A diverse array of gene products are effectors of the type I interferon antiviral response (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 1.
Published in final edited form as: Nature. 2011 Apr 10;472(7344):481–485. doi: 10.1038/nature09907
Abstract
The type I interferon (IFN) response protects cells from invading viral pathogens. The cellular factors that mediate this defense are the products of interferon-stimulated genes (ISGs). Although hundreds of ISGs have been identified since their discovery over 25 years ago1,2,3, only few have been characterized with respect to antiviral activity. For most, little is known about their antiviral potential, their target specificity, and their mechanisms of action. Using an overexpression screening approach, we show that different viruses are targeted by unique sets of ISGs, with each viral species susceptible to multiple antiviral genes with a range of inhibitory activities. To conduct the screen, over 380 ISGs were tested for their ability to inhibit the replication of several important viruses including hepatitis C virus (HCV), yellow fever virus (YFV), West Nile virus (WNV), chikungunya virus (CHIKV), Venezuelan equine encephalitis virus (VEEV), and human immunodeficiency virus (HIV-1). Broadly acting effectors included IRF1, C6orf150, HPSE, RIG-I, MDA5, and IFITM3, while more targeted antiviral specificity was observed with DDX60, IFI44L, IFI6, IFITM2, MAP3K14, MOV10, NAMPT, OASL, RTP4, TREX1, and UNC84B. Combined expression of two-ISG pairs showed additive antiviral effects similar to moderate IFN doses. Mechanistic studies revealed a common theme of translational inhibition for numerous effectors. Several ISGs, including ADAR, FAM46C, LY6E, and MCOLN2, enhanced replication of certain viruses, highlighting another layer of complexity in the highly pleiotropic IFN system.
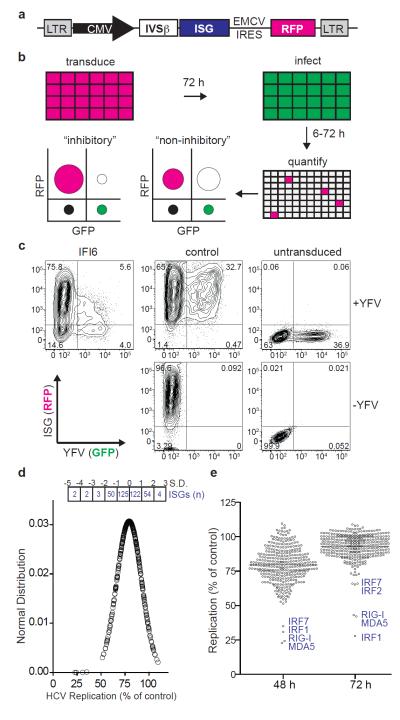
To identify novel antiviral IFN effectors, we developed a cell-based assay to determine the effects of hundreds of ISG products on viral replication. The assay relied on a bicistronic lentiviral vector co-expressing an ISG and the red fluorescent protein, TagRFP (Fig. 1a and Supplementary Fig. 1). ISG/TagRFP-expressing cells were challenged with a green fluorescent protein (GFP)-expressing virus and replication within the TagRFP-positive population was quantified by flow cytometry (FACS) (Fig. 1b). In pilot experiments, eleven ISGs (PSMB8, PSMB9, IFI6, GBP1, IFITM3, MX1, PLSCR1, RTP4, CCDC75, IFIT3, IFI27) and two control genes, Gaussia (Gluc) and Firefly (Fluc) luciferase, were tested for inhibition of vesicular stomatitis virus (VSV) and YFV in STAT 1-deficient human fibroblasts (_STAT1_−/−Fib) or in human hepatoma cells (Huh-7). _STAT1_−/−Fib produce but do not respond to IFN4, while Huh-7 respond to but are not robust producers of IFN5. Inhibition was assessed quantitatively by FACS (Fig. 1c) and qualitatively by fluorescence microscopy (Supplementary Fig. 1c). IFI6 and MX1 showed specificity for YFV and VSV, respectively. (Supplementary Fig. 1d). Both viruses were inhibited by IFITM3, which also suppresses influenza A virus, WNV, and dengue virus6. These results demonstrate the utility of this screening platform and suggest its potential to reveal antiviral ISGs targeting divergent virus families.
