The Sleeping Beauty transposon system: a non-viral vector for gene therapy (original) (raw)
Abstract
Over the past decade, the Sleeping Beauty (SB) transposon system has been developed as the leading non-viral vector for gene therapy. This vector combines the advantages of viruses and naked DNA. Here we review progress over the last 2 years in vector design, methods of delivery and safety that have supported its use in the clinic. Currently, the SB vector has been validated for ex vivo gene delivery to stem cells, including T-cells for the treatment of lymphoma. Progress in delivery of SB transposons to liver for treatment of various systemic diseases, such as hemophilia and mucopolysaccharidoses types I and VII, has encountered some problems, but even here progress is being made.
INTRODUCTION
As reviewed by several others in this issue, there has been considerable progress made over the past decade in developing viral vectors for human gene therapy. However, there are complications with viruses as gene-delivery vectors including their integration-site preferences that may increase chances of adverse effects (1,2), the need for extensive purification and quality control to prevent replication-competent virus and the costs associated with their production and handling (3,4). Advantages of non-viral vectors include the ease and relatively low cost of producing sufficient amounts required to meet the entire patient population, stability during storage and lack of immunogenicity once inside host cells (4–6). There are two major problems with non-viral gene therapy approaches that are almost always based on delivery and expression of genes carried on an engineered plasmid produced in E. coli. First, expression of the transgene in most mammalian cells is brief due to intracellular breakdown and epigenetic responses that recognize the prokaryotic origin of the plasmid. Secondly, delivery of DNA molecules into cells of a specific organ is inefficient. Because of these problems, non-viral vectors have not yet been tested for therapy of systemic inherited diseases. However, application of the Sleeping Beauty transposon system (SBTS) has changed this view (7). This non-viral vector, which combines the advantages of viruses and naked DNA (8), has experienced the most rapid of development from birth to application in humans of all vectors now in clinical trials (Fig. 1). The SBTS consists of two components: (i) a transposon containing a gene-expression cassette and (ii) a source of transposase enzyme. By transposing the expression cassette from a plasmid into the genome, sustained transcription of a transgene can be achieved (Fig. 2).
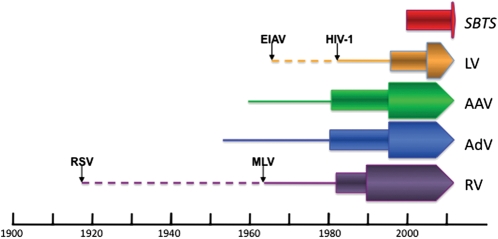
Figure 1.
History of some vectors used for human gene therapy. For each type of vector, the lines indicate the period between its discovery and its use for gene transfer (in the case of retroviruses and lentiviruses, the initial virus discovered and the virus actually used for gene therapy are indicated by dashed and solid lines, respectively). Rectangles show the period of use for each vector as a gene transfer agent and the arrowheads indicate its use in humans. SB transposons were synthetically made and therefore validated by their use for gene transfer into cultured cells. Hence, they have no associated line; the first clinical trial has been initiated. LV, lentiviruses; AAV, adeno-associated viruses; AdV, adenoviruses; RV, retroviruses.
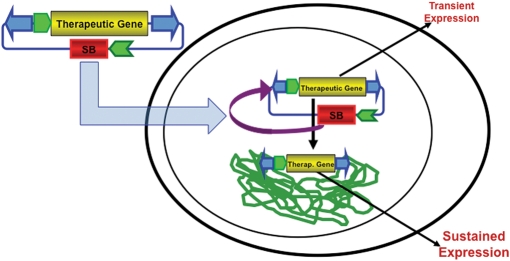
Figure 2.
SB transposon-mediated gene transfer into chromosomal DNA for long-term expression of a therapeutic gene. An SB transposon (the inverted arrowheads) in a plasmid provides only transient expression of a transgene from a promoter unless transposed into a host genome. There are several methods for delivery of the transposon system into a cell based on whether the cell is in culture (electroporation or transfection) or in a tissue (e.g. hydrodynamic injection). In most studies, the source of the SB transposase is a gene on either the same (shown here) or a different plasmid as that harboring the transposon.
The power of the SB system to treat disease models was first demonstrated by Yant et al. (9), who showed sustained expression of α1-antitrypsin in normal mice and of clotting Factor IX (FIX) in FIX-deficient hemophilic mice. This achievement was followed by successful treatment of other mouse models of genetic disease, including inherited tyrosinemia (10), hemophilia A (11–13) and mucopolysaccharidosis (MPS) types I and VII (14,15). In addition, SB transposons have been used to treat epidermolysis bullosa (16), glioblastoma multiforme (17), sickle cell anemia (18) and B-cell lymphoma (19–21). In rats, SB transposons have been used to treat pulmonary hypertension (22) and jaundice (23).
