Meta-Analysis Increases to 71 the Tally of Confirmed Crohn’s Disease Susceptibility Loci (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 12.
Published in final edited form as: Nat Genet. 2010 Dec;42(12):1118–1125. doi: 10.1038/ng.717
We undertook a meta-analysis of six Crohn’s disease (CD) genome-wide association studies (GWAS) comprising 6,333 cases and 15,056 controls, and followed up the top association signals in 15,694 cases, 14,026 controls and 414 parent/offspring trios. Thirty new susceptibility loci meeting genome-wide significance (P-value <5×10−8) were identified. A series of in silico analyses highlighted particular genes within these loci and, together with manual curation, implicated functionally interesting candidate genes including SMAD3, ERAP2, IL10, IL2RA, TYK2, FUT2, DNMT3a, DENND1B, BACH2 and TAGAP. Combined with previously confirmed loci, the results described here identify a total of 71 distinct loci with genome-wide significant evidence for association with Crohn’s disease.
Crohn’s disease (OMIM #266600) results from the interaction of environmental factors, including the intestinal microbiota, with host immune mechanisms in genetically susceptible individuals. Along with ulcerative colitis (UC), it is one of the main subphenotypes of inflammatory bowel disease (IBD). GWAS have highlighted key CD pathogenic mechanisms, including autophagy and Th17 pathways. A meta-analysis of these early scans implicated 32 susceptibility loci, but only accounted for 20% of the genetic contribution to disease risk - suggesting that more loci await discovery1. Recognizing that an increased sample size would be required to detect these, we have expanded the International IBD Genetics Consortium (IIBDGC), approximately doubling the discovery panel size in comparison with the first meta-analysis.
The discovery panel for the current study comprised 6,333 CD subjects and 15,056 controls, all of European descent, with data derived from six index GWAS studies (for overview see Supplementary Table 1)2-6. Imputation using HapMap3 reference data allowed us to test for association at 953,241 autosomal SNPs. Our discovery panel had 80% power to detect variants conferring odds ratios ≥1.18 at the genome-wide significance level of P<5×10−8, assuming a minor allele frequency ≥20% in healthy controls. Under the same conditions, the sample size of our original meta-analysis had only 11% power1.
A quantile-quantile plot of the primary meta-statistic, using single-SNP Z-scores combined across all sample sets, showed a marked excess of significant associations (Supplementary Figure 1). A total of 2,024 SNPs within 107 distinct genomic loci, including all previously defined significant hits from our earlier meta-analysis, demonstrated association with P-values <10−5. A Manhattan plot is shown in Supplementary Figure 2. 51 of the regions, representing new loci associated at P<5×10−6, were followed up by genotyping the most significant SNPs in an independent panel of 15,694 CD cases, 14,026 controls and 414 parent/offspring trios (see Table 1 and Supplementary Table 2).
Table 1. Association results and in silico analyses for all 71 confirmed CD loci.
The upper tier lists new Crohn’s disease susceptibility loci (beyond the first international meta-analysis1) confirmed in the current study with a genome-wide significant P-value (P<5×10−8) in the combined analysis (discovery + replication sample.) and P<0.05 on replication. Results for replication are listed for all 39 loci which at the time of study design had not met P<5×10−8 plus at least nominal evidence of replication in an independent sample set. The 7 loci identified in subsequent studies are identified (see footnotes). The lower tier lists new data for SNPs/loci confirmed in the earlier meta-analysis1. Genomic positions were retrieved from NCBI’s dbSNP build v130. Linkage disequilibrium (LD) regions around focal SNPs were defined by extending the region to the left for 0.1 cM or until another SNP with P<10−5 was reached, in which case the process was repeated from this SNP. Right-hand boundaries were defined in the same way. We identified loci previously associated with other relevant traits by a manual literature search and using the NIH catalog of published Genome-wide association studies and the HuGe database (version 1.4) (accessed on May 28th 2010) 43,44.
UC – ulcerative colitis, AS – ankylosing spondylitis, Ps – psoriasis, PBC – primary biliary cirrhosis, T1D – type 1 diabetes, RA – rheumatoid arthritis, SLE – systemic lupus erythematosus, celiac – celiac disease, T2D – type 2 diabetes, MS – multiple sclerosis, Graves – Graves disease, AD – Alzheimer’s disease, MCV – mean corpuscular volume, ALL – acute lymphocytic leukemia, Lepr. – leprosy, SpA – spondyloarthritis, PD – Parkinson’s disease, CRC – colorectal cancer, CRP – C-reactive protein, TGs – triglycerides, PC – prostate cancer, HSV – human simplex virus, CAD – coronary artery disease, CLL – chronic lymphocytic leukemia, BD – bone density, B12 – serum vitamin B12 levels, HP – Helicobacter pylori, AA – alopecia areata, AITD – autoimmune thyroid disease, BC – breast cancer, BD – Behcet’s disease, GC – gastric cancer, Hep.C – hepatitis C susceptibility, SSc – systemic sclerosis, Myelo. – myeloproliferative disease, TB – tuberculosis, GvHD – Graft versus host disease, WBC – white blood cell count, HIES – hyper immunoglobulin E syndrome. Regional association plots for all 71 loci are shown in Supplementary Figure 4 and genotype data is shown in Supplementary Tables 3 and 4.