Figure 1. FACS-based screen for identifying antiviral ISGs.
a. Gateway-compatible bicistronic lentiviral vector. b. Schematic of overexpression screen showing hypothetical FACS plots of inhibitory versus non-inhibitory genes. Overlap of RFP (magenta) and GFP (green) is depicted as white. c. FACS plots showing _IFI6_-mediated inhibition of YFV in _STAT1_−/−Fib. d. Distribution of HCV replication levels at 48 h normalized to Fluc control. The number of ISGs within standard deviation (S.D. or Z-score) ranges are shown in boxes. e. Dot plots of HCV replication levels at 48 h and 72 h normalized to Fluc control. Select ISGs are denoted in blue. Black line indicates population mean.
To conduct a large-scale antiviral ISG screen, we analyzed published microarray datasets from IFN-treated cells or tissues and established inclusion criteria1,7,8,9,10,11,12,13,14,15 (Supplementary Table 1). We compiled a list of 389 ISGs, inserted each into the bicistronic lentiviral vector, and produced high-titer lentiviral stocks (Supplementary Methods). Approximately 50 of these lentiviral stocks failed to mediate high-level transduction in target cells (Supplementary Fig. 2a), perhaps resulting from ISG-mediated toxicity, auto-downregulation of IRES-driven TagRFP expression, poor packaging of the vector genome, or activity against the HIV-1 based vector. We consider the latter possibility likely since several known antiretroviral genes (EIF2AK2/PKR, APOBEC3G, BST2/tetherin, MOV10) were included in this subset16,17,18. The potential cytotoxicity of ISG overexpression was inferred from FACS-based cell counts (Supplementary Fig. 3a) and validated for a number of genes by luminescence-based cell viability assay (Supplementary Fig. 3b). While two genes were toxic (CDKN1A and TNFSFR10A), the vast majority of ISGs did not strongly impact cell viability.
The lentivirus-expressed ISG collection was screened for inhibition of HCV expressing Ypet GFP (Supplementary Fig. 4) in Huh-7.5, a RIG-I defective derivative of Huh-7 cells, at 48 h and 72 h post-infection. Data are represented as a normal distribution (Fig. 1d) or dot plots (Fig. 1e). Most genes affected HCV infection to some extent, with the mean (Z-score = 0) level of inhibition at 20%. Four genes (MDA5, RIG-I, IRF1, IRF7) had Z-scores below −3.0, while 53 genes had Z-scores between −1.0 and −3.0 (Supplementary Tables 2 and 3). At 72 h post-infection, HCV replication approaches a maximum (Supplementary Fig. 4d) and the overall ISG profile constricted (Fig. 1e), likely reflecting the ability of the virus to overcome inhibition. Transient transfection was used to screen the subset of ISGs that were poorly expressed via lentiviral transduction, and several genes (including DDX60, MOV10, and MS4A4A) were found to have anti-HCV activity (Supplementary Fig. 5). To investigate the impact of the host cell environment on ISG activity, we rescreened HCV in a less-permissive cell type (Huh-7)19. The strongest effectors were consistently active in the two cell lines (Fig. 1e and Fig. 2a), while several additional genes (including MAP3K14 and SLC1A1) inhibited HCV replication in Huh-7 cells (Z-score below −2.0, Supplementary Table 4). Taken together, these data suggest at least two categories of anti-HCV ISGs: i) strong inhibitors that likely feed back into IFN or other signaling pathways and ii) modest inhibitors that may have targeted effector functions.
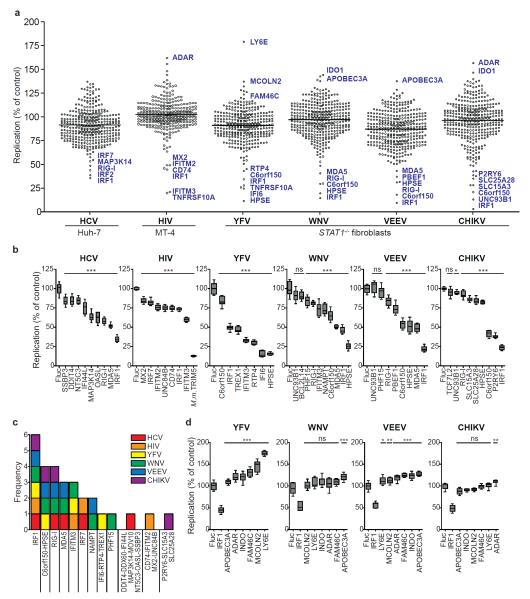
Figure 2. Identification of ISGs that inhibit or enhance virus replication.
a. Dot plots of large-scale ISG screens against six viruses. Replication levels were normalized to Fluc control. Select ISGs are denoted in blue. Black line indicates population mean. b, d. Confirmation assays of selected ISGs. For HIV, Macaca mulatta (M.m.) TRIM5 was included as a control. Replication levels were normalized to Fluc control. Data are represented by box and whisker plots. n=8 for HCV, n=9 for other viruses. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant). c. Frequency of validated antiviral ISG prevalence across six screens.
