esBAF Facilitates Pluripotency by Conditioning the Genome for LIF/STAT3Signalingand by Regulating Polycomb Function (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 1.
Published in final edited form as: Nat Cell Biol. 2011 Jul 24;13(8):903–913. doi: 10.1038/ncb2285
Abstract
Signaling by the cytokine LIF and its downstream transcription factor, STAT3, prevents differentiation of pluripotent embryonic stem cells (ESCs) by opposing MAP kinase signaling. This contrasts with most cell types where STAT3signaling induces differentiation. We find that STAT3binding across the pluripotent genome is dependent upon Brg, the ATPase subunit of a specialized chromatin remodeling complex (esBAF) found in ESCs. Brg is required to establish chromatin accessibility at STAT3 binding targets, in essence preparing these sites to respond to LIF signaling. Moreover, Brg deletion leads to rapid Polycomb (PcG) binding and H3K27me3-mediated silencing of many Brg-activated targets genome-wide, including the target genes of the LIF signaling pathway. Hence, one crucial role of Brg in ESCs involves its ability to potentiate LIF signaling by opposing PcG. Contrary to expectations, Brg also facilitates PcG function at classical PcG target including all four Hox loci, reinforcing their repression in ESCs. These findings reveal that esBAF does not simply antagonize PcG, but rather, the two chromatin regulators act both antagonistically and synergistically with the common goal of supporting pluripotency.
Pluripotency requires an intricate interplay between a specialized transcriptional circuitry, signaling pathways, and non-redundant mechanisms of ATP-dependent chromatin remodeling 1, 2. In mammals, Brg is one member of a large family of DNA-dependent ATPases that are homologs of the yeast SWI2/SNF2 gene 3 and is assembled with 11 other subunits into BAF (Brg/Brahma Associated Factors, also known as mSWI/SNF) complexes4. In ESCs, BAF complexes have a unique subunit composition (termed esBAF) not found in other cell types 5,6, 7. Altering this subunit composition causes a reduction in self renewal and pluripotency 5. Conversely, adding esBAF subunits to fibroblasts facilitates reprogramming to pluripotent cells 8. Because Brg is always found only within BAF complexes 9 and provides the essential catalytic activity of the complex, and because its homolog Brm is not expressed in ESCs 5, the functions of esBAF can be inferred through studies of the role of Brg in ESCs. Recent studies in mouse ESCs (mESCs)indicate that esBAF is functionally and biochemically specialized, and facilitates the pluripotent state by interacting with pluripotency transcription factors and participating in the core transcriptional network of ESCs 5, 10–14,. However, the mechanisms used by esBAF in facilitating the pluripotent state remain unclear.
The cytokine LIF and its downstream signaling/transcriptional effector, STAT3, are both essential for the establishment and maintenance of pluripotent cells, and withdrawal of LIF leads to rapid differentiation of mESCs. Recent studies have shown “naïve” human embryonic stem cells (hESCs)and reprogramming of human fibroblast into naïve hESCs are also LIF dependent15–17,indicating that the importance of LIF/STAT3extends to pluripotency in humans. STAT3 is a ubiquitous transcription factor involved in diverse developmental processes. However, in contrast to the role of STAT3in other cell types, where it is often dedicated to differentiation, STAT3signaling in ESCs opposes differentiation signals provided by the ERK/MAPK (Mitogen Activated Protein Kinase) pathways 18. The observed genome-wide colocalization of Brg and STAT311 suggests that the LIF signaling pathway is integrated with ATP-dependent remodeling, and raises the intriguing possibility that the specialized outcome of STAT3signaling in ESCs (i.e. pluripotency) is dependent on esBAF.
Polycomb and Trithorax complexes play classical antagonistic roles in development. The SWI/SNF ATPase in Drosophila, Brahma, is known to antagonize the function of Polycomb (PcG)during fly development and is hence classified as a Trithorax protein. PcG proteins play essential roles in stem cells by repressing differentiation genes 19 and by regulating genes involved in metabolism and cell proliferation 20. Consistent with the studies of PcG and Brm in Drosophila, we observed that esBAF avoids cobinding with PcG components genome-wide in mESCs11, suggesting that like in flies, they have extensive antagonistic functions in ESCs. However, whether the critical functions of PcG and BAF in ESCs involve their mutual antagonism is unknown. The mechanism underlying the antagonism between PcG and mSWI/SNF or BAF has also been controversial 21–23.
In this study, we present evidence that esBAF prepares the ESC genome to receive LIF/STAT3 signals by opposing PcG repression of these targets. In contrast to expectations Brg acts both synergistically and antagonistically with PcG, with the common goal of sustaining a transcription network that is compatible with pluripotency.
Results
Coordinate Regulation of esBAF and LIF Target Genes
Binding sites for Brg and the transcription factor STAT3overlap extensively throughout the ES cell genome11.This reflects some degree of dedication of Brg to STAT3, since the observed/expected overlap for Brg and STAT3 are substantially higher than for Brg and RNA polymerase II (PolII) (Figure S1a). To investigate the functional significance of the observed co-occupancy of Brg and STAT3, we determined if they transcriptionally regulate a similar set of genes in ESCs. In order to completely remove the Brg protein, we prepared an ESC line from Brglox/lox;Actin-CreER mice, which we will refer to as (Brgcond),in which the ATPase Brg is acutely deleted by homologous recombination following 4-hydroxytamoxifen (4-OHT)treatment. Depletion of Brg leads to near complete loss of Oct4, Sox2 and Nanog expression and pluripotency after about 10 days11. However, by studying acutely deleted ESCs before the pluripotency transcriptional circuit is shut down (Figure 1a), we defined a set of Brg-dependent genes by comparing wildtype Brgcond with 4-OHT-treated Brgcond ESCs. We also defined a set of LIF-dependent genes in the Brgcond ESC line by performing microarray analysis of cells starved of LIF for 48 hours compared to cells grown with LIF. Surprisingly, Brg and LIF corepress and coactivate a large number of common genes (Figure S1b, p-value<5E-212, hypergeometic distribution). This extensive transcriptional co-regulation of target genes is depicted using a 2D gene density heatmap in Figure 1b, which shows that Brg-deleted or LIF-starved ESCs undergo very similar changes in the global transcriptional profile, suggesting that Brg could be required for LIF-dependent STAT3-mediated gene regulation. Of these, the majority of direct cotarget genes(i.e. genes cobound by Brg and STAT3, see Methods) were in fact coactivated by the two proteins (Figure 1c; genes in the bottom-right corner, BR1, see Methods and Table S1). Furthermore, this coactivated group includes a number of genes essential for pluripotency including Esrrb, Tcl1, Tbx3, c-Kit and others. Our previous studies using shRNA-mediated depletion of Brg showed that esBAF is predominantly represses transcription and tonically suppresses ESC-specific genes to maintain the pluripotent transcriptional circuitry11. Our current studies confirm the role of esBAF as a tonic repressor (Figure S1c), but the complete depletion of Brg by conditional genetic deletion revealed that esBAF acts equally as a transcriptional activator (Figure S1c), revealing possible differences in the dose dependency of Brg-mediated activation versus repression.