| No. | dbSNP ID | Chr. | Left - right (Mb) | Risk allele - Allele frequency in control population | P-value meta | P-value repl. | P comb. | OR (95% CI); *Loci with evidence of > than 1 independent association | Reported association | Positional candidate genes of interest: some additionally highlighted by 1kG cSNP(s) in LD; GRAIL (bold) ;eQTL (LOD score) 1kG cSNP(s) in LD; GRAIL (bold) ;eQTL (LOD score) |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) New Loci meeting Genome–wide significance (P-value <5.0×10−8) in this study | ||||||||||
| 1 | rs2797685 | 1p36 | 7.66 - 7.89 | A - 0.190 | 2.69×10−10 | 1.40×10−2 | 7.10×10−9 | 1.05 (1.01-1.10) | Celiac | VAMP3 |
| 2 | rs3180018 | 1q22 | 153.24 - 154.39 | A - 0.250 | 1.29×10−9 | 2.70×10−5 | 2.30×10−13 | 1.13 (1.06-1.19)* | T2D, Asthma, PD | SCAMP3, MUC1 |
| 3 | rs1998598 | 1q31 | 195.58 - 196.21 | G - 0.302 | 4.90×10−9 | 1.60×10−2 | 8.70×10−9 | 1.04 (1.00-1.09) | Asthma | DENND1B |
| 4 | rs3024505 | 1q32 | 204.87 - 205.10 | T - 0.157 | 8.32×10−9 | 1.50×10−7 | 1.60×10−14 | 1.12 (1.07-1.17) | T1D, UC, SLE, BD, Hep. C, | IL10, IL19 |
| 5 | rs13428812 | 2p23 | 25.30 - 25.46 | G - 0.326 | 1.41×10−8 | 5.90×10−4 | 8.50×10−10 | 1.06 (1.03-1.10) | DNMT3A | |
| 6 | rs780093 | 2p23 | 27.24-27.71 | T - 0.418 | 1.10×10−4 | 3.30×10−8 | 4.70×10−11 | 1.15 (1.10-1.21) | CRP, Glucose, TGs |  GCKR GCKR |
| 7 | rs10495903 | 2p21 | 43.30 - 43.80 | T - 0.129 | 7.70×10−8 | 2.90×10−8 | 1.60×10−14 | 1.14 (1.09-1.20)* | T2D, PC |  THADA THADA |
| 8§ | rs10181042 | 2p16 | 60.77 - 61.74 | T - 0.420 | 6.61×10−9 | N/A | N/A | 1.14 (1.09-1.19) | RA, UC, Celiac | C2orf74 (9.6), REL |
| 9† | rs2058660 | 2q12 | 102.17 - 102.67 | G - 0.231 | 1.58×10−12 | N/A | N/A | 1.19 (1.14-1.26) | Celiac, Asthma, T1D, HSV | IL18RAP, IL12RL2, IL18R1, IL1RL1 |
| 10 | rs6738825 | 2q33 | 197.85 - 198.67 | A - 0.473 | 1.82×10−7 | 1.60×10−3 | 3.50×10−9 | 1.06 (1.02-1.11) | CAD |  PLCL1 PLCL1 |
| 11 | rs7423615 | 2q37 | 230.76 - 230.94 | T - 0.187 | 4.57×10−9 | 7.40×10−6 | 3.10×10−13 | 1.12 (1.07-1.18) | CLL | SP140 (8.8) |
| 12 | rs13073817 | 3p24 | 18.58 - 18.86 | A - 0.322 | 8.20×10−7 | 1.00×10−3 | 6.70×10−9 | 1.08 (1.03-1.13) | ||
| 13 | rs7702331 | 5q13 | 72.49 - 72.62 | A - 0.600 | 2.00×10−6 | 6.40×10−7 | 5.90×10−12 | 1.12 (1.07-1.17) | TMEM174 | |
| 14 | rs2549794 | 5q15 | 96.11 - 96.45 | C - 0.409 | 4.47×10−11 | 2.00×10−3 | 1.10×10−10 | 1.05 (1.02-1.09) | AS, PD, T1D |  ERAP2, LRAP (47.2) ERAP2, LRAP (47.2) |
| 15 | rs11167764 | 5q31 | 141.39 - 141.62 | C - 0.796 | 1.10×10−9 | 4.20×10−3 | 2.00×10−9 | 1.06 (1.02-1.11) | NDFIP1 | |
| 16 | rs359457 | 5q35 | 173.15 - 173.47 | T - 0.571 | 5.25×10−8 | 3.30×10−6 | 2.50×10−12 | 1.08 (1.04-1.12) | CPEB4 (6.1) | |
| 17 | rs17309827 | 6p25 | 3.35 - 3.41 | T - 0.639 | 6.16×10−7 | 3.10×10−4 | 6.70×10−9 | 1.10 (1.05-1.16) | C6orf85 | |
| 18 | rs1847472 | 6q15 | 90.86 - 91.14 | G - 0.658 | 3.63×10−6 | 1.40×10−4 | 5.10×10−9 | 1.07 (1.03-1.11) | T1D, Celiac | BACH2 |
| 19 | rs212388 | 6q25 | 159.26 - 159.46 | G - 0.393 | 1.41×10−7 | 2.40×10−5 | 2.30×10−11 | 1.10 (1.05-1.14) | RA, Celiac, T1D↕ | TAGAP |
| 20 | rs6651252 | 8q24 | 129.56 - 129.67 | T - 0.865 | 2.29×10−6 | 2.40×10−13 | 3.90×10−18 | 1.23 (1.17-1.30) | ||
| 21† | rs4077515 | 9q34 | 138.27 - 138.54 | T - 0.411 | 4.37×10−19 | 1.50×10−19 | 1.30×10−36 | 1.18 (1.13-1.22) | UC, AS | CARD9 (12.4),  CARD9, CARD9,  SNAPC4 SNAPC4 |
| 22 | rs12722489 | 10p15 | 6.07 - 6.21 | C - 0.852 | 8.51×10−6 | 5.20×10−5 | 2.90×10−9 | 1.11 (1.05-1.16) | MS, T1D, Vitiligo, RA, AA, Asthma, AITD | IL2RA |
| 23 | rs1819658 | 10q21 | 59.50 - 59.81 | C - 0.774 | 1.41×10−7 | 1.10×10−10 | 9.10×10−17 | 1.19 (1.13-1.25) | AD | UBE2D1 |
| 24‡ | rs1250550 | 10q22 | 80.67 - 80.77 | G - 0.669 | 2.00×10−10 | 7.30×10−22 | 1.10×10−30 | 1.19 (1.15-1.23) | Celiac, MS, Vitiligo, BC | ZMIZ1 |
| 25 | rs102275 | 11q12 | 61.28 - 61.44 | C - 0.341 | 7.24×10−8 | 1.70×10−5 | 2.30×10−11 | 1.08 (1.04-1.12) | CAD; Dyslipidemia | FADS1 (5.0) |
| 26 | rs694739 | 11q13 | 63.58 - 64.05 | A - 0.626 | 3.38×10−7 | 3.50×10−4 | 6.00×10−10 | 1.10 (1.05-1.16) | AA | PRDX5, ESRRA |