We also screened the ISG collection with other medically important viruses from three different families, Flaviviridae (YFV, WNV), Togaviridae (CHIKV, VEEV), and Retroviridae (HIV-1) (Fig. 2a). We assayed YFV, WNV, VEEV, and CHIKV in _STAT1_−/−Fib and HIV-1 in MT-4 cells (an HTLV-1-transformed T cell line). Global ISG inhibition profiles were similar for all viruses, with 10-25 genes exhibiting Z scores of −1.5 or less (Fig. 2a and Supplementary Tables 7-10). YFV was also screened in Huh-7 cells (Supplementary Fig. 6a) and this gave fewer hits, with five genes showing a Z-score below −1.5 (Supplementary Tables 5 and 6). The strongest hits in Huh-7 cells (HPSE, IRF1, IRF7, IFITM3, IFI6) were also among the top inhibitors in _STAT1_−/−Fib, suggesting these effectors function independently of cell type. Differences in data spread between the two cell lines may be due to higher levels of YFV replication in Huh-7 cells (Supplementary Fig. 6b) and/or increased ISG expression in _STAT1_−/−Fib (Supplementary Fig. 2b), and indicate that _STAT1_−/−Fib provide a more sensitive environment for detecting ISG-mediated inhibition.
The strongest antiviral ISGs from each primary screen were validated by generating independent lentiviral stocks and measuring inhibition from at least eight replicates (Fig. 3b). Forty-five hits were confirmed to significantly reduce virus replication compared to a control (Fluc) (p<0.001 in 44 assays and p<0.05 in 1 assay), and five genes were false positives. Further validation was conducted for a subset of ISGs. Inhibition of YFV by IFI6, IFITM3, and RTP4 was dose-dependent (Supplementary Fig. 7a), and both IFITM3 and IFI6 inhibited production of the parental, non-GFP virus (YFV-17D) (Supplementary Fig. 7b, c). We also measured the effects of anti-HCV ISGs when they were introduced after HCV infection. In contrast to the 20-70% range of inhibition observed when ISGs were expressed prior to infection (Fig 2b), ISG expression after infection impacted replication by either less than 20% (DDIT4, NT5C3, IFI44L, MAP3K14, IRF2) or greater than 80% (IRF7, IRF1, MDA5, RIG-I). Only one gene, OASL, showed an intermediate phentoype (Supplementary Fig 8). These data demonstrate that the overexpression platform is highly reliable for identifying antiviral ISGs and sensitive enough to detect even modest levels of inhibition in both pre- and post-infection scenarios.
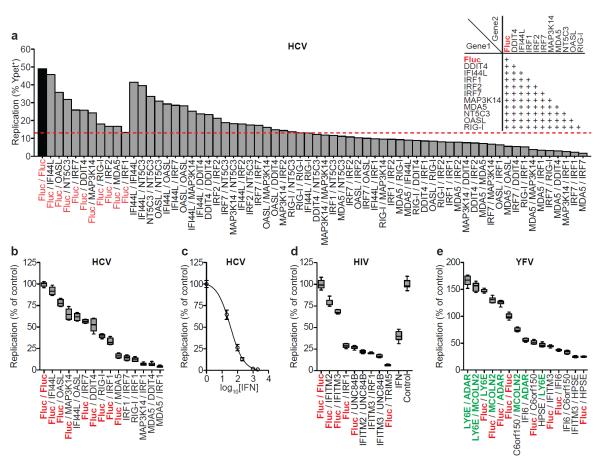
Figure 3. Combinatorial action of inhibitory and enhancing ISGs.
a. Anti-HCV ISGs were tested in 2-gene combinations (inset). Dashed red line denotes the strongest single-gene inhibitor, IRF1. b, d, e. Confirmation assays from two-gene screens for HCV (b), HIV(d), YFV(e). Data were normalized to Fluc control. n=8 for HCV, YFV, n=9 for HIV. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant). Inhibition of HIV IFNα is plotted for comparison. ISGs are color-coded: control (red), inhibitory (black), enhancing (green) c. Inhibition of HCV by IFNβ. Results are mean ± s.d., n=3.