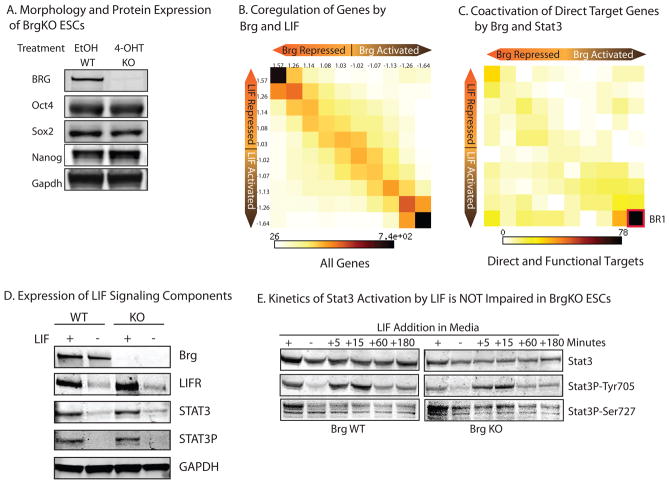
Figure 1. esBAF Is Dedicated to the LIF/STAT3 Signaling Pathway.
A) Western blot showing protein levels of Brg and pluripotent markers after 72hours of treatment of Brgcond ESCs with 4-OHT (BrgKO) or EtOH (BrgWT) vehicle control. Brg protein is completely absent only after 48hours of 4-OHT treatment (data not shown). Full length blots are presented in Figure S9a.
(B) 2D matrix and heatmap depicting gene expression changes in BrgKO ESCs and 48hr LIF-starved ESCs compared to WT ESCs for all genes (N=17030). Axes indicate degree of fold change, from nil (middle of axis) to greater than 1.5-fold (outermost square). Numbers indicate the median fold change of genes in each column or row. Intensity of each square represents the number of genes that fall in that square.
(C) 2D matrix and heatmap of direct STAT3and Brg targets (binding sites detected from TSS to TES of the same gene) depicting changes in their expression in BrgKO or LIF-withdrawn ESCs. BR1= bottom right 1 corner.
(D) STAT3Protein levels in WT versus KO ESCs (72 hours 4OHT
STAT3P=Phosphotyrosine705 STAT3; LIFR=LIF receptor)in the presence of LIF (+) or after 18 hours of LIF starvation (−). Full length blots are presented in Figure S9b.
(E)Timecourse of STAT3activation in BrgWT and KOESCs. Cells were starved for 18 hours from LIF (−), followed by LIF restimuation for the indicated durations. Full length blots are presented in Figure S9c.
Because Brg and BAF are generally thought to function at active promoters, the ability of Brg to coactivate genes with STAT3 might be non-specific and true of Brg and all other ESC transcriptional regulators. However, this is not the case, since we did not observe the same trend of transcriptional coregulation when we performed the same analysis using ESCs depleted of Oct424, 25 or Sox221. Rather, Brg functions with Oct4 and Sox2 to both repress and activate a large number of common and direct targets, but also in opposition to Oct4 and Sox2 at a different subset of targets (Figure S1d). The highly overlapping gene expression pattern between Brg KO and LIF starvation is also not due to impaired LIF/STAT3signal transduction in Brg KO ESCs, since Brg KO ESCs express normal levels of the LIF chimeric receptor consisting of gp130 and LIFR (Figures 1d and S1e). Brg KO ESCs have normal levels of both total and phosphorylated (i.e. activated) STAT3(Figures 1d and S1e), normal kinetics of STAT3phosphorylation following LIF stimulation (Figure 1f), and normal nuclear localization of phosphorylated STAT3(Figure S1f). Lastly, Brg KO ESCs are not merely undergoing spontaneous differentiation because we did not observe a positive correlation of gene expression between BrgKO ESCs and differentiated embryoid bodies (Figure S1g).
esBAF participates in STAT3-mediated activation of genes on target chromatin
To further investigate the mechanism of STAT3and Brg coactivation, we examined the genes most dramatically and directly regulated by Brg and LIF(i.e. BR1 genes). While mostBR1genes showed LIF-responsive reactivation in WT ESCs following LIF starvation, this response is generally lost in Brg KO ESCs(Figure S2a). Hence, STAT3-dependent gene activation is impaired in the absence of functional esBAF complexes.
We expanded these findings genome-wide by performing microarray analyses of LIF-starved WT and KO ESCs, followed by a 30 minute LIF restimulation (Figure S2b). Indeed, a large number of genes showed defective upregulation in response to LIF in Brg KO ESCs. More strikingly, many genes that are not LIF-responsive in WT ESCs are instead activated in response to LIF in BrgKO ESCs (blue box, “ectopic STAT3 activation”), suggesting that Brg is essential for ensuring that the correct complement of STAT3 targets involved with pluripotency are activated in ESCs in response to LIF.
esBAF is Crucial for Genome-Wide STAT3Binding to Target Chromatin
The loss of LIF responsiveness at genes coactivatedbySTAT3and Brg prompted us to examine the ability of STAT3to bind its target sites on chromatin in the absence of Brg. ChIP studies showed that STAT3binding is severely impaired at many of the coactivated gene targets from BR1group in Brg KO ESCs compared to WT (Figure 2a). Moreover, genome-wide ChIP-Seq analysis of STAT3in Brg WT and Brg KO ESCs revealed that out of 2730 STAT3binding sites in Brg WT ESCs, only 442 (16%) are preserved in the KO (Figure 2b),with an emergence of 103 new sites in Brg KO (Figure S3a). Hence, Brg is required to shape STAT3-responsiveness in ESCs by enabling it to bind to its appropriate targets. Even among the 442 sites judged to be BRG-independent, we found reduced STAT3 binding after Brg deletion (Figure 2b) indicating that Brg-independence is a quantitative and not qualitative difference.
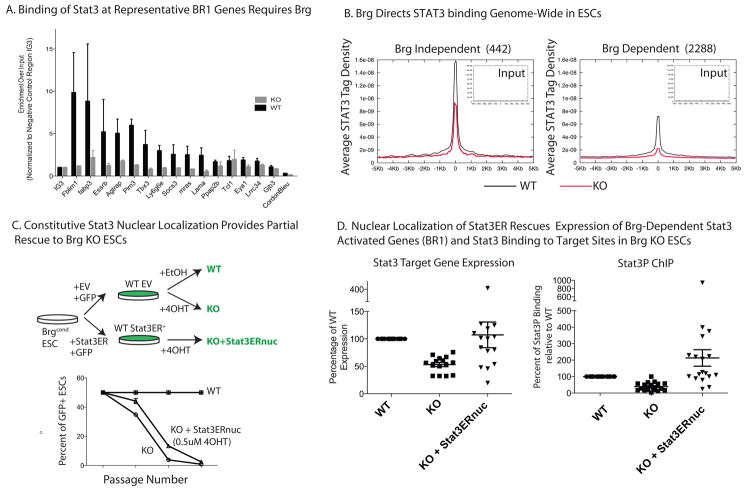
Figure 2. STAT3Binding Genome-Wide Is Brg-Dependent.
(A) ChIP assay of STAT3 target regions within BR1 genes in BrgKO and WT ESCs. Y-axis represents input enrichment over input normalized to negative control IG3 region. Error bars = SEM of 3 experiments. See text and SI for gene selection criteria.
(B) High resolution ChIP-Seq for total STAT3 levels in BrgWT and KO ESCs. Average tag density (y-axis) of each site called with p<0.01 is plotted against distance in Kb from the center of each STAT3binding sites for WT (black) and KO (grey).
(C) Experimental scheme to generate GFP+ Brg WT and KO ESCs expressing the STAT3ER fusion protein. GFP+ cells of the indicated genotype were mixed with GFP- WT ESCs at a 1:1 ratio and the GFP ratio of cultures grown in the presence or absence of 4-OHT was measured at each passage by FACS. Error bars = SEM of 3 technical replicates. Results are representative of 2 independent experiments.