| 27 | rs2062305 | 13q14 | 41.72 - 42.00 | G - 0.346 | 2.00×10−6 | 5.70×10−5 | 4.90×10−10 | 1.10 (1.05-1.15) | BD, RA | TNFSF11,TNFSF11 (5.9) |
| 28 | rs4902642 | 14q24 | 68.23 - 68.39 | G - 0.584 | 2.00×10−7 | 4.50×10−5 | 1.60×10−10 | 1.07 (1.11-1.04)* | Celiac, T1D | ZFP36L1 |
| 29 | rs8005161 | 14q35 | 87.28 - 87.71 | T - 0.119 | 1.29×10−8 | 5.90×10−11 | 4.20×10−18 | 1.23 (1.16-1.31)* |  GALC, GALC,  GPR65, GPR65 GPR65, GPR65 |
|
| 30 | rs17293632 | 15q22 | 65.20 - 65.27 | T - 0.233 | 1.41×10−13 | 2.00×10−8 | 2.70×10−19 | 1.12 (1.07-1.16) | CAD, T2D | SMAD3 |
| 31‡ | rs151181 | 16p11 | 28.20 - 28.94 | G - 0.386 | 1.10×10−10 | 1.20×10−3 | 1.50×10−11 | 1.07 (1.03-1.12) | T1D, obesity, Asthma, CRC, SLE, RA |  APOB48R, APOB48R,  IL27, IL27,  SULT1A2, SULT1A2,  SULT1A1, SULT1A1,  SH2B1, EIF3C (11.3), IL27, LAT, CD19, NFATC2IP SH2B1, EIF3C (11.3), IL27, LAT, CD19, NFATC2IP |
| 32§ | rs3091315 | 17q12 | 29.51 - 29.70 | A - 0.723 | 1.70×10−13 | N/A | N/A | 1.20 (1.14-1.26) | HIV resistance | CCL2, CCL7 |
| 33 | rs12720356 | 19p13 | 10.26 - 10.50 | G - 0.084 | 9.20×10−10 | 1.90×10−5 | 1.40×10−12 | 1.12 (1.06-1.19)* | T1D, SLE, MS, HIES |  TYK2, TYK2, ICAM1, ICAM3 TYK2, TYK2, ICAM1, ICAM3 |
| 34‡ | rs736289 | 19q13 | 38.42 - 38.47 | T - 0.612 | 2.69×10−7 | 2.00×10−3 | 8.70×10−9 | 1.06 (1.02-1.11) | ||
| 35‡ | rs281379 | 19q13 | 53.78 - 53.97 | A - 0.487 | 8.60×10−10 | 5.20×10−5 | 7.40×10−12 | 1.07 (1.04-1.11) | B12, Norovirus, HP |  FUT2, FUT2,  RASIP1 RASIP1 |
| 36 | rs4809330 | 20q13 | 61.65 - 61.95 | G - 0.709 | 2.51×10−12 | 4.60×10−5 | 2.70×10−15 | 1.12 (1.06-1.18) | Glioma |  RTEL1, TNFRSF6B, SLC2A4RG RTEL1, TNFRSF6B, SLC2A4RG |
| 37 | rs181359 | 22q11 | 20.14 - 20.39 | T - 0.203 | 6.31×10−13 | 2.30×10−6 | 4.80×10−16 | 1.10 (1.06-1.15) | RA, Celiac, SLE, MCV |  YDJC YDJC |
| 38‡ | rs713875 | 22q12 | 28.23 - 29.00 | C - 0.471 | 5.70×10−9 | 8.30×10−5 | 7.30×10−12 | 1.08 (1.04-1.13) | T1D | MTMR3 |
| 39 | rs2413583 | 22q13 | 38.00 - 38.14 | C - 0.830 | 1.70×10−10 | 9.50×10−18 | 1.10×10−26 | 1.23 (1.17-1.29) | MAP3K7IP1 | |
| (b) Loci that met Genome–wide significance (P-value <5.0×10−8) in Barrett et al. | ||||||||||
| 1 | rs11209026 | 1p31 | 67.13 - 67.54 | G - 0.932 | 1.00×10−64 | N/A | N/A | 2.66 (2.36-3.00) | UC, AS, Ps, PBC, GC, BD |  IL23R, IL23R IL23R, IL23R |
| 2 | rs2476601 | 1p13 | 113.66 - 114.42 | G - 0.907 | 4.47×10−9 | N/A | N/A | 1.26 (1.17-1.37) | T1D↕, RA, SLE, Ps, Vitiligo↕, AITD |  PTPN22, PTPN22 PTPN22, PTPN22 |
| 3 | rs4656940 | 1q23 | 158.96 - 159.20 | A - 0.801 | 6.17×10−7 | N/A | N/A | 1.15 (1.09-1.21) | SLE, RA | CD244 (7.7), CD244, ITLN1 |
| 4 | rs7517810 | 1q24 | 170.92 - 171.21 | T - 0.246 | 1.51×10−15 | N/A | N/A | 1.22 (1.16-1.28) | Hep.C, SLE, SSc, T2D | TNFSF18, TNFSF4, FASLG |
| 5 | rs7554511 | 1q32 | 199.11 - 199.32 | C - 0.726 | 1.58×10−7 | N/A | N/A | 1.14 (1.08-1.19) | UC, celiac, MS |  C1orf106, KIF21B C1orf106, KIF21B |
| 6 | rs3792109 | 2q37 | 233.81 - 234.23 | A - 0.529 | 6.76×10−41 | N/A | N/A | 1.34 (1.29-1.40) | UC |  ATG16L1 ATG16L1 |
| 7 | rs3197999 | 3p21 | 48.16 - 51.73 | A - 0.297 | 6.17×10−17 | N/A | N/A | 1.22 (1.16-1.27) | UC |  MST1, MST1,  GPX1, GPX1,  BSN BSN |
| 8 | rs11742570 | 5p13 | 39.88 - 41.00 | C - 0.606 | 7.08×10−36 | N/A | N/A | 1.33 (1.27-1.39) | MS | PTGER4 |
| 9 | rs12521868 | 5q31 | 129.41 - 132.05 | T - 0.422 | 1.41×10−20 | N/A | N/A | 1.23 (1.18-1.28) | Ps, Fibrinogen, Asthma,TB, UC |  SLC22A4, SLC22A5 (5.4),IRF1, CSF2, IL3 SLC22A4, SLC22A5 (5.4),IRF1, CSF2, IL3 |
| 10 | rs7714584 | 5q33 | 150.01 - 150.38 | G - 0.088 | 7.76×10−19 | N/A | N/A | 1.37 (1.28-1.47) | TB | IRGM |
| 11 | rs6556412 | 5q33 | 158.43 - 158.88 | A - 0.332 | 5.37×10−14 | N/A | N/A | 1.18 (1.13-1.24) | Ps, SLE, Malaria, Asthma | IL12B |
| 12 | rs6908425 | 6p22 | 20.60 - 21.25 | C - 0.784 | 1.41×10−8 | N/A | N/A | 1.17 (1.11-1.23) | T2D, Ps, UC | CDKAL1 |
| 13 | rs1799964 | 6p21 | 31.49 - 32.98 | C - 0.209 | 3.98×10−11 | N/A | N/A | 1.19 (1.13-1.25) | Multiple including UC |  MCCD1, MCCD1,  LTA, HLA-DQA2, TNF, LST1, LTB, LTA, NCR3 LTA, HLA-DQA2, TNF, LST1, LTB, LTA, NCR3 |