In the primary screens, several genes enhanced viral replication (Fig 2a). Six of these (ADAR, APOBEC3A, FAM46C, IDO1, LY6E, and MCOLN2) were confirmed with YFV, WNV, VEEV, and CHIKV in _STAT1_−/−Fib (Fig 2d). For most genes, we found an increase in the number of YFV or VEEV-infected cells, whereas WNV and CHIKV were largely unaffected. Consistent with the primary YFV screen, LY6E conferred a striking 75% enhancement in infection frequency (Fig 2d and Supplementary Fig 9), a phenotype that may correlate with studies implicating LY6E in susceptibility of chickens to Marek’s disease virus20 and mice to adenovirus21. Analysis of mean GFP intensity in infected cells revealed that ADAR significantly enhanced replication of all viruses (Supplementary Fig 9). Thus, in contrast to LY6E, which caused an increase in the number of infected cells, ADAR affected the level of replication within the cell. ADAR encodes an RNA-specific adenosine deaminase that has been reported to stimulate HIV-1 infection by RNA editing22. ADAR also promotes measles virus infection23, supporting a pan-viral mechanism of enhancement.
The screening and validation data revealed both broad-acting and specific effectors. IRF1, a transcription factor in the IFN pathway, inhibited all viruses tested (Fig. 2c), even in a _STAT1_−/− background. Given that over 200 IRF1 binding sites reside in the human genome24, our data suggest that IRF1 activates a unique antiviral program. Indeed, microarray analysis of IRF1-transduced Huh-7 or _STAT1_−/−Fib revealed induction of numerous ISGs (Supplementary Fig 10), but no IFN genes were induced (data not shown). The majority of genes induced greater than 3-fold (p<0.05) in Huh-7 were similarly upregulated in _STAT1_−/−Fib, and both cells lines had a subset of uniquely regulated transcripts. Several of our antiviral hits were among the IRF1-induced transcripts, supporting the observation that IRF1 effector mechanisms partially overlap with IFN, but do not require its action. Genes C6orf150, HPSE, RIG-I, MDA5, IFITM3, IRF7, and NAMPT/PBEF1 were also active against more than one virus species, while other genes showed more restricted antiviral specificities. Gene ontology analysis was performed to classify confirmed antiviral ISGs. Compared to the entire ISG collection, the validated list showed a statistically significant overrepresentation of three molecular functions (nucleic acid binding, hydrolase, and helicase activity) and three biological processes (signal transduction, transcription initiation, and small molecule transport) (Supplementary Fig 11 and Supplementary Table 11).
The identification of modest to strong inhibitors suggests that an effective IFN response requires the combinatorial action of numerous ISGs. This hypothesis has been inferred from microarray1 and knockout studies25, but experimental data showing combinatorial ISG function is limited. To detemine whether ISG effects were greater when expressed in combination, selected hits from the HCV, HIV-1, and YFV screens were tested in all two-gene combinations (Fig 3a and Supplementary Fig 12). ISG effects were generally additive, with some combinations reducing replication greater than 90%, an efficacy similar to moderate doses of IFN (Fig 3b, c, d, e). For YFV, when one inhibitory and one enhancing ISG were tested together (e.g. C6orf150+MCOLN2), the magnitude of viral replication was more strongly influenced by the inhibitory gene.
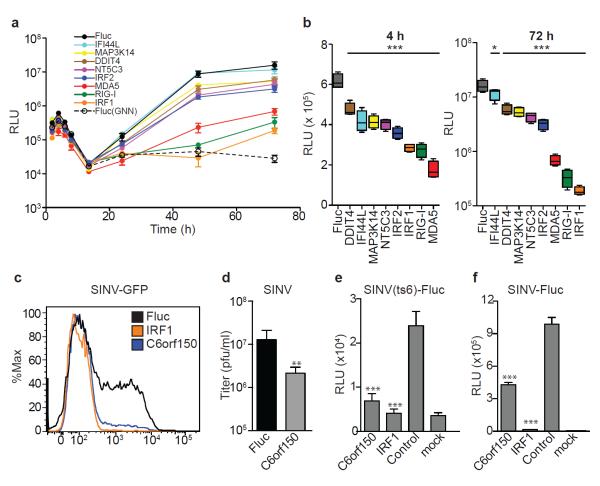
To probe antiviral mechanisms of action, select ISGs were tested for inhibition at various life cycle stages. Using HCV pseudoparticles (HCVpp), none of the eight anti-HCV ISGs significantly impaired viral entry (Supplementary Fig 13a). In contrast, analysis of subgenomic HCV replicons expressing Gluc revealed that all ISGs tested inhibited primary translation 25-70 % at 4 h post-transfection (Fig 4a, b and Supplementary Fig 13b, c). Notably, the strongest translational inhibitors were also the most potent suppressors of replication, as measured by Gluc production at 72 h (Fig 4c) and confirmed by quanitfying HCV genome copy number (Supplementary Fig 13d). The finding that eight genes with a range of predicted molecular functions (Supplementary Table 11) each block initial translation suggests the importance of this host strategy for HCV suppression. IRF1 and RIG-I were also associated with a kinetic delay in the onset of replication, suggesting multiple blocks in the HCV life cycle.