Thus, early proof-of-principle studies stressing efficacy of sustained gene expression have demonstrated the significant potential of the SBTS for gene therapy. Other important considerations with respect to gene therapy include efficacy in larger animals and safety. Here, we review these issues and recent progress in turning the SBTS from a gene transfer vector into a gene therapy vector that can be used in the clinic during the era of personalized medicine.
Improvements have involved all aspects of the SBTS that are necessary for its use in the clinic (Fig. 3). These include (i) efficacy—transpositional efficiency and engineered cassettes to achieve appropriate expression of therapeutic or reporter genes, as well as SB transposase; (ii) delivery—methods and routes of plasmid delivery as well as cell-culture conditions used for ex vivo gene transfer to therapeutic target cells and (iii) safety—the integration preferences of SB transposase and duration of the transposition reaction in transduced cells. Along the way, we have gained insights into the effect of genetic background on transposition, targeting tissues, gender effects on gene expression and transposon dose–response. Some of these issues are specific to the SBTS (e.g. over-production inhibition), while others are associated with all non-viral vectors (e.g. efficiency of delivery), with integrating vectors in general (e.g. insertional mutagenesis and post-integrative promoter silencing), or with any type of gene therapy vector (e.g. host immune responses to transgenes and/or their products).
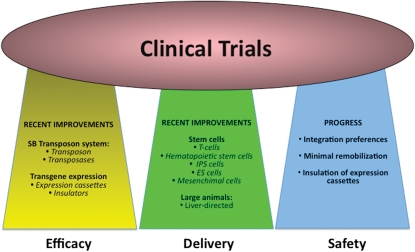
Figure 3.
Success in clinical trials depends on three aspects of a vector such as its efficacy, its delivery and its safety. Recent developments in each of these areas support the application of the SBTS in clinical trials.
EFFICACY: IMPROVEMENT OF THE SBTS
The issue of low transposition levels has been addressed over the past decade by modifying the transposon (24,25) and the transposase. Hyperactive transposases have been derived from the original SB10 by mutagenesis of the catalytic and DNA-binding domains (26–29). SB100X, engineered using a combination of in vitro molecular evolution and selection (30), is the most active of the transposases generated so far, and was named molecule of the year for 2009 (http://iscspm.webnode.com/). This powerful transposase directs the highest levels of transposon integration yet demonstrated, presumably due to increased stability (30,31).
Appropriate expression cassettes are critical for attaining high and sustained levels of transgene expression in a target organ such as the liver. Tissue-specific transcriptional regulators are thought to help counteract host immune responses (32) by preventing transgene expression in antigen-presenting cells. Accordingly, the choice of transcriptional motifs for driving both therapeutic transgene and transposase expression should be considered to reduce or prevent silencing of the therapeutic transgene, allow expression of the transposase for only a very short period of time while avoiding over-production inhibition of transposition that can occur at excessive transposase levels (26,33). There are multiple implications of improvements in the expression cassette. If the goal is to achieve strong, long-term expression of the transgene, promoter strength is not the only issue to consider. Gender can influence gene expression from some promoters, as we unexpectedly found in experiments using the CAGS (sometimes called CAGGS) promoter to drive human α-l-iduronidase (IDUA): males expressed transgenic IDUA at 30–50-fold higher levels than females (15). On the other hand, promoter silencing can be usefully employed in the SBTS; the cytomegalovirus early promoter is shut down within a few days in many mammalian cells including liver (34) and therefore is a good choice for transient expression of SB transposase to prohibit sustained re-mobilization of transposons after their integration into the genome (35).
Immune responses often curtail gene expression (32). Host immune responses can be expected, particularly when there is over-expression of a secretable transgenic protein. For example, after hydrodynamic tail vein injection 99% of the transgene expression is observed in the liver (15,36). However, antigen-presenting cells both in the liver and in the spleen may express the foreign protein. Substituting a hepatocyte-specific promoter, such as ApoEhAAT (37,38), for the ubiquitously expressed mini (m)CAGGS promoter to regulate IDUA resulted in prolongation of IDUA expression as well as a lack of significant gender bias in C57BL/6 mice (39). Substitution of the SB100X transposase (30) for SB11, in combination with the liver-specific promoter to drive IDUA expression and transient immunosuppression with cyclophosphamide resulted in significantly prolonged expression of transgenic IDUA at levels ∼100-fold higher than the endogenous level, which is therapeutically relevant for treating MPS I (40).
In addition to regulating desired levels of transgene expression, different promoters have been found to demonstrate unexpected features. For example, the CAGGS promoter yielded an exceptionally high level of eGFP expression specifically in differentiated cardiomyocytes following SB-mediated gene transfer to human embryonic stem cells (41). Moreover, in stem cells, the transcriptional motifs may not be equally active following differentiation (42,43).