| 14 | rs6568421 | 6q21 | 106.50 - 106.67 | G - 0.301 | 4.37×10−8 | N/A | N/A | 1.13 (1.07-1.18)* | SLE, RA | PRDM1 |
| 15 | rs415890 | 6q27 | 167.26 - 167.47 | C - 0.522 | 2.51×10−12 | N/A | N/A | 1.17 (1.12-1.22) | RA, Graves | CCR6 |
| 16 | rs1456896 | 7p12 | 50.22 - 50.34 | T - 0.69 | 1.20×10−8 | N/A | N/A | 1.14 (1.09-1.20) | AD, SLE, MCV, ALL | IKZF1, ZPBP, FIGNL1 |
| 17 | rs4871611 | 8q24 | 126.54 - 126.65 | A - 0.609 | 1.51×10−12 | N/A | N/A | 1.17 (1.12-1.23) | ||
| 18 | rs10758669 | 9p24 | 4.93 - 5.29 | C - 0.349 | 1.00×10−13 | N/A | N/A | 1.18 (1.13-1.23) | UC, Myelo. | JAK2 |
| 19 | rs3810936 | 9q32 | 116.47 - 116.74 | C - 0.682 | 1.00×10−15 | N/A | N/A | 1.21 (1.15-1.27) | UC, Lepr., SpA | TNFSF15, TNFSF8 |
| 20 | rs12242110 | 10p11 | 35.22 - 35.94 | G - 0.315 | 1.10×10−09 | N/A | N/A | 1.15 (1.10-1.20) | UC | CREM (6.4) |
| 21 | rs10761659 | 10q21 | 63.97 - 64.43 | G - 0.538 | 4.37×10−22 | N/A | N/A | 1.23 (1.18-1.29) | BC | ZNF365 |
| 22 | rs4409764 | 10q24 | 101.26 - 101.33 | T - 0.492 | 2.29×10−20 | N/A | N/A | 1.22 (1.17-1.27) | UC | NKX2-3 |
| 23 | rs7927997 | 11q13 | 75.70 - 76.04 | T - 0.389 | 5.62×10−13 | N/A | N/A | 1.17 (1.12-1.22) | Atopy↕ | C11orf30 |
| 24 | rs11564258 | 12q12 | 38.42 - 39.31 | A - 0.025 | 6.17×10−21 | N/A | N/A | 1.74 (1.55-1.95) | PD, Lepr. |  MUC19, LRRK2 MUC19, LRRK2 |
| 25 | rs3764147 | 13q14 | 43.13 - 43.54 | G - 0.245 | 1.41×10−10 | N/A | N/A | 1.17 (1.12-1.23) | Lepr. |  C13orf31 C13orf31 |
| 26 | rs2076756 | 16q12 | 49.02 - 49.41 | G - 0.26 | 3.98×10−69 | N/A | N/A | 1.53 (1.46-1.60) | Lepr., Atopy, Blau, GvHD | NOD2 |
| 27 | rs2872507 | 17q21 | 34.62 - 35.51 | A - 0.458 | 1.51×10−9 | N/A | N/A | 1.14 (1.09-1.19) | Asthma, UC, PBC, T1D, RA, WBC |  GSMDL, GSMDL,  ZPBP2, ORMDL3 (20.3),IKZF3 ZPBP2, ORMDL3 (20.3),IKZF3 |
| 28 | rs11871801 | 17q21 | 37.57 - 38.25 | A - 0.756 | 2.51×10−8 | N/A | N/A | 1.15 (1.10-1.21) | MS↕, obesity, HIES |  MLX, STAT3 MLX, STAT3 |
| 29 | rs1893217 | 18p11 | 12.73 - 12.92 | G - 0.153 | 1.29×10−14 | N/A | N/A | 1.25 (1.18-1.32) | T1D↕, celiac | PTPN2 |
| 30 | rs740495 | 19p13 | 1.04 - 1.13 | G - 0.247 | 8.13×10−12 | N/A | N/A | 1.16 (1.10-1.21) | GPX4, SBNO2 | |
| 31 | rs1736020 | 21q21 | 15.62 - 15.77 | C - 0.579 | 9.33×10−12 | N/A | N/A | 1.16 (1.11-1.21) | UC | |
| 32 | rs2838519 | 21q22 | 44.41 - 44.52 | G - 0.391 | 2.09×10−14 | N/A | N/A | 1.18 (1.13-1.23) | Celiac, UC | ICOSLG |
Variants within 30 distinct new loci met a genome-wide significance threshold of P<5×10−8 for association with CD in the combined discovery plus replication panel, with at least nominal association in the replication panel (see Table 1). Two additional loci, encompassing the CARD9 and IL18RAP genes, had previously been reported as associated with CD in a candidate gene study7 and were here both replicated and confirmed at P<5×10−8. Five loci were identified at genome-wide significance in GWAS studies published subsequent to our replication experiment being designed. One, the FUT2 locus, was from a recent adult CD GWAS6. Four more (ZMIZ1, IL27 at 16p11, 19q13 and 22q12) were identified in a pediatric IBD population5, these replicating here in our current sample set. Two further loci had produced “suggestive” evidence of association with replication in our earlier study1. Here, these clearly exceeded the genome-wide significance threshold in the meta-analysis alone and, given the previous replication evidence, were not followed up further (see Table 1). Thus cumulatively, 39 additional loci can now be added to the 32 confirmed CD susceptibility loci identified at the time of the Barrett et al. study. We did not observe statistically significant heterogeneity of the odds ratios (Breslow Day test P-value <0.05 after Bonferroni correction; Supplementary Table 4) between the panels from our 15 different countries (Supplementary Tables 1 and 2) for any of the 71 loci. Nor was any evidence of interaction between the associated loci observed (Supplementary Figure 3).