Figure 4. Translational inhibition as a common mechanism of ISG-mediated antiviral action.
a. ISG-expressing Huh-7 cells were transfected with HCV subgenomic replicon RNA and Gluc levels in cell supernatants were measured b. Inhibition of primary translation (left panel) or replication (right panel) was inferred from 4 h and 72 h data, respectively. Results are mean ± s.d., n=4. Statistical significance was determined by one-way ANOVA. (***,P<0.001, **,P<0.01, *,P<0.05, ns-not significant) d-g. C6orf150-mediated inhibition of SINV in _STAT1_−/−Fib. C6orf150 inhibits SINV-GFP replication (d) and non-GFP SINV production(e). C6orf150 and IRF1 inhibit SINV(ts6) primary translation (f) and SINV-Fluc replication (g). Results are mean ± s.d., n=3.
We also investigated the mechanism of C6orf150, which encodes a putative nucleotidyltransferase26 and contains a mab-21 domain, but is otherwise uncharacterized. Since C6orf150 inhibited alphaviruses CHIKV and VEEV, we tested it against the related Sindbis virus (SINV). C6orf150 inhibited SINV-GFP replication (Fig 4d) and SINV (non-GFP) infectious virus production (Fig 4e). We next assessed the activity of C6orf150 against a Fluc-expressing temperature-sensitive virus, SINV(ts6), which is competent for primary translation but does not replicate at the nonpermissive temperature (see Supporting Methods). SINV(ts6) was susceptible to C6orf150 inhibition (Fig 4f), indicating an early viral translation block. Interestingly, the pan-viral inhibitor IRF1 also blocked SINV(ts6) translation, suggesting this effector mechanism is reinforced by IFN-induced transcriptional factors. Similar to the HCV life cycle studies, the importance of translational inhibition is highlighted by a corresponding reduction in replication of wild type SINV-Luc (Fig 4g).
The antiviral effects of type I IFN were first described over 50 years ago27. Since then, great strides into the mechanisms governing virus-mediated IFN production, IFN gene regulation, and IFN signaling have been made28,29. Insight into downstream IFN effector mechanisms, however, have largely been limited to a select group of ISGs, likely due to difficulties involved in systematically overexpressing hundreds of genes. With the strategy presented here, we have overcome these technical barriers and identified multiple novel antiviral ISGs, many of which target early steps in the viral life cycle. Combined with well-known effectors such as PKR and IFIT proteins, life-cycle characterization of these newly identified ISGs suggests that the IFN system may use multiple strategies to engender a single outcome such as translational inhibition. Clinically, IFN is used to treat several viral infections and is a principle component of HCV treatment. Given the clinical side effects of IFN, and the natural ability of viruses to thwart its activity30, exploiting the actions of individual ISGs may be a preferable therapeutic strategy. Further insight into the biochemical mechanisms of these effectors may provide a platform for future studies on the development of alternatives to IFN-based therapies.
Methods
Viruses, replicons, and cells
Huh-7, Huh-7.5, and 293T cells were maintained in DMEM (Invitrogen) with 10% FBS and 0.1mM non-essential amino acids. _STAT1_−/−Fib (an SV40 large T antigen immortalized skin fibroblast line) and MT-4 (an HTLV-1 immortalized T cell line) were grown in RPMI (Invitrogen) with 10% FBS. BHK-J cells were grown in MEM (Invitrogen) with 7.5% FBS. Infectious HCV-Ypet31 (derived from Bi-Ypet-Jc1FLAG2), YFV-Venus31 (derived from YF17D-5′C25Venus2AUbi), WNV-GFP32 (derived from pBELO-WNV-GFP-RZ ic) and CHIKV-GFP33 (derived from pCHIKV-LR 5′GFP), SINV/SINV-GFP34 (derived from pS300/pS300-GFP), SINV-Fluc and the temperature sensitive variant SINV(ts6)Fluc were derived from pToto1101/Luc and pToto11-1/Luc:ts6. SINV(ts6)Fluc carries mutations that prevent replication at the nonpermissive temperature, but the virus retains the ability to translate incoming genomes35. VEEV-GFP is a double subgenomic EGFP reporter virus derived from the TC83 vaccine strain of Venezuelan equine encephalitis (kindly provided by I. Frolov).