(D) mRNA levels of STAT3/Brg cobound and coactivated targets were measured in WT, KO and KO;STAT3ERnuc (nuclear) and expressed as a percentage of WT levels. Each data point represents a distinct gene from BR1. Error bars = SEM of data points. (E) ChIP assay for STAT3P-Tyr705 in WT, KO and KO;STAT3ERnuc. ChIP levels are measured as percent of input, normalized to that of a negative intergenic control IG3, and expressed as a percentage of WT levels.
Consistent with early studies of SWI/SNF recruitment, the levels of esBAF components, Brg, BAF155 and BAF57 are reduced at STAT3/Brg cobinding sites upon LIF withdrawal, albeit only moderately (Figure S3b). Hence, we do not rule out the possibility that STAT3 plays a minor role in recruiting esBAF. This genome-wide analysis revises the commonly accepted view that SWI/SNF-like complexes are solely recruited by transcription factors. Rather, we favor an interdependent model of recruitment, where STAT3 and Brg binding are mutually reinforced. However, the binding of STAT3 is strictly dependent on Brg.
Enhancement of LIF/STAT3Provides Partial Rescue to Brg KO ESCs
Brg regulates a large number of genes that are unrelated to LIF signaling and indeed Brg cobinds many target genes with Oct4, Sox2, Nanog and Smad111. Hence, to determine if the observed contribution of Brg to STAT3 recruitment is physiologically relevant, we tested the ability of increasedSTAT3 activity to partially rescue the self-renewal defects observed in Brg KO ESCs. We constructed a Brgcond ESC line expressing GFP and STAT3-ER, which is inducibly localized to the nucleus in the presence of 4OHT 26 (Figure 2c) and is sufficient to maintain WT ESCs in an undifferentiated state in the absence of LIF (Figure S3c). In Brgcond STAT3ER ESCs, addition of 4OHT simultaneously induces Brg deletion andSTAT3ERnuclear localization. When we put these GFP-positive Brg KO STAT3ER ESCs and Brg KO GFP-empty vector control ESCs in competitive growth with WT GFP-negative ESCs in the presence of 4-OHT, we noticed a small but significant and reproducible increase in the proportion of Brg KO ESCs expressing STAT3ER compared to the empty vector control (Figure 2c), indicating that STAT3ER expression partially alleviates the self-renewal defect in BrgKO ESCs. In addition, the transcription levels of many Brg-dependent STAT3-activated targets and phospho-STAT3 binding was rescued by the enforced nuclear localization of STAT3ER (Figure 2d). These results confirm that one of Brg’s functions in ESCs is to facilitate STAT3-mediated transcriptionunder physiological conditions. However, the incomplete rescue by STAT3 implies that Brg is also involved in other pathways required for ESC maintenance in addition to LIF/STAT3signaling, as evidenced by its extensive cobinding with other regulators of pluripotency, such as Oct4 and Sox2.
Brg-Dependent STAT3 Sites Require Brg-Mediated Chromatin Accessibility for Binding
Our ChIP-Seq studies reveal that not all STAT3 sites are equally dependent on Brg for binding. Why doSTAT3sites display a variable dependence on Brg for STAT3 binding? Brg-dependent STAT3sites have a lower average tag density compared to Brg independent sites (Figure 3a). Tag density of a protein reflects the average protein occupancy of a site, and a lower tag density can indicate lower binding affinity to the chromatinized DNA binding site27. This observation suggests that lower affinity or less stable STAT3sites are more dependent on Brg for STAT3binding. What then is the underlying difference between high-affinity versus low-affinitysites? Previous studies in yeast found that SWI/SNF-dependency of a transcription factor at a particular site inversely correlates with how closely the site matches the consensus binding motif 28. The STAT3consensus motif derived from our dataset, TTCCNGGAA (Figure 3b), matches the published consensus motif 29. 65% of Brg-dependent STAT3 binding sites contain the consensus motif, as does 66% of the Brg-independent sites. Hence, the observed difference in affinity is not simply due to differences in the underlying sequence of the STAT3binding site.
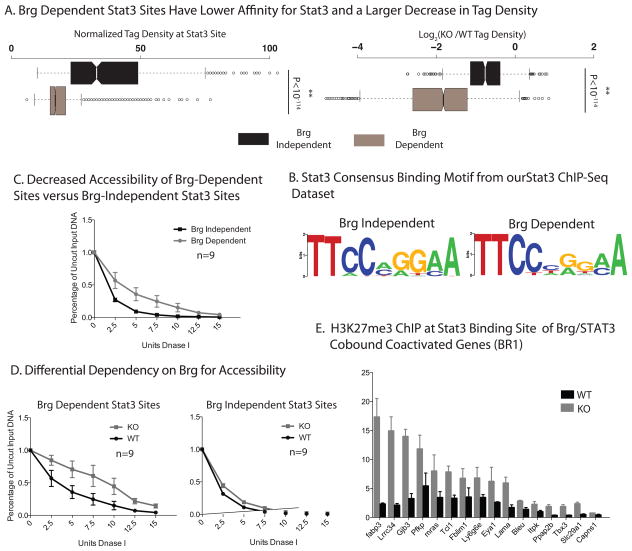
Figure 3. Brg is essential to enhance accessibility at STAT3target genes.
(A)Brg Dependency Corelates with Tag Density of STAT3 Sites. (Left) Box whisker plot of ChIP-seq tag numbers of each STAT3 site in Brg WT ESCs, grouped according to Brg dependency. (Right) Fold change of tag density of each STAT3site in BrgKO ESCs compared to WT ESCs, grouped according to Brg dependency. P-values are calculated using a hypergeometric distribution.
(B) Consensus STAT3 binding MOTIFS were calculated by MEME49 using STAT3 ChIP-Seq dataset from both Brg WT and KO ESCs.
(C)DnaseI hypersensitivity assay of Brg-dependent and Brg-Independent STAT3binding sites (n=9 each) (See text and SI for gene selection criteria.). Error bars, s.e.m. of data for nine sites obtained in two experiments.
(D)DnaseI assay of Brg dependent and independent sites (n=9 each) in WT ESCs or Brg KO ESCs. Error bars, s.e.m. of data for nine sites obtained in two experiments.
(E) H3K27me3 ChIP at the STAT3 binding site of representative Brg-and LIF coactivated genes in WTand BrgKO ESCs. Y-axis represents ChIP/Input ratio for each region, normalized with the ratio at the GAPDH promoter. Error bars = SEM of 3 independent experiments.
One of the functions of chromatin remodeling complexes is to regulate DNA accessibility. Indeed, in murine ESCs, more than 80% of Brg binding regions coincide with a DNaseI hypersensitive site 30 (Figure S4a). We hypothesized that Brg-dependent STAT3sites have lower affinity due to lower intrinsic chromatin accessibility. Hence, we tested the accessibility of Brg-dependent versus Brg-independent STAT3sites using a DNaseI hypersensitivity assay coupled to QPCR amplification of the target region to quantitatively assess the “openness” of a region 31. Indeed, Brg-dependent STAT3sites are less accessible compared to Brg-independent STAT3sites, as shown by their greater resistance to digestion by DNaseI (Figure 3c), Brg-dependent STAT3 sites show a reproducibly higher decrease in accessibility in BrgKO ESCs than Brg-independent STAT3 sites (Figure 3d and Figure S4b). The observed decreased accessibility is a direct consequence of the loss of Brg and not loss of STAT3 and transcription of the nearest target gene, since these sites do not lose accessibility in WT ESCs starved of LIF (Figure S4c top), which results in a loss of STAT3 binding and attenuation of transcription (Figure S4c bottom). Hence, Brg prepares most STAT3 sites for phospho-STAT3 dimers entering the nucleus in response to LIF by making these sites accessible.