Regional association plots of all 71 susceptibility loci including the underlying genes are shown in detail in Supplementary Figure 4, and complete genotype data including odds ratios and allele frequencies are shown in Supplementary Tables 3 and 4. Five loci had evidence for more than one independently associated variant (Table 1). While 6 of the 30 novel regions contain just a single gene, which is thereby strongly implicated in CD pathogenesis _(_e.g. SMAD3, NDFIP1 and BACH2), 22 include more than one gene within the associated interval (Table 1; two regions without any gene or gene prediction). We thus applied additional in silico analyses to refine the list of functional candidate genes further. These were:
- Interrogation of a publicly available expression quantitative trait loci (eQTL) database8. These analyses identified genes for which expression correlates with genotype at our most associated SNP (see Supplementary Results).
- Use of 1000 Genomes Project Pilot sequence data and HapMap3 to identify genes containing non-synonymous variants in strong LD (r2>0.5) with the focal SNP within each region (for details on coding SNP see Supplementary Table 5).
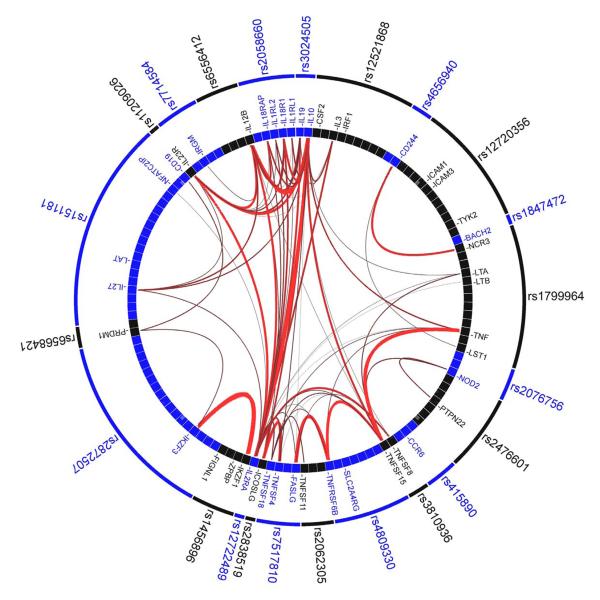
- Use of GRAIL9, to identify non-random and evidence-based connectivity between the genes in the 71 confirmed CD loci. Specifically, GRAIL evaluates each gene in a CD-associated locus for non-random correlation with genes in the other 70 loci via word-usage in PubMed abstracts related to the gene (see Figure 1).
Figure 1. Gene Relationships Across Implicated Loci (GRAIL) pathway analysis.
Links between genes at 23 of 71 Crohn’s disease associated loci which scored P<0.01 using GRAIL. Specifically, of the 71 CD-associated SNPs, 69 are in LD intervals containing or within 50 kb of at least one gene. In total, there are 355 genes implicated by proximity to these 69 SNPs. Each observed CD-association was scored with GRAIL, which takes all genes mapping within CD-associated intervals and evaluates for each whether it is non-randomly linked to the other genes, via word-usage in PubMed abstracts. 23 SNPs shown in the outer circle are P<0.01 hits - indicating that the regions which they tag contain genes which are more significantly linked to genes in the other 68 regions than expected by chance at that level. The lines between genes represent individually significant connections that contribute to the positive signal, with thickness of lines inversely proportional to the probability a literature-based connection would be seen by chance.
To accurately assess the statistical significance of this set of connections, we conducted simulations where we selected 1000 sets of 69 SNPs implicating in total 355 genes ±18 (5%) (selecting the SNPs randomly and using rejection sampling - only taking lists that implicated the same number of genes). Each of those 1000 sets were scored with GRAIL. The mean number of P<0.01 hits in a simulated list was 0.91 with a range in the 1000 sets from 0 to 11, suggesting that the likelihood of observing 23 hits with P<0.01 is far less than 0.1%.
Summary results of these analyses are shown in the rightmost column of Table 1. The highlighted genes are described briefly in Box 1, as are genes that constitute particularly noteworthy candidates from intervals containing one or few genes. While we believe that these evidence-based approaches are helpful in identifying likely functional candidates, in some instances the different techniques highlight different genes. This reflects uncertainty as to which is causal, and highlights the need for functional studies.
30 new signals were identified here beyond those described in the earlier meta-analysis1 and other subsequent publications. The new associations were driven primarily by increased power arising from the expanded sample size rather than improved imputation, as more than two-thirds of the novel loci identified here have good proxies (r2>0.8) on both earlier generation arrays (Illumina 300K and Affymetrix 500k Set). Extending this argument beyond the current analysis, it seems likely that many more loci of modest effect size still await discovery.
For many of the novel loci, associations have been reported previously in other complex diseases, comprising mostly chronic inflammatory disorders (Table 1). Such diseases can cluster both within families and individuals, reflecting shared genetic risk factors. For example, IBD and ankylosing spondylitis can co-segregate and both are associated with IL23R2,10 and TNFSF1511,12. The IL10 locus was previously associated with UC13 and was identified as a novel CD locus in the present study. Thus IL10 is a generic IBD locus, which is a functionally intuitive finding of potential therapeutic significance.
For loci previously associated with other inflammatory diseases the direction of effect in CD is usually the same, but in five cases the risk allele for one disease appears to be protective in another disease (see arrow symbol in “Reported association” column in Table 1). In most such instances, functional annotation suggests modulation of T cell and other immune pathways. Indeed, GRAIL highlights a number of such genes. These inverse associations may reflect overlap in the pathways by which the host regulates effector functions in defense and regulatory functions in self-tolerance. This is a delicate balance and, in the face of competing requirements, selection pressures may have conferred advantage for divergent alleles in a cell- and environmentally dependent manner.
The associated SNP rs281379 at 19q13, recently also identified by McGovern et al.6 is highly correlated (r2>0.80) with a common nonsense variant (rs601338 also known as G428A or W142X) at FUT2. This is classically referred to as the non-secretor variant, as individuals homozygous for this null-allele do not secrete blood group antigens at epithelial surfaces. Recently, non-secretors were identified as having near-complete protection from symptomatic GII.4 norovirus infection14 and the same null allele is identified here as a CD risk factor. This suggests one potential elusive link between infection and immune-mediated disease.