HCV subgenomic replicon RNAs were derived from Bi-Gluc-JFH(SG) and Bi-Gluc-JFH-GNN(SG), which are bicistronic constructs expressing Gluc from the HCV IRES and genotype 2a JFH-1 nonstructural proteins from the EMCV IRES. The GNN variant encodes polymerase mutations and is not competent for replication. Viral stocks were generated by electroporation of in vitro transcribed RNAs into Huh-7.5 (HCV) or BHK-J (YFV, WNV, VEEV, CHIKV, SINV) as previously described32,33,35, 36,37,38. Single cycle HIV-1-GFP was generated by transfection of HEK 293T cells with a modified env- proviral plasmid encoding GFP in place of Nef (pR7/Δ_env_/GFP)39. HIV particles were pseudotyped with VSV-G glycoprotein. Lentiviral pseudoparticles with no envelope or carrying HCV glycoprotein (HCVpp) were generated and assayed for HCV entry as previously described40. Experiments with WNV, CHIKV, and HIV-1 were carried out in biosafety level 3 containment in compliance with institutional and federal guidelines.
DNA constructs
To generate a lentiviral-based, Gateway-compatible destination vector for ISG expression, a Gateway expression cassette (containing the tetracyline-inducible-hybrid CMV promoter, chloramphenicol resistance gene, and ccdB suicide gene) from pLenti4.TO.V5-DEST (Invitrogen) was subcloned into the XhoI-NdeI sites of pTRIP-EGFP41. Overlap extension PCR followed by a three-fragment ligation was used to insert the EMCV IRES and TagRFP (Evrogen) downstream of the Gateway module
(EMCV IRES 5′ oligo:
5′-GATATCTCGAGGCCCCTCTCCCTCCCCCCCCCCTAA-3′,
EMCV IRES 3′ oligo:
5′-CACGATGATAATATGGCCACAACCCCGCGGATATG-3′,
TagRFP 5′ oligo:
5′-CATAGCTAGCATGGTGTCTAAGGGCGAAGAGCTG,
TagRFP 3′ oligo:
5′-CTAGCAAACTGGGGCACAAACTTAATTGACCGCGGGGTACCTGCG-3′.
To enhance gene expression, the β-globin intron (IVSβ) was subcloned from pLenti6-TR (Invitrogen) into the SpeI site between the CMV promoter and the Gateway cassette
(IVSβ 5′ oligo:
5′-GACCCACTAGTGTGAGTTTGGGGACCCTTGATTG-3′,
IVSβ 3′ oligo:
5′-CATGCCTTCTTCTTTTTCCTACAGACTAGTCCCAG-3′).
All DEST vector variants were grown in DB3.1 cells (Invitrogen) under ampicillin-chloramphenicol double selection. The final vector was named pTRIP.CMV.IVSb.ires.TagRFP-DEST.
Sequence-validated, Gateway-compatible ORFEXPRESS shuttle clones corresponding to 387 ISGs were obtained from Genecopoeia. Two additional ISG entry clones encoding RSAD2 and ZC3HAV1 and two control clones encoding Firefly luciferase (Fluc) or Gaussia luciferase (Gluc) were generated as follows. Genes encoding RSAD2, ZC3HAV1/ZAP, Fluc, and Gluc were PCR-amplified with oligos containing attB sites flanking gene-specific sequences
(RSAD2 5′ oligo:
5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGTGGGTGCTTACACCTGC-3′,
RSAD2 3′ oligo:
5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTACCAATCCAGCTTCAGAT-3′,
ZCHAV1 5′ oligo:
5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGCGGACCCGGAG GTGTG-5′,
ZCHAV1 3′ oligo:
5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACTCTGGCCCTCTCTTCATCT-3′,
Fluc 5′ oligo:
5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGAAGATGCCAAAAACATTAAGAA-3′,
Fluc 3′ oligo:
5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACACGGCGATCTTGCCGCCCTTC-3′,
Gluc 5′ oligo:
5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGGGAGTCAAAGTTCTGTTTGCCC-3′,
Gluc 3′ oligo:
5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGTCACCACCGGCCCCCTTGATC-3′.