DELIVERY IN VIVO TO THE LIVER AND EX VIVO TO HEMATOPOIETIC CELLS
Although a variety of different DNA-conjugating materials and delivery vehicles have been explored for the purpose of promoting non-viral gene transfer into different animal tissues, the most effective method to be developed is the hydrodynamics-based procedure (44–46), which targets the liver for sustained gene expression. At present, this is the most efficient method of plasmid delivery, when a liver-directed approach is an option for therapy. The hydrodynamics-based procedure has successfully been adapted to rats (47). We have shown that with this method, transgene expression 2 weeks following hydrodynamic delivery to mouse liver is 100–1000-fold higher than after delivery using polyethylenimine–DNA conjugate (36,48). If the transgene expresses a secretable protein at a high level, a low percentage of genetically modified cells can provide therapeutic levels of gene product sufficient to treat an entire animal.
Based on its success in treating mouse models of hemophilia (9,11,12), we used this approach in gene therapy studies of the lysosomal storage diseases (14,15). Using the improved pT2 transposon and hyperactive transposase SB11, we demonstrated adequate delivery of therapeutic transgenes to mouse liver for treatment of MPS I and VII, systemic metabolic diseases caused by deficiency of the lysosomal enzymes IDUA and β-glucuronidase, respectively. In our experience, the efficiency of hydrodynamic delivery in mice is ∼10% of that reported for retroviral delivery (14,49). In the case of MPS I, sustained IDUA activity in the liver after hydrodynamic infusion was ∼100 times higher than wild-type levels, which was necessary to restore enzymatic activity in other organs by enzymatic cross-correction. IDUA levels and sustainability of enzymatic activity were comparable with those observed with viral vectors and, as a consequence, several clinical manifestations of the disease, including skeletal malformations, were significantly ameliorated (15).
When adapting the hydrodynamic delivery method to larger animal models, several parameters must be considered: DNA dose, injection volume, rate of injection and species-specific variations in the anatomy of the target organ. From the outset, it was clear that in animals larger than 1 kg, hydrodynamic infusion would have to be limited to an isolated organ or tissue region because injections of >100 ml (10% of the animal's weight) in <10 s would not be practical. Catheter-mediated infusions to liver and other tissues, often employing balloons to limit direct hydrodynamic effects, have been conducted in larger animals, including rabbits (50,51), swine (52–59) and dogs (60,61,62,63). Although we have recently been able to achieve extended expression after hydrodynamic injection of SB transposons into dog liver using balloon catheters (64), reporter gene (canine secreted alkaline phosphatase) expression nonetheless was extinguished after 6 weeks. In mice, we have observed more effective expression when the DNA solution is injected with greater rapidity. Based on these studies, we have hypothesized that impulse, rather than pressure, may be the critical determinant for effectiveness of hydrodynamic delivery (65); this has several implications in the details of hydrodynamic delivery in larger animals.
An alternative delivery approach that combines the power of viruses to penetrate cellular membranes and SB transposons to deliver defined expression cassettes more randomly in the genome has been developed using chimeric viruses containing SB transposons. Chimeras include lentivirus/SB (66–68), adenovirus/SB (69) and herpes simplex virus/SB (70). A hybrid adenovirus/SB vector has been used for canine FIX gene delivery to a hemophilia B dog (69). However, the benefit provided by the hybrid in terms of delivery must be weighed against the loss of simplicity, low cost of production, long and stable shelf life, as well as other aspects involved with storage and quality control of purified DNA compared with virus preparations.
A primary target for integrative gene transfer has been hematopoietic stem cells (HSC), for which retroviruses, and more recently lentiviruses, have been the vector of choice for ex vivo gene transfer (71). Gene transfer into HSC requires stable integration, and the advent of hyperactive SB transposases in combination with efficient plasmid DNA loading by electroporation has resulted in effective transposition of primary human cord blood CD34+ cells that can be engrafted with the potential for differentiation into multiple cell lineages in vivo for non-viral gene therapy of blood disorders (43,72).