In contrast to the implication of coding variation in the FUT2 gene, our previous data demonstrated that most CD-associated SNPs were not in LD with coding polymorphisms1, suggesting that regulatory effects are likely to be a more common mechanism of disease susceptibility. Providing further direct evidence for this, a number of new eQTL effects were identified here (see Table 1 and Supplementary Results Section) – notably including CARD9 (LOD=12.4), ERAP2 (LOD=47.2) and TNFSF11 (RANKL) (LOD=5.9). The latter maps adjacent to but outside the associated recombination interval, suggesting another potential long-range cis-regulatory effect as previously described for PTGER4 in CD4. RANKL has pleiotropic immunological effects and also stimulates osteoclast activity. This finding may be relevant to the osteoporosis clinically associated with CD.
Given the importance of regulatory effects, it is intriguing that variants within the gene encoding a key mediator of epigenetic regulation, DNA methyltransferase 3a (DNMT3A), should be associated with CD. By inducing transcriptional silencing, DNMT3a is known to play an important role in immunoregulation. For example, it methylates IL-4 and IFN-γ promoters following T cell receptor stimulation, hence regulating T cell polarization15, and induces dynamic regulation of TNF-α transcription following lipopolysaccharide exposure in leukocytes16. Genetically determined alterations in DNMT3a activity could thus have far-reaching effects.
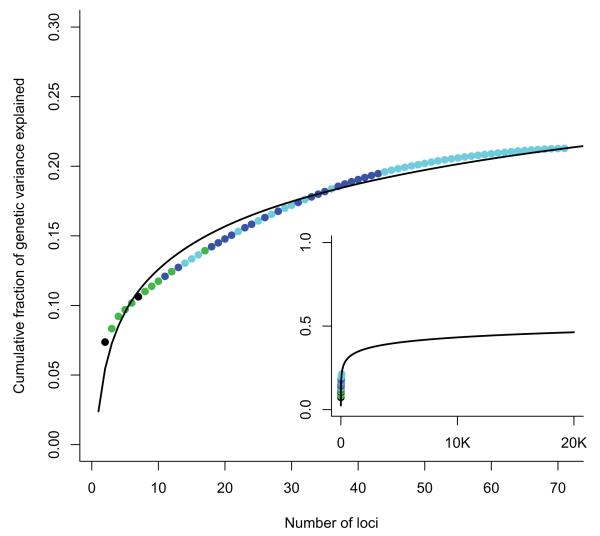
The 32 loci described up to 2008 explained approximately 20% of CD heritability. Adding the 39 loci described since increases the proportion of heritability explained to just 23.2%. This pattern of common alleles, explaining a logarithmically decreasing fraction of heritability (Figure 2), is consistent with a recent model of effect size distribution17, which predicted (based on the previous CD meta-analysis) that our current sample size would likely identify 48 new loci. Furthermore, it is likely that more high-frequency CD risk alleles of even smaller effect size remain unidentified: The same model predicts that 140 loci would be identified by a sample size of 50,000, but these would explain only a few more percent of CD heritability. It is clear, therefore, that larger GWAS alone will not explain all of the missing heritability in CD.
Figure 2. Cumulative fraction of genetic variance explained by 71 CD loci.
Cumulative fraction of genetic variance explained by the 71 CD loci reported here, ordered from largest to smallest individual contribution. Black points were identified pre-GWAS, green in first generation GWAS, blue in an earlier meta-analysis and cyan in this analysis. Inset shows a logarithmic fit to these data extrapolated to an extreme scenario where 20,000 independent common alleles are associated with disease. Even in this situation less than half of the genetic variance would be explained. This demonstrates that other types of effect (e.g. less common and rare alleles with higher penetrance) must also exist.
One key shortcoming of our current model of heritability explained by these loci is a direct consequence of the extent to which GWAS tag SNPs are often imperfect proxies for causal alleles, and thus substantially underestimate the true attributable risk. For example, the best tag SNP at the NOD2 locus in our meta-analysis appears to explain just 0.8% of genetic variance, whereas the three NOD2 coding mutations themselves account for 5%. If an analogous situation applies to even a small fraction of the other 70 CD susceptibility loci, the proportion of overall heritability explained will increase significantly. Indeed, one study of LD between tag SNPs and causal variants in the heritability of human height18 suggests that this effect might double the total fraction of heritability explained by GWAS SNPs. Coding variants identified here from the 1000 Genomes Project which are in strong LD with the focal SNPs in several of our regions (see Supplementary Table 4) thus now require direct assessment in order to explore this possibility.
Other factors will also account for the heritability gap, including uncertain epidemiological estimates of disease prevalence and total heritability, as well as our observation that several of the new regions contain more than one independent risk allele. The likelihood is that many more such effects will be identified. Indeed, detailed future analyses will play a key role in helping us to understand the absolute contribution of common causal alleles, as well as identifying less common variants and rare (even family-specific) mutations. By contrast, our lack of evidence for epistasis among the loci described here suggests that non-additive interactions among common risk alleles do not play an important role in the genetic architecture of CD.
The current study has approximately doubled the number of confirmed CD susceptibility loci. For many of these loci we have identified potentially causal genes, accepting that confirmation of their role must await detailed fine mapping, expression and functional studies. While the alleles detected only modestly affect disease risk, they continue to enhance our understanding of the genetic etiology of CD. Analysis for evidence of sub-phenotype associations represents an important future goal for the consortium. Thus, we are working towards sharing of detailed genotype and clinical data to allow this. In the meantime, extensive resequencing, together with large-scale fine mapping exercises using custom array-based technologies, are already underway and will further elucidate the pathogenic mechanisms of IBD.
Supplementary Material
Supplementary Text and Figures
Supplementary Table 3
Supplementary Table 4
Supplementary Table 6
Box 1.
Noteworthy genes within loci newly implicated in Crohn’s disease pathogenesis. N.B. Although we highlight these as interesting genes, we do not yet have data to confirm causality
- ▪ VAMP3 (1p36) encodes vesicle-associated membrane protein 3: following bacterial stimulation of TNF-α production within macrophages, VAMP3 interacts with SNARE proteins first on the trans-Golgi network, where TNF-alpha is taken up, and then on the cell membrane, where TNF-α is released19. VAMP3 also plays a role in cell migration and adhesion, by trafficking molecules such as beta1 integrin to the cell surface, and has been implicated in autophagy20.