PCR products were purified over GFX columns (GE Healthcare) and cloned into pDONR (Invitrogen) with BP Clonase. BP reactions were transformed into OMNIMax competent E. coli (Invitrogen) and colonies were screened by restriction digest and sequencing.
The ISG-encoding sequences from pENTR clones were moved into pTRIP.CMV.IVSb.ires.TagRFP-DEST using LR Clonase II (Invitrogen) according to manufacturer’s instructions. LR reaction products were transformed into MDS42Rec reduced genome E. coli. (Scarab Genomics). One or two colonies for each clone were grown in 3 ml Luria broth (LB) plus ampicillin and transfection-quality plasmid DNA was purified over anion-exchange columns (Qiagen). All pTRIP.CMV.IVSb.ISG.ires.TagRFP constructs were sequenced at the 5′ end of the expression cassette to verify gene insertion (sequencing oligo: 5′-CCTGCCTTTCTCTTTATGG-3′).
Generation of ISG-expressing lentiviral pseudoparticles
Lentiviral pseudoparticles were generated by co-transfecting 4×105 293T cells in 6 well plates or 1.2×104 cells in 96 well plates with plasmids expressing: 1) the pTRIP.CMV.IVSb.ISG.ires.TagRFP proviral DNA, 2) HIV gag-pol, and 3) the vesicular stomatitis virus glycoprotein (VSV-G) in a ratio of 1:0.8:0.2, respectively. For each transfection, 6 μl Fugene (Roche) was combined with 2.0 μg total DNA in 100 μl Optimem (Gibco). Transfections were carried out for 6 h followed by a media change to DMEM with 3%FBS. Supernatants were harvested at 48 h and 72 h, pooled, cleared by centrifugation, and stored at −80°C.
Lentiviral transduction and viral replication assays
Transduction assays
Huh-7, Huh-7.5, _STAT1_−/−Fib were seeded into 24 well plates at a density of 7 ×104 cells/well and transduced with lentiviral pseudoparticles by spinoculation at 1000-1500 x g for 1 h at 37°C in media containing 3% FBS, 20 mM Hepes, and 4 μg/ml polybrene. For HIV studies, suspension MT-4 cells were transduced in a 96 well format. For monolayer experiments with Huh-7, Huh-7.5, _STAT1_−/−Fib, cells were split 1:2 or 1:3 at 48 h post-transduction.
Replication assays
Tansduced cells were infected with the indicated virus at a dose that gave approximately 50% infected (GFP+) cells, which had been previously determined by FACS-based infectivity assays. Infected cells were harvested at various time points, within the first viral replication cycle when possible: HCV (48 h or 72 h), HIV (48 h), YFV (24 h), WNV (6 h), VEEV (5.5 h), CHIKV (10 h). Adherent cells were harvested into 200 μl Accumax cell dissociation medium (eBioscience) and transferred to a 96 well plate. Cells were pelleted at 2,000 RPM for 5 min at 4°C and resuspended in 1% paraformaldehyde fixation solution for at least 1 h. Cells were pelleted by centrifugation at 2,000 RPM for 5 min at 4°C, resuspended in cold 1X PBS+3% FBS, and stored at 4°C until FACS analysis. Samples were analyzed in a 96 well-based high throughput manner using the LSRII/HTS flow cytometer (BD Biosciences) equipped with a 561nm laser for detection of TagRFP. Data was analyzed using Flowjo software (Treestar) with a 0.1% compensation matrix.
For interferon titrations, Huh-7 or MT-4 T cells were treated with varying doses of IFNβ (PBL InterferonSource) or 1000 U/mL IFNα (Sigma), respectively, for 24 h prior to infection. Cells were infected with HCV (Huh-7) or HIV-1 (MT-4) and replication levels were monitored by FACS as described above.
For SINV-Fluc and SINV(ts6)-Fluc infection, cells were infected at 28°C (ts6) or 37°C (wild type) and harvested 4 h post-infection. Intracellular Fluc levels were assessed by the Luciferase Assay System reporter assay (Promega) according to the manufacturer’s instructions.