Brg prevents active silencing of STAT3 and Brg Coactivated target Genes by H3K27me3
The observation that esBAF is required to mediate accessibility of chromatin for STAT3binding prompted us to examine histone modifications in BrgKO ESCs. Brg deletion did not result in a significant change in eitherH3K4me3 or H4acetylationmarks at the promoters of Brg-dependent STAT3-activated targets, indicating that activation marks are not generally affected by the absence of esBAF (Figure S5a). However, we observed a general increase of the repressive modification H3K27me3 at STAT3sites (Figure 3e) and promoters (Figure S5b) of many Brg-dependent STAT3-activated targets after Brg deletion, suggesting that many STAT3 target genes, in addition to requiring Brg for continuous STAT3-mediated transactivation, are actively silenced in the absence of Brg.
Brg Regulates Levels of H3K27me3 at its Target Genes
H3K27me3 is deposited by Polycomb Repressive Complex 2 (PRC2). Hence, these results are consistent with our earlier studies where we found that Brg and the PRC2 component, Suz12, avoided cobinding over the genome of ES cells11. We were initially skeptical of the lack of colocalization between Brg and Suz12/PRC2, since Brg extensively colocalizes with Oct4/Sox2, while Suz12/PRC2also colocalizes with Oct4/Sox2 in human ESCs 32. However, when we compared the target gene overlap of Suz12/PRC2 29, 33, Oct4/Sox2 29 and Brg in mESCs, we found that while Suz12/PRC2does co-occupy some Oct4/Sox2 targets, the overlap is far less than that between Oct4/Sox2 and Brg (Figure S5c), providing an explanation for the apparent discrepancy between reports. Since the functional interaction between BAF and Polycomb is an outstanding question, we expanded our findings to a genome-wide scale by ChIP-seq mapping of the histone mark H3K27me3 in Brg WT and KO ESCs. Examination of functional Brg binding regions (i.e. Brg sites that can be assigned to a nearby target gene whose expression is Brg-dependent, see Methods), revealed significant changes in the levels of H3K27me3 at Brg binding regions and also at the transcription start sites(TSS) of corresponding Brg-regulated genes (Figure 4). These changes occur directly over genomic regions with highest levels of Brg binding (Figure S5d), suggesting that Brg directly influences local levels of H3K27me3. Surprisingly, Brg deletion does not lead solely to an increase in H3K27me3 at Brg target genes, as would be expected if Brg and PRC2 solely antagonized each other. Rather, we observed both a significant decrease in H3K27me3 at Brg-repressed genes (Figure 4a), and an even more significant increase in H3K27me3 at Brg-activated genes (Figure 4b). In both cases, the extent of the change in H3K27me3 is directly proportional to the degree of change in expression of Brg-activated genes upon Brg deletion. Unlike in Snf-5 deficient tumors23, acute deletion of Brg does not lead to upregulation of PRC2 component Ezh2 or Suz12 (Figure S5e). Consistent with this finding, bulk levels of H3K27me3 levels are not increased in BrgKO ESCs (Figure S5f). By comparing the total number of normalized ChIP-Seq tags recovered from WT and KO ESCs in contiguous 0.5Mb windows, we confirmed that there is no overall genome-wide increase in H3K27me3 levels in BrgKO ESCs (Figure S5g). Hence, PRC2 activity does not generally increase in BrgKO ESCs, in contrast to the general increase in PRC2 expression and activity in _Snf5-_deficient tumors, revealing inherent differences in BAF and PcG antagonism in non-transformed versus transformed cell types.
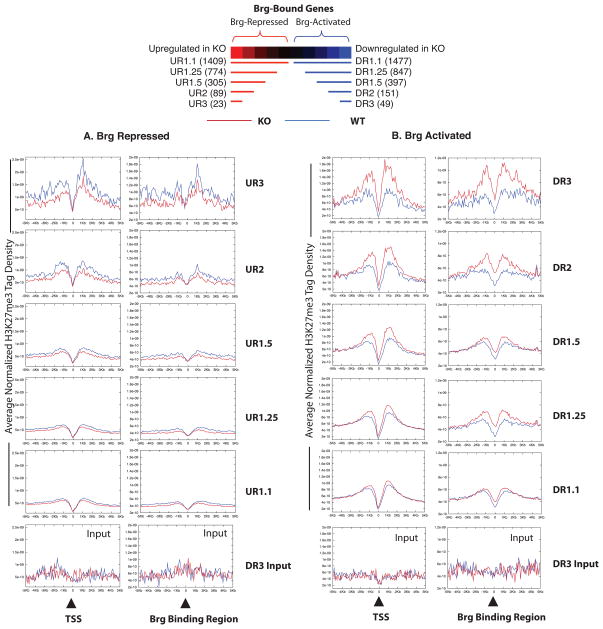
Figure 4. Brg Deletion Leads to Genome-wide Increased H3K27Me3 at Brg –Activated Genes and Reduced H3K27Me3 at Brg-Repressed Genes.
Average H3K27me3 tag density at transcriptional start site (TSS) of Brg-repressed (A) or Brg-activated (B)genes (defined as Brg-bound genes that undergo transcriptional changein BrgKO ESCs) and atthe corresponding Brg binding regions in WT (blue) versus Brg KO (red) ESCs. These genes were grouped according to the fold changein BrgKO ESCs (i.e. DR3 = 3-fold DOWNregulated, UR3 = 3-fold UPregulated etc.) The number within parentheses besides each set identifier (top panel) denotes the number of genes within that set. Bottom panel illustrates average input tag density of UR3 (for panel A) or DR3(for panel B)genes and is representative of other subsets.
Brg Acts Synergistically with Polycomb at all 4 Hox Loci
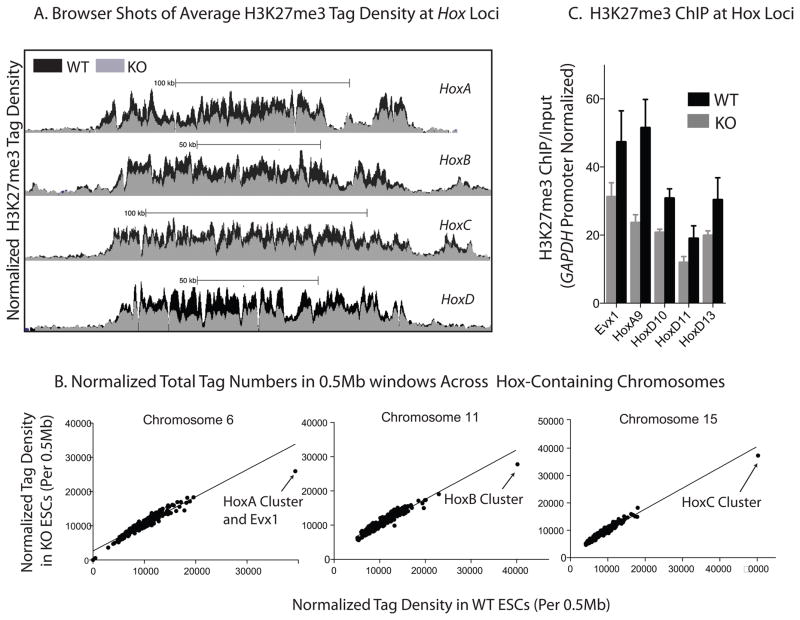
Our results suggest that in contrast to expectations, Brg and PRC2 act both synergistically and antagonistically. Surprisingly, Brg synergizes with PRC2 over all four Hox chromosomal loci (Figure 5a). Although there is no global decrease in H3K27me3 in BrgKO ESCs, we found a dramatic decrease in total normalized tag numbers derived from genomic regions containing the Hox clusters in BrgKO compared to WT ESCs (Figure 5b). This was confirmed by ChIP assays (Figure 5c)at representative Hox genes, suggesting that Brg aids in the silencing of PcG-repressed genes in ESCs in order to prevent premature differentiation. Consistent with this finding, deletion of Brg results in the transcriptional derepression of these representative Hox genes (Figure S5h),
Figure 5. Synergistic Interaction Between Brg and PRC2at Hox Genes.