The most advanced use of the SBTS for gene therapy has been to transduce progenitor T-cells ex vivo to treat lymphoma. Retroviruses, including lentiviruses and spumaviruses, have been the preferred vector of choice for ex vivo gene transfer. Until recently, any non-viral gene therapy approach, including ex vivo transfer of the SBTS to stem cells_,_ was hampered by low efficacy and/or high toxicity of transfection, lipofection or electroporation in combination with inefficient gene transfer catalyzed by the original SB10 transposase. However, non-viral, ex vivo gene delivery is now feasible with improved transposases and expression cassettes, refined methods of electroporation of appropriate ratios of transposon-to-transposase genes and advanced cell-culture conditions. As a result, genes can be introduced into primary human CD34+ cells to obtain populations of stem cells that can be engrafted and differentiated into multilineage cell types in vivo for non-viral gene therapy of HSC (43,72). Stable transposition has been achieved in induced pluripotent stem cells (iPS), which retained their ability to differentiate along neural, cardiac and hepatic lineages without causing cytogenetic abnormalities (42). Efficient SB-mediated integration and expression in human embryonic stem cells has been achieved while preserving both the undifferentiated state and the potential to be directed down specific pathways of differentiation (41,73). The SBTS was employed to insert T-cell receptor genes into the genomes of peripheral blood lymphocytes for targeting against antigens presented on melanoma cells. Transgene expression and anti-tumor activity were comparable with those attained by γ-retroviral transduction in a clinical trial (74).
The first clinical application of the SBTS will be to provide a new specificity for T-cells to treat B-lineage malignancies (6). The SBTS has been used effectively to generate T cells that express a chimeric antigen receptor recognizing CD19, a lineage-specific tumor antigen (19,20). When the genetically altered T-cells are expanded on CD19+ artificial antigen-presenting cells, there is a rapid outgrowth of CD4+ and CD8+ T-cells that express the anti-tumor chimeric antigen receptor. An early-phase human trial has been approved to assess the safety and feasibility of this approach.
SAFETY
There are several safety issues facing the SBTS for gene therapy. The first is common to all integrating vectors, insertional mutagenesis. The SBTS has the most random integration preference of the vectors currently in use for gene therapy (2,75,76). Loci into which the transposon integrates can influence the expression of the gene it carries, often leading to transcriptional silencing (77–79). Likewise, there is the possibility that the promoters in the transgenic expression cassette will influence genes in the vicinity of the integration site. Accordingly, some SB transposons have been engineered to contain expression cassette flanked by insulators such as the cHS4 locus control region. This results in a significant increase in stable transfection rates in vitro (80) as well as theoretical prevention of accidental transactivation of genes residing close to the transposon insertion site (81).
A second safety issue is the presence of the SB transposase gene and the potential for remobilization of transposons already sited in the recipient genome. We have found that by using a promoter that is active only over a few days, the chances of remobilization are below minimal detection (35). Moreover, were any transposon to remobilize, it would not have any increased probability of causing an adverse event per insertion than the original integration. Nevertheless, these two aspects represent fields for future development, such as the use of RNA as a source of SB transposase (82,83).
Increasing safety of the SBTS by targeting ‘safe havens' in the genome is a prime future goal. The first attempts to combine the targeted integration ability of zinc fingers with SB transposons consisted of adding a zinc finger DNA-binding domain to SB transposase (84,85). These approaches resulted in diminished SB activity and marginal targeting. However, other strategies remain untested and represent one of the most important future directions to pursue.
CONCLUDING REMARKS
Several other DNA integrase/transposon systems providing efficient integration in mouse and human genomes have been developed during the past decade. One of these, the piggyBac (PB) transposon of insect origin (86), has shown promise for regenerative medicine as a tool for reprogramming iPS (87). Side-by-side comparison of SB and PB (31) showed that the SBTS has several advantages for human gene therapy application. SB100X has superior transduction rates for human hematopoietic cells (31,72). The near-random integration profile of SB would seem safer than that of PB, which tends to target genes and their upstream regulatory regions (88,89). Moreover, SB vectors have negligible inherent enhancer/promoter activity compared with PB transposons (8). However, PB is less sensitive than SB to over-production inhibition (31), which suggests greater care must be taken with the SBTS to achieve optimal rates of gene transfer. In contrast to SB and PB transposons, which integrate throughout the genome, the gene transfer system derived from phiC31 bacteriophage has an advantage of semi-targeted integration (90). However, phiC31-mediated recombination is not always precise and is associated with chromosomal rearrangements (91–93).
In summary, although the efficiency of naked DNA delivery is sometimes an issue, the SBTS has particular advantages for treatment of large populations of patients in terms of its relatively low cost and near-random integration profile, especially for treatment of lymphoma by modification of T-stem cells. However, there remain significant hurdles for the SBTS, primarily efficient delivery and maintenance of gene expression in the liver, before it will be an efficacious vector to treat hemophilia and select metabolic diseases.
FUNDING
We acknowledge the financial support of NIH grant 1R01DK082516.
ACKNOWLEDGEMENT
We thank our colleagues in the Center for Genome Engineering and on the Gene Therapy for Metabolic Diseases Program Project Grant for many insightful discussions.
Conflict of Interest statement. P.B.H. and R.S.M. are co-founders and have equity in Discovery Genomics, Inc., a startup biotech company that receives funding from the NIH to explore the feasibility of using SB transposons for commercial gene therapy.