- ▪ MUC1/SCAMP3 (1q22) MUC1 encodes a key constituent of mucus, which is the physical barrier that protects the intestinal epithelium from gut bacteria. MUC1 overexpression and hypoglycosylation have been reported in IBD21 and Muc1 knockout mice exhibit increased small intestinal damage after C. jejuni infection22. Secretory carrier membrane protein 3 (SCAMP3) regulates EGFR trafficking within endosomal membranes23. It is manipulated by intracellular salmonellae to acquire nutrients and influence host immune responses24.
- ▪ DENND1B (1q31) has been recently associated with asthma25, and is expressed in dendritic and effector memory T cells. Among other functions it modulates inflammatory signaling pathways, including Th1-Th2 cytokines, through repression of TNFR1 signaling.
- ▪ IL10 (1q32): association with CD follows its recent implication in UC13 and reporting of mutations in the IL10 receptors in extreme CD of infancy26. Known to inhibit synthesis of pro-inflammatory cytokines within macrophages and Th1 cells, IL10 also suppresses antigen presenting cell activity. Knockdown of IL10 in mice presents one of the best animal models of IBD.
- ▪ DNMT3A (2p23): DNA methyltransferase 3a is one of three key methyltransferase genes in humans - effecting epigenetic regulation of gene transcription by methylating cytosine residues within CpG islands. Among many other roles this is known to determine dynamic regulation of both adaptive and innate immune mechanisms15,16.
- ▪ GCKR (2p23) encodes an inhibitor of glucokinase, with the focal SNPs at this locus also correlating with both fibrinogen and CRP levels27.
- ▪ THADA (2p21) is expressed in small intestine and appears to encode a death receptor-interacting protein suggesting an apoptotic function28.
- ▪ ERAP2 (5q15): regulated by NF-κB, this gene encodes one of two human endoplasmic reticulum aminopeptidases, which work in concert to trim peptides for presentation on MHC class I and hence critically affect antigen presentation to T cells29. Ankylosing spondylitis is associated with this locus but with a different pattern of associated variants30. Given the close clinical relationship between CD and AS, and the strong association of HLA-B27 with the latter but not the former, the divergent association of these closely related molecules is intriguing and will refocus interest on the MHC class I associations in CD.
- ▪ NDFIP1 (5q31): Nedd4 family interacting protein 1 is a membrane protein involved in maintenance of the Golgi complex31. It is important for protein trafficking via exosomes and may play a role in rapid sequestration and removal of proteins during stress32.
- ▪ CPEB4 (5q35) is a regulator of protein translation and cell division, and a transcriptional target of RORγt. Mouse work suggests that the product of CPEB4 is the effector by which RORγt (a key determinant of Th17 cell differentiation) inhibits proliferation of thymocytes33.
- ▪ TAGAP (6q25): T-cell Activation GTPase-Activating Protein, associated with multiple autoimmune diseases, was originally identified through its involvement in human T cell activation and co-regulation with IL-234.
- ▪ IL2RA (10p15) encodes part of the IL2 receptor complex, thus mediating IL2 signalling in host defence and regulating response to autoantigens by Tregs. The associated variants correlate with differential expression of IL2RA (CD25) on CD4+ naïve and memory T cells35 possibly affecting Foxp3+ Treg homeostasis36.
- ▪ FADS 2 (11q12): fatty acid desaturase 2 is predominantly located in the endoplasmic reticulum. Fads2 knockout mice develop duodenal and ileocecal ulceration37.
- ▪ TNFSF11 (13q14) (synonyms RANKL - receptor activator of nuclear factor kappa B; ODF osteoclast differentiation factor) encodes a member of the TNF cytokine family. RANKL stimulation of dendritic cells leads to proliferation of naive T cells and inducible Tregs38; it also regulates osteoclast activity / bone loss. Previous studies have demonstrated increased plasma levels in Crohn’s disease39.
- ▪ SMAD3 (15q22): phosphorylated following TGF-β signalling through its receptor, the SMAD3 protein complexes with SMAD4 and is then translocated to the nucleus to modulate target gene expression. SMAD3 plays a key role in the TGF-β-mediated induction of Foxp3+ regulatory T cells40, with SMAD3 deficiency reciprocally enhancing Th17**.** Reduced SMAD3 phosphorylation has been observed in IBD, and may impair the immunosuppressive effect of TGF-β.
- ▪ TYK2 (19p13) encodes tyrosine kinase 2, a member of the JAK-signal transduction family. It is involved in cytokine signaling by IFN-γ, IL-12 and IL-23 among others – hence affecting Th1 and Th17 lineage development. TYK2 also plays an important role in TLR-mediated responses in dendritic cells, including IL-12 and IL-23 production, and TYK2 mutations predispose to opportunistic infection41.
- ▪ FUT2 (19q13) encodes alpha-(1,2)fucosyltransferase which regulates expression of the Lewis AB0(H) histo-blood group antigens on the surface of epithelial cells and in body fluids. Strongly associated with Norovirus infection, also with Helicobacter pylori infection and serum vitamin B12 levels14,42.