(A) Browser snapshots of average normalized H3K27me3 ChIP-Seq tag density in KO (grey) and WT (black) ESCs at the four Hox loci.
(B) Scatter plots of H3K27me3 levels on _Hox-_containing chromosomes. Each point represents the total number of tags in a particular 0.5Mb window in Brg KO (y-axis) and the total number of tags in the corresponding 0.5Mb window in Brg WT (x-axis) ESCs. If a point falls on the diagonal of the plot, there is a similar overall tag number in that window in Brg KO ESCs compared to WT. The datapoints corresponding to the window containing Hox genes are labeled.
(C) H3K27me3 ChIP at the transcriptional start site of Brg-repressed Hox genes in WTand BrgKO ESCs. Y-axis represents ChIP/Input ratio for each region, normalized with the ratio at the GAPDH promoter, which is not H3K27me3 modified in WT or KO ESCs and serves as a negative baseline internal control. Error bars = SEM of 3 independent experiments.
Brg Antagonizes Polycomb at STAT3 and other targets
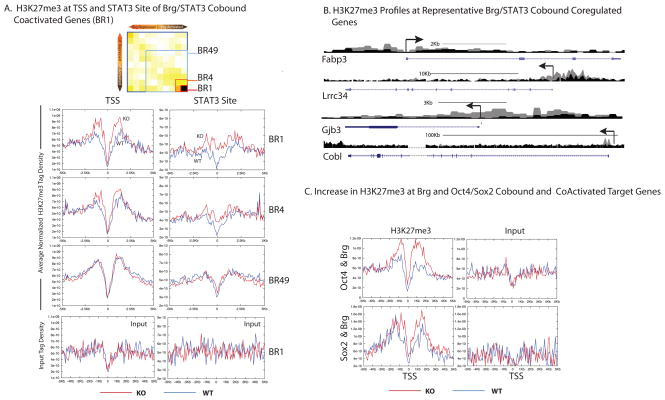
To examine the genes where BRG and PRC2 appear to act antagonistically, we expanded the above analysis of STAT3 target genes in Figure 3f to include all genes in BR1. Indeed, at the TSS (Figure 6a, top left) and STAT3 site (Figure 6a top right) of such genes, we found a significant overall increase in the H3K27me3 levels in Brg KO ESCs compared to WT ESCs. Once again, the changes in H3K27me3 occur over genomic regions containing high levels of Brg binding, suggesting a direct effect of Brg on H3K27me3 (Figure S6a). The extent of increase in H3K27me3 appears to be proportional to the degree of activation by Brg (Figure 6a). Hence, the increase in H3K27me3 is tightly correlated to the transcriptional status of a particular gene in BrgKO ESCs. The increase in H3K27me3 does not simply reflect reduced transcription through these areas, since a time-course analysis of the increase in H3K27me3 after Brg deletion at representative BR1 genes (Figure S6b) revealed that the gain of H3K27me3 mark occurs concurrently with transcriptional downregulation, and might be a cause rather than an indicator of silencing. Morever, H3K27me3 does not increase at these genes when LIF is withdrawn from WT ESCs (Figure S6c), again indicating that the gain of H3K27me3 is not simply reflective of a loss of transcription. These observations suggest that in the absence of Brg, H3K27me3 expands into STAT3 and Brg-coactivated target genes, leading to the formation of repressive chromatin and transcriptional silencing. Representative browser shots of BR1 genes (Figures 6b and S7) show that the increase in H3K27me3 is usually focused at or around the promoter (usually the TSS, shaded boxes) but extends on average over a stretch of 2-4Kb.
Figure 6. Increased H3K27me3 At Brg and STAT3 Coactivated Genes in BrgKO ESCs.
(A) High resolution ChIP-Seq for H3K27me3 levels in BrgWT (blue) and KO (red) ESCs. Stat3 and Brg cobound target genes were grouped according to their degree of coactivation by Brg and Stat3. BR1=highly co-activated to BR49 = not coactivated. Average normalized H3K27me3 tag density across the TSS and over the STAT3 sites of BR1, BR4 and BR49 genes were plotted (y-axis) against the distance in Kb (x-axis) from the TSS or STAT3 site. Lowest panel depicts average input tag density of BR1 genes, and is representative of all subsets.
(B) UCSC genome browser shots of H3K27me3 profiles at representative BR1 genes in WT (black) and KO (grey) ESCs.
(C) Average H3K27me3 tag desity at TSS of all Brg and Oct4 (top, n=70) and Brg and Sox2 (bottom, n=13) co-bound coactivated genes in WT ESCs (blue) and BrgKO ESCs (red). Oct4 and Sox2 ESCs sites are from ChIP-Seq datasets from46.
Although we have so far used STAT3 as a specific example of Brg and PRC2 antagonism, the increase in H3K27me3 at Brg-activated genes is not limited to STAT3 target genes. Although Brg and Oct4/Sox2 act mostly antagonistically11, there are a small number of genes where these two factors cobind and coactivate the nearest target gene. At the TSS of such genes, we also find an increase in H3K27me3 in BrgKO ESCs compared to WT ESCs (Figure 6c). Hence, PRC2-mediated silencing is not limited to STAT3-activated target genes, but extends to genes coactivated by Brg and Sox2 or Oct4. Brg generally prevents silencing of its actively transcribed target genes by preventing the encroachment of PRC2. Because STAT3 and Brg coactivate a large number of genes, the relevance of this antagonism to the STAT3 pathway is more pronounced and significant, even if it is not exclusive to STAT3 targets. Therefore, we continued to use the STAT3 pathway as a model for functional validation of antagonism between esBAF and PRC2.