REFERENCES
- 1.Kohn D., Sadelain M., Dunbar C., Bodine D., Kiem H.P., Candotti F., Tisdale J., Riviere I., Blau C.A., Richard R.E., et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Mol. Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- 2.Hackett C.S., Geurts A.M., Hackett P.B. Predicting preferential DNA vector insertion sites: implications for functional genomics and gene therapy. Genome Biol. 2007;8(Suppl. 1):S12. doi: 10.1186/gb-2007-8-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., Reeves L., Sanburn N., Croop J., Williams D.A., Cornetta K. Packaging cell line DNA contamination of vector supernatants: implication for laboratory and clinical research. Virology. 2001;282:186–197. doi: 10.1006/viro.2001.0826. [DOI] [PubMed] [Google Scholar]
- 4.Hodges B.L., Cheng S.H. Cell and gene-based therapies for the lysosomal storage diseases. Curr. Gene Ther. 2006;6:227–241. doi: 10.2174/156652306776359522. [DOI] [PubMed] [Google Scholar]
- 5.Essner J.J., McIvor R.S., Hackett P.B. Awakening of gene therapy with Sleeping Beauty transposons. Curr. Opin. Pharmacol. 2005;5:513–519. doi: 10.1016/j.coph.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Hackett P.B., Largaespada D.A., Cooper L.J.N. A transposon and transposase system for human application. Mol. Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivics Z., Hackett P.B., Plasterk R.H., Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 8.Izsvak Z., Hackett P.B., Cooper L.J.N., Ivics Z. Translating Sleeping Beauty transposition to molecular therapy: victories and challenges. BioEssays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yant S.R., Meuse L., Chiu W., Ivics Z., Izsvak Z., Kay M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 10.Montini E., Held P.K., Noll M., Morcinek N., Al-Dhalimy M., Finegold M., Yant S.R., Kay M.A., Grompe M. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol. Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- 11.Ohlfest J.R., Frandsen J.L., Fritz S., Lobitz P.D., Perkinson S.G., Clark K.J., Nelsestuen G., Key N.S., McIvor R.S., Hackett P.B., et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Mah C., Fletcher B.S. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice. Mol. Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Kren B.T., Unger G.M., Sjeklocha L., Trossen A.A., Korman V., Diethelm-Okita B.M., Reding M.T., Steer C.J. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J. Clin. Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aronovich E.L., Bell J.B., Belur L.R., Gunther R., Koniar B., Erickson D.C., Schachern P.A., Matise I., McIvor R.S., Whitley C.B., et al. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J. Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronovich E.L., Bell J.B., Kahn S.A., Belur L.R., Gunther R., Koniar B., Schachern P.A., Parker J., Carlson C.S., Whitley C.B., et al. Systemic correction of storage disease in MPS I NOD/SCID mice using the Sleeping Beauty transposon system. Mol. Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortiz S., Lin Q., Yant S.R., Keene D., Kay M.A., Khavari P.A. Sustainable correction of junctional epidermollysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]
- 17.Ohlfest J.R., Lobitz P.D., Perkinson S.G., Largaespada D.A. Integration and long-term expression in xenografted human glioblastoma cells using a plasmid-based transposon system. Mol. Ther. 2004;10:260–268. doi: 10.1016/j.ymthe.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Belcher J.D., Vineyard J.V., Bruzzone C.M., Chen C., Beckman J.D., Nguyen J., Steer C.J., Vercellotti G.M. Heme oxygenase-1 gene delivery by Sleeping Beauty inhibits vascular stasis in a murine model of sickle cell disease. J. Mol. Med. 2010;88:665–675. doi: 10.1007/s00109-010-0613-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh H., Manuri P.R., Olivares S., Dara N., Dawson M.J., Huls H., Hackett P.B., Kohn D.B., Shpall E.J., Champlin R.E., et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;15:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X., Guo H., Kang J., Choi S., Zhou T.C., Tammana S., Lees C.J., Li Z.Z., Milone M., Levine B.L., et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19(+) lymphoid malignancies. Mol. Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jena B., Dotti G., Cooper L.J. Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood. 2010;116:1035–1044. doi: 10.1182/blood-2010-01-043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Liu H., Visner G., Fletcher B.S. Sleeping Beauty-mediated eNOS gene therapy attenuates monocrotaline-induced pulmonary hypertension in rats. FASEB J. 2006;20:2594–2596. doi: 10.1096/fj.06-6254fje. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Sarkar D.P., Mani P., Steer C.J., Chen Y., Guha C., Chandrasekhar V., Chaudhuri A., Roy-Chowdhury N., Kren B.T., Roy-Chowdhury J. Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of Sleeping Beauty transposon. Hepatology. 2009;50:815–824. doi: 10.1002/hep.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izsvak Z., Ivics Z., Plasterk R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 25.Cui Z., Guerts A.M., Liu G., Kaufman C.D., Hackett P.B. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 26.Geurts A.M., Yang Y., Clark K.J., Cui Z., Dupuy A.J., Largaespada D.A., Hackett P.B. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 27.Zayed H., Izsvak Z., Walisko O., Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Yant S.R., Park J., Huang Y., Mikkelsen J.G., Kay M.A. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baus J., Liu L., Heggestad A.D., Sanz S., Fletcher B.S. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 30.Mátés L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 31.Grabundzija I., Irgang M., Mátés L., Belay E., Matrai J., Gogol-Döring A., Kawakami K., Chen W., Ruiz P., Chuah M.K., et al. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arruda V.R., Favaro P., Finn J.D. Strategies to modulate immune responses: a new frontier for gene therapy. Mol. Ther. 2009;17:1492–1503. doi: 10.1038/mt.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackett P.B. Integrating DNA vectors for gene therapy. Mol. Ther. 2007;15:10–12. doi: 10.1038/sj.mt.6300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilber A.C., Frandsen J.L., Wangensteen K.J., Ekker S.C., Wang X., McIvor R.S. Dynamic gene expression following systemic delivery of plasmid DNA as determined by in vivo bioluminescence imaging. Hum. Gene Ther. 2005;16:1325–1332. doi: 10.1089/hum.2005.16.1325. [DOI] [PubMed] [Google Scholar]
- 35.Bell J.B., Aronovich E.A., Schreifels J.M., Beadnell T.C., Hackett P.B. Duration of expression of Sleeping Beauty transposase in mouse liver following hydrodynamic DNA delivery. Mol. Ther. 2010;18:1792–1802. doi: 10.1038/mt.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podetz-Pedersen K., Bell J.B., Steele T.W., Wilber A., Shier W.T., Belur L.R., McIvor R.S., Hackett P.B. Gene expression in lung and liver after intravenous infusion of polyethylenimine complexes of Sleeping Beauty transposons. Gene Ther. 2010;21:210–220. doi: 10.1089/hum.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuyama T., Huber R.M., Bowling W., Pearline R., Kennedy S.C., Flye M.W., Ponder K.P. Liver-directed gene therapy: a retroviral vector with a complete LTR and the ApoE enhancer-alpha 1-antitrypsin promoter dramatically increases expression of human alpha 1-antitrypsin in vivo. Hum. Gene Ther. 1996;7:637–645. doi: 10.1089/hum.1996.7.5-637. [DOI] [PubMed] [Google Scholar]
- 38.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 39.Aronovich E.L., Bell J.B., Koniar B.L., Hackett P.B. A liver-specific promoter enhances expression of alpha-l-iduronidase in Sleeping Beauty-mediated gene therapy for murine MPS I. Mol. Ther. 2010;18:S284. [Google Scholar]
- 40.Aronovich E.L., Bell J.B., Podetz-Pedersen K., Koniar B.L., McIvor R.S., Hackett P.B. Immunosuppression with cyclophosphamide significantly prolongs high-level expression of IDUA mediated by the Sleeping Beauty transposon system. (submitted) [Google Scholar]
- 41.Orbán T.I., Apáti A., Németh A., Varga N., Krizsik V., Schamberger A., Szebényi K., Erdei Z., Várady G., Karászi E., et al. Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells. 2009;27:1077–1087. doi: 10.1002/stem.45. [DOI] [PubMed] [Google Scholar]
- 42.Belay E., Mátrai J., Acosta-Sanchez A., Ma L., Quattrocelli M., Mátés L., Sancho-Bru P., Geraerts M., Yan B., Vermeesch J., et al. Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: a nonviral paradigm for coaxed differentiation. Stem Cells. 2010;28:1760–1771. doi: 10.1002/stem.501. [DOI] [PubMed] [Google Scholar]
- 43.Sumiyoshi T., Holt N.G., Hollis R.P., Ge S., Cannon P.M., Crooks G.M., Kohn D.B. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells using the Sleeping Beauty transposon system. Hum. Gene Ther. 2009;20:1607–1626. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 46.Bell J.B., Podetz-Pedersen K., Aronovich E.L., Belur L.R., McIvor R.S., Hackett P.B. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat. Protocols. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruyama H., Higuchi N., Nishikaw Y., Kameda S., Iino N., Kazama J.J., Takahashi N., Sugawa M., Hanawa H., Tada N., et al. High-level expression of naked DNA delivered to rat liver via tail vein injection. J. Gene Med. 2002;4:333–341. doi: 10.1002/jgm.281. [DOI] [PubMed] [Google Scholar]
