol. 27, 8466–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couture J. F., Dirk L. M., Brunzelle J. S., Houtz R. L., Trievel R. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20659–20664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng X., Collins R. E., Zhang X. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 267–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrd K. N., Shearn A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11535–11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y., Katagiri Z., Kawahashi K., Kioussis D., Kitajima S. (2007) Gene 397, 161–168 [DOI] [PubMed] [Google Scholar]
- 15.Simpson R. J. (2006) Cold Spring Harb Protoc., pdb.prot5545 [DOI] [PubMed] [Google Scholar]
- 16.Terwilliger T. C. (2003) Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 17.Owen-Hughes T., Workman J. L. (1996) EMBO J. 15, 4702–4712 [PMC free article] [PubMed] [Google Scholar]
- 18.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 19.Couture J. F., Collazo E., Brunzelle J. S., Trievel R. C. (2005) Genes Dev. 19, 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R. (2009) Mol. Cell 33, 181–191 [DOI] [PubMed] [Google Scholar]
- 21.Chen L., Jiao Z. H., Zheng L. S., Zhang Y. Y., Xie S. T., Wang Z. X., Wu J. W. (2009) Nature 459, 1146–1149 [DOI] [PubMed] [Google Scholar]
- 22.Deindl S., Kadlecek T. A., Brdicka T., Cao X., Weiss A., Kuriyan J. (2007) Cell 129, 735–746 [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama N., Ischenko I., Hayman M. J., Miller W. T. (2005) J. Biol. Chem. 280, 8893–8900 [DOI] [PubMed] [Google Scholar]
- 24.Li Y., Trojer P., Xu C. F., Cheung P., Kuo A., Drury W. J., 3rd, Qiao Q., Neubert T. A., Xu R. M., Gozani O., Reinberg D. (2009) J. Biol. Chem. 284, 34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao Q., Li Y., Chen Z., Wang M., Reinberg D., Xu R. M. (2011) J. Biol. Chem. 286, 8361–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data