esBAF Antagonizes the Activity of PRC2 Complex at STAT3Target Genes
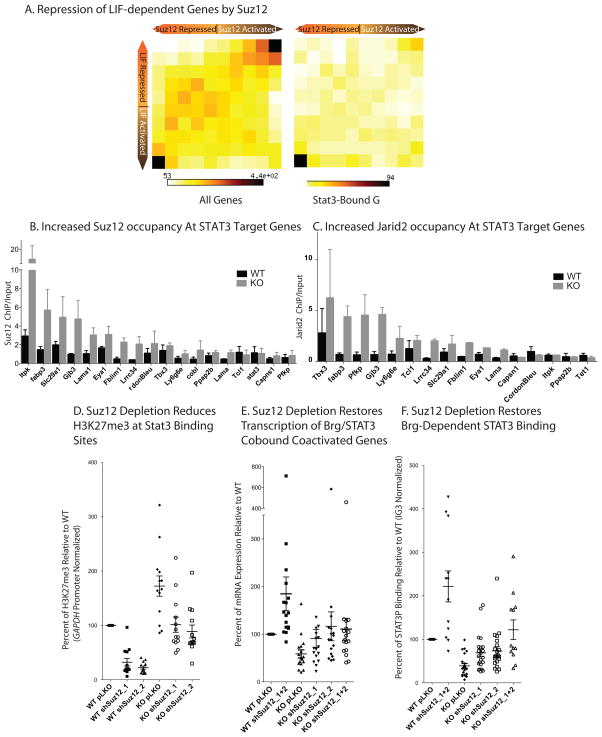
Since the only enzymes known to catalyze the formation of H3K27me3 are Ezh1 and Ezh2 of PRC2, we hypothesized that PRC2is responsible for silencing of STAT3-dependent transcription in BrgKO ESCs. Although PRC2 is not known to regulate LIF signaling, microarraydataofSuz12KO ESCs from Helin and colleagues 34 revealed, surprisingly, that over 70% of Suz12-repressed genes (i.e. up-regulated in Suz12KO ESCs) are LIF-activated (i.e. down-regulated after 48 hours of LIF withdrawal). A 2D gene density heatmap visualization of the genes regulated by Suz12 and LIF revealed that LIF signaling appears to be opposed by Suz12 (Figure 7a, left). In particular, Suz12 appears to repress direct STAT3 targets (Figure 7a bottom right), which contrasts dramatically with the role of Brg. This strongly suggests that Suz12 has previously unappreciated roles in modulating LIF signaling, perhaps by providing transient or basal antagonism of LIF-responsive genes, which might be critical for mediating exit from the pluripotent state. Consistent with these findings, we observed a significant increase in the levels of Suz12 at STAT3 target genes in Brg KO ESCs (Figure 7b) and increased binding of Jarid2 (Figure 7c), which has been implicated in recruiting PRC2to its targets 35–38. Again, this is not due to the loss of STAT3binding, and does not reflect spontaneous differentiation by LIF withdrawal (Figure S8a)suggesting that the increased recruitment of PRC2and the deposition of H3K27me3 precedesthe loss of STAT3binding. Hence, at Brg/STAT3 cobound and co-activated genes, Brg either antagonizes theactivity of low levels of PRC2complex already present at the site,or prevents aninappropriate increase in PRC2recruitment which would otherwiselead to silencing of STAT3target genes.
Figure 7. Opposing activity and localization of esBAF and PRC2 complexes.
(A) 2D Matrices depicting the gene expression changes comparing Suz12 KO ESCs (Pasini et al., 2007) and 48hr LIF-starved ESCs for all genes (left) and STAT3 bound direct targets (right).
(B) Suz12 ChIP at STAT3 binding sites of BR1 genes in WT (black) and BrgKO (grey) ESCs. Error bars = SEM of 3 biological replicates. Y-axis represents ChIP/Input ratio for each region, normalized with the ratio at the GAPDH promoter.
(C) ChIP assay of PRC2 component Jarid2 at Brg-dependent STAT3 sites in BrgWT versus BrgKO (grey) ESCs. Y-axis represents ChIP/Input ratio for each region, normalized with the ratio at the GAPDH promoter. Error bars = SEM of 2 biological replicates.
(D) WT ESCs were infected with control (pLKO) or 2 distinct anti-Suz12 shRNA (shSuz12_1 and shSuz12_2) expressing lentiviruses either separately or together (shSuz12_1 +2). Brg deletion was induced with 4OHT after stable knockdown of Suz12 was achieved. 72 hours post 4OHT treatment, cells of the indicated genotype were harvested for H3K27me3 ChIP assay (D), transcript levels (E), and STAT3P ChIP assay (F) at Brg-dependent STAT3 binding sites in BR1. Each point represents a distinct STAT3 target gene ortarget site.
According to this model, removing PRC2activity in BrgKO ESCs should prevent silencing of Brg and STAT3 cobound and coactivatedgenes and rescue their transcription. To this end, we peformed shRNA-mediated knockdown of the PRC2component Suz12 using two distinct shRNA constructs (Figure S8b). We chose Suz12 because of its role in maintaining the stability and activityof the PRC2complex 39 and because of its apparent role in antagonizing LIF signaling. In BrgKO ESCs, Suz12 depletion was sufficient to reduceH3K27me3 levels back to WT levels at affected Brg-dependent STAT3 sites (Figure 7d), indicating that increased H3K27me3 levels at these STAT3 binding sites is PRC2-dependent. As our observations would predict, Suz12 depletion in BrgKO ESCs resulted in partial to complete transcriptional rescue of Brg-dependent STAT3 target genes (Figure 7e). Concurrently, the levels of STAT3binding to its target sites, previously lost in the BrgKOESCs, were partially to fully restored (Figure 7f). These results suggest that Brg is required to prevent PRC2 and H3K27me3-mediated heterochromatinization of STAT3 target sites in ESCs so that these sites remain accessible to STAT3 and responsive to LIF signaling.
Discussion
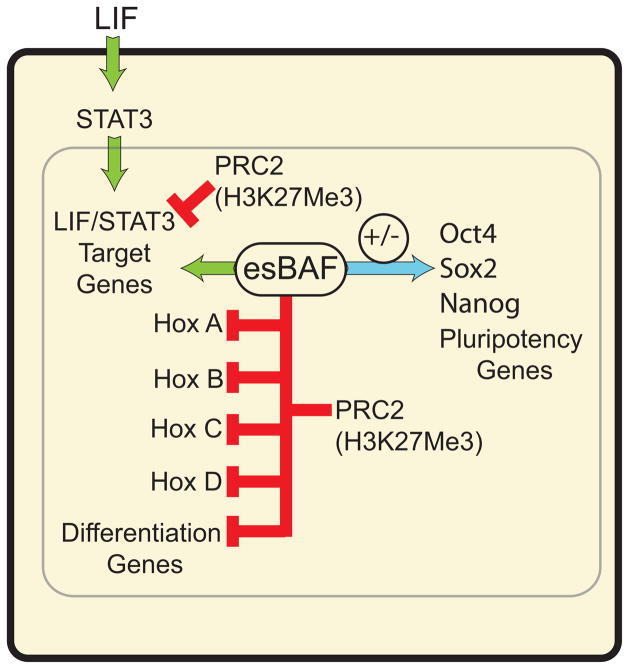
Our studies indicate that esBAF determines the pattern of STAT3 binding across the pluripotent genome. STAT3 signalingactivates a very different group of genes in other cell types with distinct compositions of BAF complexes. We propose thatESC genome is conditioned by the esBAF complex, which is present on chromatin prior to STAT3binding, and can therefore contribute to determining which STAT3 target genesare activated in ESCs. In WT ESCs, Brg (and by inference esBAF complexes) mediates the accessibility of functional STAT3binding sites, thereby facilitating robust STAT3binding and STAT3-activated transcriptionin response to LIF (Figure 8). Without esBAF, STAT3is still activated and localized to the nucleus but is unable to bind to the vast majority of its legitimate target sites in ES cells but is instead targeted to inappropriate sites. esBAF also prevents the inappropriate silencing of STAT3 target genes by Polycomb complexes, ensuring that ESCs maintain LIF competency required for pluripotency. Unexpectedly, esBAF also synergizes with Polycomb at a subset of genes, including all four Hox clusters, ensuring their firm repression in ESCs.
Figure 8. esBAF both antagonizes and synergizes with PRC2 to promote pluripotency.
esBAF antagonizes PRC2 action at LIF target genes preparing them to be activated by phospho-STAT3 entering the nucleus. In contrast, esBAF works with PRC2 to enforce the H3K27Me3 repressive mark at all 4 Hox loci and over many differentiation genes. The levels of pluripotency genes are both repressed and activated by Brg (esBAF) as indicated by the blue arrow, a context-dependent function that we have called “refinement”.