- 48.Putnam D. Polymers for gene delivery across length scales. Nat. Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 49.Ma X., Liu Y., Tittiger M., Hennig A., Kovacs A., Popelka S., Wang B., Herati R., Bigg M., Ponder K.P. Improvements in mucopolysaccharidosis I mice after adult retroviral vector-mediated gene therapy with immunomodulation. Mol. Ther. 2007;15:889–902. doi: 10.1038/sj.mt.6300112. [DOI] [PubMed] [Google Scholar]
- 50.Eastman S.J., Baskin K.M., Hodges B.L., Chu Q., Gates A., Dreusicke R., Anderson S., Scheule R.K. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum. Gene Ther. 2002;13:2065–2077. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- 51.Dulak J., Schwarzacher S.P., Zwick R.H., Alber H., Millonig G., Weiss C., Hugel H., Frick M., Jozkowicz A., Pachinger O., et al. Effects of local gene transfer of VEGF on neointima formation after balloon injury in hypercholesterolemic rabbits. Vasc. Med. 2005;10:285–291. doi: 10.1191/1358863x05vm630oa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aliño S.F., Herrero M.J., Noguera I., Dasí F., Sánchez M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- 53.Yoshino H., Hashizume K., Kobayashi E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006;13:1696–1702. doi: 10.1038/sj.gt.3302833. [DOI] [PubMed] [Google Scholar]
- 54.Suda T., Suda K., Liu D. Computer-assisted hydrodynamic gene delivery. Mol. Ther. 2008;16:1098–1104. doi: 10.1038/mt.2008.66. [DOI] [PubMed] [Google Scholar]
- 55.Kamimura K., Suda T., Xu W., Zhang G., Liu D. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol. Ther. 2008;17:491–499. doi: 10.1038/mt.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fabre J.W., Grehan A., Whitehorne M., Sawyer G.J., Dong X., Salehi S., Eckley L., Zhang X., Seddon M., Shah A.M., et al. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008;15:452–462. doi: 10.1038/sj.gt.3303079. [DOI] [PubMed] [Google Scholar]
- 57.Khorsandi S.E., Bachellier P., Weber J.C., Greget M., Jaeck D., Zacharoulis D., Rountas C., Helmy S., Helmy A., Al-Waracky M., et al. Minimally invasive and selective hydrodynamic gene therapy of liver segments in the pig and human. Cancer Gene Ther. 2008;15:225–230. doi: 10.1038/sj.cgt.7701119. [DOI] [PubMed] [Google Scholar]
- 58.Sawyer G.J., Zhang X., Fabre J.W. Technical requirements for effective regional hydrodynamic gene delivery to the left lateral lobe of the rat liver. Gene Ther. 2010;17:560–564. doi: 10.1038/gt.2009.167. [DOI] [PubMed] [Google Scholar]
- 59.Fabre J.W., Whitehorne M., Grehan A., Sawyer G.J., Zhang X., Davenport M., Rela M. Critical physiological and surgical considerations for hydrodynamic pressurization of individual segments of the pig liver. Hum. Gene Ther. 2011 doi: 10.1089/hum.2010.144. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 60.Chapman G.D., Lim C.S., Gammon R.S., Culp S.C., Desper J.S., Bauman R.P., Swain J.L., Stack R.S. Gene transfer into coronary arteries of intact animals with a percutaneous balloon catheter. Circ. Res. 1992;71:27–33. doi: 10.1161/01.res.71.1.27. [DOI] [PubMed] [Google Scholar]
- 61.Zhang G., Vargo D., Budker V., Armstrong N., Knechtle S., Wolff J.A. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum. Gene Ther. 1997;8:1763–1772. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- 62.Huang G.M., Li G.S., Zhu G.Y., Lai Y.M., Zhang H.X., Wang J., Wang H.R. Safety and bioactivity of intracoronary delivery of naked plasmid DNA encoding human atrial natriuretic factor. Acta Pharmacol. Sin. 2002;23:609–611. [PubMed] [Google Scholar]
- 63.Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L., Herweijer H., Wolff J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Hackett P.B., Urness M., Bell J.B., Aronovich E.L., Olson E.R., Gunther R., Hunter D., McIvor R.S. Long term gene expression in dogs after liver-directed hydrodynamic DNA delivery using a double-balloon catheter system. doi: 10.1089/hum.2017.003. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hackett P.B., Aronovich E.L., Hunter D., Urness M., Bell J.B., Kass S.J., Cooper L.C., McIvor R.S. Efficacy and safety of Sleeping Beauty transpson-mediated gene transfer in preclinical animal studies. Curr. Gene Ther. 2011 doi: 10.2174/156652311797415827. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nightingale S.J., Hollis R.P., Pepper K.A., Petersen D., Yu X.J., Yang C., Bahner I., Kohn D.B. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Staunstrup N.H., Moldt B., Mátés L., Villesen P., Jakobsen M., Ivics Z., Izsvák Z., Mikkelsen J.G. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol. Ther. 2009;17:1205–1214. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vink C.A., Gaspar H.B., Gabriel R., Schmidt M., McIvor R.S., Thrasher A.J., Qasim W. Sleeping Beauty transposition from nonintegrating lentivirus. Mol. Ther. 2009;17:1197–1204. doi: 10.1038/mt.