Acknowledgements
We thank all subjects who contributed samples, and physicians and nursing staff who helped with recruitment globally. This study was supported by the German Ministry of Education and Research through the National Genome Research Network and infrastructure support through the DFG cluster of excellence “Inflammation at Interfaces”. Also the Italian Ministry for Health GR-2008-1144485, with case collections supported by the Italian Group for IBD and the Italian Society for Paediatric Gastroenterology, Hepatology and Nutrition. We acknowledge funding provided by Royal Brisbane and Women’s Hospital Foundation; University of Queensland (Ferguson Fellowship); National Health and Medical Research Council, Australia and by the European Community (5th PCRDT) and by the European Crohn’s and Colitis Organization. UK case collections were supported by the National Association for Colitis and Crohn’s disease, Wellcome Trust, Medical Research Council UK and Peninsular College of Medicine and Dentistry, Exeter. We also acknowledge the NIHR Biomedical Research Centre awards to Guy’s & St Thomas’ NHS Trust / King’s College London and to Addenbrooke’s Hospital / University of Cambridge School of Clinical Medicine. The NIDDK IBD Genetics Consortium is funded by the following grants: DK062431 (S.R.B.), DK062422 (J.H.C.), DK062420 (R.H.D.), DK062432 & DK064869 (J.D.R.), DK062423 (M.S.S.), DK062413 (D.P.B.M.), DK76984 (MD), and DK084554 (MD and DPBM), and DK062429 (J.H.C.). J.H.C. is also funded by the Crohn’s and Colitis Foundation of America; and SLG by DK069513 and Primary Children’s Medical Center Foundation. Cedars Sinai supported by NCRR grant M01-RR00425; NIH/NIDDK grant P01-DK046763; DK 063491; and Cedars-Sinai Medical Center Inflammatory Bowel Disease Research Funds. RW is supported by a clinical fellow grant (90700281) from the Netherlands Organization for Scientific Research; EL, DF and SV are senior clinical investigators for the Funds for Scientific Research (FWO/FNRS) Belgium. SB was supported by the “Deutsche Forschungsgemeinschaft” (DFG; BR 1912/5-1). JCB is supported by Wellcome Trust grant WT089120/Z/09/Z. Replication genotyping was supported by unrestricted grants from Abbott Laboratories Ltd and Giuliani SpA. We acknowledge the Wellcome Trust Case Control Consortium. We thank the 1958 British Birth Cohort and Banco Nacional de ADN, Salamanca, Spain who supplied control DNA samples. The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. Other significant contributors: K. Hanigan, Z.-Z. Zhao, N. Huang, P. Webb, N. Hayward, A. Rutherford, R. Gwilliam, J. Ghori, D Strachan, W. McCardle, W. Ouwehand, M. Newsky, S. Ehlers, I. Pauselius, K. Holm, C. Sina, L. Baidoo, A. Andriulli and M.C. Renda.
Footnotes
Contribution of authors AF, DPBM, GRS, TA, JL, RR, JB, TH, AL, CGM, NP, JIR, PS, YS, LS, KDT, DW, CW, GKU, JDR, MD’A, RW, SV, RHD, JS, SS, VA, HH were involved in establishing DNA collections, and/or assembling phenotypic data; AF, DE, JCB, KW, TG, SR, CAA, LJ, MJD performed statistical analyses; DPBM, GRS, CWL, EMF, RNB, MB, TMB, SB, CB, AC, J-FC, MC, SC, TD, MdV, RD’I, MD, CE, TF, DF, RG, JG, AVG, SLG, JH, DH, J-PH, DL, IL, ML, AL, CL, EL, CM, WN, JP, AP, DDP, MR, PR, JS, MS, FS, AHS, PCFS, SRT, LT, TW, SRB, RW, SK, AMG, JCM, SV, RHD, MSS, JS, SS, JHC, VA recruited patients; AF, DPBM, TB, SB, KT, MG, GM supervised laboratory work; AF, DPBM, JCB, KW, SB, RHD, JS, SS, JHC, MJD, MP contributed to writing the manuscript. All authors read and approved the final manuscript before submission.
All authors declare no financial interest.
References
- 1.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imielinski M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–40. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGovern DP, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 19:3468–76. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhernakova A, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–10. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rueda B, et al. The IL23R Arg381Gln non-synonymous polymorphism confers susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2008;67:1451–4. doi: 10.1136/ard.2007.080283. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki K, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 12.Zinovieva E, et al. Comprehensive linkage and association analyses identify haplotype, near to the TNFSF15 gene, significantly associated with spondyloarthritis. PLoS Genet. 2009;5:e1000528. doi: 10.1371/journal.pgen.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 14.Carlsson B, et al. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 Norovirus infection. PLoS One. 2009;4:e5593. doi: 10.1371/journal.pone.0005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamper CJ, Agoston AT, Nelson WG, Powell JD. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol. 2009;183:2267–76. doi: 10.4049/jimmunol.0802960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Gazzar M, et al. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283:32198–208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 42:570–5. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science. 2005;310:1492–5. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- 20.Luftman K, Hasan N, Day P, Hardee D, Hu C. Silencing of VAMP3 inhibits cell migration and integrin-mediated adhesion. Biochem Biophys Res Commun. 2009;380:65–70. doi: 10.1016/j.bbrc.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell BJ, Yu LG, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J. 2001;18:851–8. doi: 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- 22.McAuley JL, et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–24. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20:1816–32. doi: 10.1091/mbc.E08-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mota LJ, Ramsden AE, Liu M, Castle JD, Holden DW. SCAMP3 is a component of the Salmonella-induced tubular network and reveals an interaction between bacterial effectors and post-Golgi trafficking. Cell Microbiol. 2009;11:1236–53. doi: 10.1111/j.1462-5822.2009.01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleiman PM, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 26.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danik JS, et al. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association study of fibrinogen in 17 686 women: the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2:134–41. doi: 10.1161/CIRCGENETICS.108.825273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rippe V, et al. Identification of a gene rearranged by 2p21 aberrations in thyroid adenomas. Oncogene. 2003;22:6111–4. doi: 10.1038/sj.onc.1206867. [DOI] [PubMed] [Google Scholar]
- 29.Saveanu L, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–97. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 30.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem. 2002;277:9307–17. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- 32.Putz U, et al. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J Biol Chem. 2008;283:32621–7. doi: 10.1074/jbc.M804120200. [DOI] [PubMed] [Google Scholar]
- 33.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–26. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Mao M, et al. T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics. 2004;83:989–99. doi: 10.1016/j.ygeno.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Dendrou CA, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41:1011–5. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burchill MA, Yang J, Vang KB, Farrar MA. Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol Lett. 2007;114:1–8. doi: 10.1016/j.imlet.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stroud CK, et al. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J Lipid Res. 2009;50:1870–80. doi: 10.1194/jlr.M900039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loser K, et al. Epidermal RANKL controls regulatory T-cell numbers via activation of dendritic cells. Nat Med. 2006;12:1372–9. doi: 10.1038/nm1518. [DOI] [PubMed] [Google Scholar]
- 39.Moschen AR, et al. The RANKL/OPG system is activated in inflammatory bowel disease and relates to the state of bone loss. Gut. 2005;54:479–87. doi: 10.1136/gut.2004.044370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu L, et al. Role of SMAD and non-SMAD signals in the development of Th17 and regulatory T cells. J Immunol. 184:4295–306. doi: 10.4049/jimmunol.0903418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–87. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazra A, et al. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat Genet. 2008;40:1160–2. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 26:145–6. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text and Figures
Supplementary Table 3
Supplementary Table 4
Supplementary Table 6