The role of SWI/SNF-like complexes in signaling is not surprising, since the yeast SWI/SNF complex was discovered in screens for regulators of signal-dependent gene activation. However, in yeast, a transcription factor SWI5 appears to recruit SWI/SNF to its target genes40. In mammalian systems, STAT3 was shown in one case to be recruited to a target gene by an undefined Brg-containing complex41, however other studies suggest the opposite,i.e. that STAT3 recruits Brg42. Our studies provide genome-wide evidence that STAT3 recruitment is dependent upon the prior actions of Brg. In our proteomic analysis of esBAF complexes we did not detect STAT3 peptides indicating that it is unlikely that a stable interaction exist between esBAF and STAT3 in solution.However, esBAF occupancy also appears to be partially dependent on STAT3, indicating that the current models of unidirectional recruitment, either of STAT3 by Brg or of Brg by STAT3, are incomplete. Rather, we favor a model of interdependent recruitment. While STAT3 binding is dependent on Brg preassembly on target chromatin, steady-state occupancyof esBAF is reinforced by bound STAT3.
The nature of the antagonistic roles between PcG and Brg has been a matter of much debate 21–23. Our studies demonstrate that genome-wide, Brg excludes PcG function because Brg deletion leads to the invasion of PcG at the sites we have tested and most Brg-activated genes show an increase in H3K27me3 and transcriptional silencing. Thus, we favor a mechanism of opposition in which Brg prevents PcG binding to a large group of genes including LIF/esBAF targets (Figure 8). This is clearly not due to a loss of transcription, since removing LIF does not lead to PcG binding and H3K27me3 deposition. In human rabdhoid tumors caused by the deletion of BAF47 (Snf5), re-expression of BAF47 leads to eviction of PcG and reduced H3K27Me3 at the Ink4a locus22, which is consistent with the genome-wide antagonism we report.
In contrast to textbook descriptions of the antagonism between Brm and Polycomb, we find that esBAF synergizes with PRC2 at all four Hox loci (Figure 8), which must be effectively silenced in ESCs to prevent premature differentiation. Previous studies have not uncovered the presence of synergism between BAF and PcG complexesbecause only single genes were examined43, and because synergism was not expected in light of early studies in Drosophila44. This unexpected finding might be restricted to ESCs and could arise from the fact that ESCs contain a specialized SWI/SNF-like BAF complex (esBAF) not found in other cell types.
Hence, Brg appears to function in synergy with PcG to repress differentiation genes in ES cells, while preventing PcG-mediated repression of pluripotency-related genes activated by ESC transcription factors such as Stat3, Oct4 and Sox2. These findings imply that esBAF regulates the activity of Polycomb across the genome in both positive and negative modes. Clearly an important future goal will be to understand the biochemical basis of the ability of Brg to direct Polycomb function. This more complex view of Brg’s function is consistent with the observation that mutations in Brg, Brm or other components of mammalian BAF complexes have not shown homeotic-like defects in mice. Thus, during the course of evolution, the interplay between _Brm_-based chromatin remodeling complexes and PcG has expanded beyond mere antagonism. Nevertheless, the two complexes appear to use both their synergistic and antagonistic functions to facilitate and maintain the pluripotent state.
Materials and Methods
Generation of Brgcond ESCs
Day 3.5 blastocysts were flushed from oviducts of Brglox/lox female mice 45(of mixed background) to Brglox/lox; Actin-CreER male mice. Embyroes were cultured on MEFs in ES media (with 15% Knockout serum replacement and 3000U/ml ESGRO LIF). After 5-6 days of culture, inner cell mass outgrowths from the embryos were dissociated by trypsin digestion and passaged onto MEF feeders. Single colonies were picked from these bulk cultures, expanded and genotyped to obtain Brglox/lox; Actin-CreER ESC clones, or Brgcond ESCs. We performed all our experiments with line 6–14, which was karyotypically normal. Throughout the study, unless otherwise indicated, we treated the cultures grown on MEFs with 1uM 4-hydroxytamoxifen (4-OHT Sigma, dissolved in ethanol) for 24 hours, then passaged them onto gelatinized plates for 48 more hours of 4-OHT treatmentto induce Brg deletion. As a control, 6–14 Brgcond cells were treated with ethanol, which does not induce Cre expression. Brg protein levels are completely abolished only at 48 hours. Hence Brg KO ESCs experience about 24hours in the complete absence of Brg.
DNase I Accessibility Assay was performed according to31 with the following modifications: an equal amount of Drosophila genomic DNA (from S2 cells) was spiked into each reaction to add as internal loading control to minimize variations arising from the phenol-chloroform extraction procedure. The undigested input sample was then briefly sonicated to solubilize it to enable more accurate pipetting. QPCR was performed with primers spanning STAT3 binding sites or control region (see Table S2). A S2-specific primer set as used as an internal control. Target sites were chosen from genes in Table S1 and are the same gene set used in ChIP assays throughout the manuscript. DnaseI accessibility for each sample was then calculated as:
[(Test_AmpliconS2_Amplicon)Digested+(Test_AmpliconS2_Amplicon)undigested_input]×100%
Datasets used in this study
ChIP-seq datasets from mESCs were obtained from11,46,29 and 33. Microarray datasets for Suz12−/− ESCs were kindlyprovided by Kristian Helin and Diego Pasini from34
Chromatin ImmunoPrecipitation (ChIP)
ChIP experiments were performed with EZ-ChIP (Millipore) according to the manufacturer’s instructions with certain modifications: ESCs were fixed on-plate with 1% formaldehyde in PBS for 12 minutes. 10 million cells were resuspended in 1ml ChIP buffer (0.1% SDS, 1mM EDTA, 20mM Tris-HCL pH 8.1, 150mM NaCl, 0.1% Triton-X100) and sonicated for 9–10 cycles, 30 seconds per cycle with 60 seconds’ rest between each cycle, at Power 6 on the Misonix Sonicator 3000. The supernatant was then diluted to a protein concentration of 1.7mg/ml and 350–500ug of protein was used for each immunoprecipitation reaction. See Table S2 for primers used in QPCR of ChIP DNA.