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hausl M.A., Zhang W., Müther N., Rauschhuber C., Franck H.G., Merricks E.P., Nichols T.C., Kay M.A., Ehrhardt A. Hyperactive Sleeping Beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol. Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Silva S., Lotta L.T.J., Burris C.A., Bowers W.J. Virion-associated cofactor high-mobility group DNA-binding protein-1 facilitates transposition from the herpes simplex virus/Sleeping Beauty amplicon vector platform. Hum. Gene Ther. 2010;21:1615–1622. doi: 10.1089/hum.2010.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nienhuis A.W. Development of gene therapy for blood disorders. Blood. 2008;111:4431–4444. doi: 10.1182/blood-2007-11-078121. [DOI] [PubMed] [Google Scholar]
- 72.Xue X., Huang X., Nodland S.E., Mátés L., Ma L., Izsvák Z., Ivics Z., LeBien T.W., McIvor R.S., Wagner J.E., et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- 73.Wilber A., Linehan J.L., Tian X., Woll P.S., Morris J.K., Belur L.R., McIvor R.S., Kaufman D.S. Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells. 2007;25:2919–2927. doi: 10.1634/stemcells.2007-0026. [DOI] [PubMed] [Google Scholar]
- 74.Peng P.D., Cohen C.J., Yang S., Hsu C., Jones S., Zhao Y., Zheng Z., Rosenberg S.A., Morgan R.A. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yant S.R., Wu X., Huang Y., Daigle B., Garrison B.A., Burgess S.M., Kay M.A. Nonrandom insertion site preferences for the SB transposon in vitro and in vivo. Mol. Ther. 2004;9:S309. [Google Scholar]
- 76.Berry C., Hannenhalli S., Leipzig J., Bushman F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comp. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bestor T.H. Gene silencing as a threat to the success of gene therapy. J. Clin. Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garrison B.S., Yant S.R., Mikkelsen J.G., Kay M.A. Post-integrative gene silencing within the Sleeping Beauty transposition system. Mol. Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minoguchi S., Iba H. Instability of retroviral DNA methylation in embryonic stem cells. Stem Cells. 2008;26:1166–1173. doi: 10.1634/stemcells.2007-1106. [DOI] [PubMed] [Google Scholar]
- 80.Dalsgaard T., Moldt B., Sharma N., Wolf G., Schmitz A., Pedersen F.S., Mikkelsen J.G. Shielding of Sleeping Beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Mol. Ther. 2009;17:121–130. doi: 10.1038/mt.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walisko O., Izsvak Z., Szabo K., Kaufman C.D., Herold S., Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl Acad. Sci. USA. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilber A.C., Frandsen J.L., Geurts A.M., Largaespada D.A., Hackett P.B., McIvor R.S. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol. Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 83.Wilber A., Wangensteen K.J., Chen Y., Zhuo L., Frandsen J.L., Bell J.B., Chen Z.J., Ekker S.C., McIvor R.S., Wang X. Messenger RNA as a source of transposase for Sleeping Beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 2007;15:1280–1287. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
- 84.Ivics Z., Katzer A., Stuwe E.E., Fiedler D., Knespel S., Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol. Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 85.Yant S.R., Huang Y., Akache B., Kay M.A. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fraser M.J., Brusca J.S., Smith G.E., Summers M.D. Transposon-mediated mutagenesis of a baculovirus. Virology. 1985;145:356–361. doi: 10.1016/0042-6822(85)90172-2. [DOI] [PubMed] [Google Scholar]
- 87.Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M., et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Galvan D.L., Nakazawa Y., Kaja A., Ketlun C., Cooper L.J.N., Rooney C.M., Wilson M.H. Genome-wide mapping of piggyBac transposons in primary human T cells. J. Immunother. 2010;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang X., Guo H., Tammana S., Jung Y.C., Mellgren E., Bassi P., Cao Q., Tu Z.J., Kim Y.C., Ekker S.C., et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Groth A.C., Olivares E.C., Thyagarajan B., Calos M.P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chalberg T.W., Portlock J.L., Olivares E.C., Thyagarajan B., Kirby P.J., Hillman R.T., Hoelters J., Calos M.P. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 92.Ehrhardt A., Engler J.A., Xu H., Kay M.A. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum. Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- 93.Liu J., Jeppesen I., Nielsen K., Jensen T.G. phiC31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]