Antibodies Used in This Study
| Antibody | Company | Clone | Dilution |
|---|---|---|---|
| Brg | Crabtree Lab | J1 | 1:10000(WB);1ug/100ug lysate(ChIP) |
| STAT3 | Santa Cruz | C20X | 1:1000(WB);1ug/100ug lysate(ChIP) |
| STAT3-pT705 | Cell Signaling | D3A7 | 1:1000(WB);1:50(ChIP) |
| STAT3-pS727 | Santa Cruz | sc-135649 | 1:1000(WB) |
| Oct4 | Santa Cruz | sc-9081 | 1:1000(WB) |
| Sox2 | Santa Cruz | sc-17319 | 1:1000(WB) |
| Nanog | Bethyl Laboratories | A300-398A | 1:1000(WB) |
| LIFR | Santa Cruz | sc-659 | 1:1000(WB); 1:200(FACS) |
| Gp130 | Santa Cruz | sc-655 | 1:1000(WB); 1:200(FACS) |
| H3K27me3 | Millipore | 07-449 | 1:10000(WB);1ug/100ug lysate(ChIP) |
ChIP-Seq and Peak Calling
Sequenced 36-bp reads from STAT3 ChIP-Seq experiments were mapped to the mouse genome (mm8 assembly) using the Solexa Analysis Pipeline. Only those reads that mapped to unique genomic locations with at most two mismatches were retained for further analysis. This resulted in 13.4 and 12.6 million reads for STAT3 ChIP and 53.9 and 51.2 million reads for H3K27me3 in WT and Brg KO cells, respectively. For control, input DNA libraries were sequenced in both WT and Brg KO cells yielding a net 14.3 and 14.5 million reads, respectively. STAT3 and the input DNA reads were processed further using the SISSRs tool 27 to identify STAT3 binding sites in WT and Brg KO cells. SISSRs was run with -a and other default options with p-value threshold set to 0.01. A total of 2729 and 545 STAT3 binding sites were detected in WT and Brg KO cells. Of the 2729 STAT3 sites in WT, 442 were also detected to bind STAT3 in Brg KO cells (referred to as Brg-dependent STAT3 sites). The remaining 2287 STAT3 sites in WT, which were lost upon Brg KO, were considered as Brg-independent STAT3 sites. ChIP-seq reads for Brg and input DNA in E14 WT ES cells, obtained from our previous study (Ho et al., 2009a), was reanalyzed using a more stringent approach (as described in Visel et al., Nature 2009, PMID: 19212405) to define Brg binding regions (peaks). In brief, the 25-bp reads were extended to 300bp in the 3′ direction to account for the average 300bp length ChIP DNA fragments size-selected for sequencing. The read coverage at individual nucleotides at 20bp resolution was determined by counting the number of extended reads that map to 20bp intervals. Brg binding regions were identified by comparing the observed frequency of coverage depths with those expected from a random distribution of the same number of reads. The probability of observing a Brg peak with a coverage depth of atleast R by chance is given by a sum of Poisson probabilities as
where λ is the average genome-wide coverage of extended reads. The mappable genome length which was estimated to be about 80% of the genome, and the smallest coverage depth T at which the ratio of the expected number of sites with at least that coverage to the observed number is at most 0.01 was determined (FDR≤0.01). Candidate Brg peaks were identified by selecting sites at which the read coverage exceed T, and the peak boundaries were extended to the nearest flanking position at which the read coverage fell below two reads. Overlapping peaks were merged into a single peak. Consecutive Peaks separated by 2 times the average ChIP DNA fragment size were merged into a single peak as well. The latter was performed to account for regions with lack of coverage depth. Peaks mapping to satellite repeats, ribosomal RNA repeats and those that have also been reported as peaks in the control input DNA sample were discarded. The remaining 10,829 peaks were considered to be the high-confidence Brg binding regions. Although the number of Brg binding regions identified in this manner is similar in number to Brg islands we had reported previously 11, the median width of the Brg bound regions identified as described above is 700bp compared to 6.5Kb that we had reported previously. The main reason for this difference is that the strategy outlined above for identifying binding regions helped reduce the number of false-positives (compared to our previous approach) and at the same time increased the resolution of the identified binding regions by breaking up previously defined large Brg binding islands. All of our results reported in this study as well as those in our previous study 11 remain the same regardless of whether we use Brg binding regions defined in this study or Brg islands defined in our previous study (data not shown).
Target Gene Assignment
Throughout the manuscript, direct target genes of a particular factor (e.g STAT3or Brg) are defined as genes that have one more more detected factor peak(s) by ChIP-seq analysis within 5Kb upstream of the transcription start site (TSS) to the end of the gene (TES).
Direct STAT3 and Brg Target Genes
We applied several filters to obtain a set of direct target genes and functional target sites of both Brg and STAT3, because LIF activates both STAT3as well as the MAPK and PI-3K/AKT pathways [reviewed in47]; many changes we observed could be secondary targets, and many STAT3and Brg sites detected in our ChIP-seq studies might be non-functional sites that do not control expression of any target gene. First, genes that were cobound or coactivated by Brg and STAT3were identified. A gene was considered a direct target of Brg/STAT3only if a Brg/STAT3binding site is located within 5 Kb upstream of its transcription start site (TSS) or within its annotated gene body. Of the Brg and STAT3sites assigned to direct target genes, a binding site is considered functional only if the transcription of the assigned target gene is altered by at least 1.5-fold in the absence of Brg or LIF. In this way, we defined direct target genes as well as functional binding sites of Brg and STAT3to use throughout the the study. We also examined several genes with potential roles in pluripotencyfrom BR1 of the gene density heatmap derived using STAT3binding sites defined by Chen et al. (2008).
Microarray Data Analysis
Purified RNA was processed and hybridized onto Affymetrix Mouse MOE4.0 3′ expression arrays according to the manufacturer’s instructions (Affymetrix Inc.).
Generation of 2D Matricesand Gene Density Heatmap
The 17,030 UCSC known genes with expression data in ES cells, obtained from a published microarray data of wildtype E14 ES cells24, were grouped into 10 bins based on expression. To study Brg’s role in LIF/STAT3 signaling in ES cells, we first computed the fold change for 17,030 known genes in Brg KO compared to the WT ES cells. To avoid including genes with artificial fold changes (nil to negligible or vice-versa), we removed ~15% of the genes with lowest expression levels in both the WT and the KO/KD samples. The resulting genes from each dataset were then grouped into 10 bins based on the fold change. Using the same strategy, we also grouped genes into 10 bins based on the fold change in ES cells with and without LIF, embryoid body (EB) cells compared to ES cells48. The relationship between 2 datasets (10-bin sets) was assessed by performing pairwise comparisons of bins from the 2 datasets, and counting the number of genes in common between every pair of bins. The resultant matrix containing the number of common genes was plotted as a gene density heat map to infer the interplay between the functional roles between Brg and LIF/STAT3.
STAT3 Motif Analysis
MEME 49 was used toidentify sequence motifs within STAT3binding sites.
Supplementary Material
1
2
3
4
5
Acknowledgments
We thank Gill Bejerano, Aaron Wenger, Dave Bristorand Cory McLean for their computational expertise and assistance, NIEHS (NIH) core facility for their advice and expertise in performing ChIP-Seq, Paul Wade and Guang Hu for providing useful comments. We thank Jiang Wu and Lei Chen for assistance in the derivation of Brgcond ESCs.
This work was funded by grants from the HHMI and NIH grants R01NS46789, R01AI60037 and R01HD55391 to G.R.C, and Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (1ZIAES102625-02)to R.J. L.H. and W.H. are funded by A*STAR (Singapore); E.L.M. and J.L.R. are funded by the National Science Foundation.
Footnotes
Data Deposition
All ChIP-Seq and microarray data have been deposited in GEO Onmibus (NCBI) with an accession number of GSE27708.
Author Contributions
L.H. and R.J. contributed to experimental design, execution and data analysis. R.J. performed ChIP-Seq and all related data analysis. J.R. and W.H. contributed to experimental execution. E.M. performed data analysis. L.H. wrote the manuscript. G.R.C. provided the brilliance, the entertainment and the money that made all of this possible.
References
- 1.Jaenisch R, Young R. Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 4.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;105(16):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the UnitedStates of America. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhal N, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 9.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 Regulates Self-Renewal and Occupies Core Pluripotency-Related Genes in Embryonic Stem Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 11.Ho L, et al. An embryonic stem cell chromatinremodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaniel C, et al. Smarcc1/Baf155 Couples Self-Renewal Gene Repression with Changes in Chromatin Structure in Mouse Embryonic Stem Cells. Stem Cells. 2009 doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60–p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buecker C, et al. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanna J, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, et al. Bmi1 regulates mitochondrialfunction and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 22.Kia S, Gorski M, Giannakopoulos S, Verrijzer C. SWI/SNF Mediates Polycomb Eviction and Epigenetic Reprogramming of the INK4b-ARF-INK4a Locus. Molecular and Cellular Biology. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh Y, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 25.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic acids research. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns LG, Peterson CL. The yeast SWI-SNF complex facilitates binding of a transcriptional activatorto nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 30.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorschner MO, et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nature methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 32.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 38.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle-and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 41.Ni Z, Bremner R. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J Immunol. 2007;178:345–351. doi: 10.4049/jimmunol.178.1.345. [DOI] [PubMed] [Google Scholar]
- 42.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 43.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigeneticreprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamkun JW, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 45.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 46.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonicstem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okita K, Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1:103–111. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- 48.Sene K, et al. Gene function in early mouse embryonic stemcell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic acids research. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5