For HCV life cycle studies, subgenomic RNAs were generated by in vitro transcription using the RiboMax T7 transcription kit (Promega). 175ng RNA were transfected into 3.5 × 104 ISG-expressing Huh-7 cells with Mirus Reagent (Mirus Bio). Translation and replication were monitored by sampling cell supernatants and assaying for Gluc production over time (at 2, 4, 6, 8, 13.5, 24, 48, and 72 h post-transfection) with the Renilla Luciferase Assay System (Promega), or by quantifying HCV genome copy number by reverse transcription (RT)-PCR as previously described42.
DNA transfection-based ISG screen in of Huh-7 cells
Huh-7 cells were seeded into 24 well plates at a density of 7×104 cells/well. The next day, growth media was changed to DMEM with 1% FBS and 0.1mM non-essential amino acids. Lentiviral plasmid DNAs were transfected at 400 ng DNA/well using Lipofectamine 2000 (Invitrogen) in a total volume of 1 ml/well. Plates were centrifuged at 1000 x g for 30 min at 37°C. Five hours later, media was changed to DMEM with 10% FBS and two days post-transfection, cells were challenged with HCV-Ypet and assayed for replication by FACS as described above.
Immunofluorescence
_STAT1_−/−Fib expressing IRF1.ires.TagRFP, C6orf150.ires.TagRFP, or MAP3K14.ires.TagRFP were stained for protein levels using Cellomics Whole Cell Stain Kit (Thermo Scientific) according to the manufacturer’s instructions. Affinity-purified polyclonal rabbit antisera detecting human IRF1, MAP3K14 (alias: NIK), and C6orf150 were obtained from Abcam or Sigma and used at manufacturer’s recommended concentrations. An AlexaFluor-488 conjugated goat anti-rabbit antibody (Invitrogen) was used at a concentration of 1:1000 for detection, and samples were visualized by epifluorescence microscopy on an Eclipse TE300 (Nikon) inverted microscope.
Microarray analysis
Huh-7 and _STAT1_−/−Fib were transduced with lentiviruses expressing Fluc.ires.TagRFP or IRF1.ires.TagRFP. Total RNA was harvested from transduced cells 48 h later using Qiagen RNeasy Mini Kit. RNA amplification was carried out with MessageAmp™ Premier RNA Amplification Kit (Applied Biosystems). 200 ng of total RNA was used to synthesize the first strand of cDNA using ArrayScript reverse transcriptase and an oligo(dT) primer bearing a T7 promoter. The single-stranded cDNA was then converted into a double-stranded DNA (dsDNA) by DNA polymerase I in the presence of E. coli RNase H and DNA ligase. The dsDNA served as a template for in vitro transcription in a reaction containing biotin-labeled UTP, unlabeled NTPs and T7 RNA Polymerase. The amplified, biotin-labeled antisense RNA (aRNA) was purified and quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit. 750 ng of aRNA in 5 μl was mixed with 10 μl of hybridization reagents and heated at 65°C for 10min. After cooling to room temperature, 15 μl of the hybridization solution was applied to an Illumina HumanHT-12 v4 chip. The chip was incubated for about 18 hours at 58°C. After washing and staining with streptavidin-Cy3, the chip was scanned using Illumina BeadArray Reader. The scanning was done using standard DirectHyb Gene Expression protocol with the following settings: Factor=1, PMT=587, Filter=100%. The raw data was extracted using Illumina BeadStudio software without normalization. Data was analyzed using GeneSpring Software with global normalization. Students unpaired t-test was used to determine statistical significance.
Supplementary Material
1
4
Acknowledgements
We are thankful to the following investigators for contributing viral molecular clones: C. Stoyanov (The Rockefeller University-YFV), S. Higgs (UTMB-CHIKV), I. Frolov (UTMB-WNV and VEEV), M. MacDonald and J. Law (The Rockefeller University-SINV(ts6) and SINV-Fluc), M. Heise (UNC-SINV,SINV-GFP). We thank E. Jouanguy and J-L. Casanova for _STAT1_−/− fibroblasts. We acknowledge the support of C. Zhao, X. Wang, and W. Zhang at The Rockefeller University Genomics Resource Center. We are thankful for the technical advice of S. Mazel and C. Bare at The Rockefeller University Flow Cytometry Resource Center, supported by the Empire State Stem Cell Fund through NY State Department of Health (NYSDOH) contract no. C023046; opinions expressed here are solely those of the authors and do not necessarily reflect those of the Empire State Stem Cell Fund, the NYSDOH, or the State of NY. We thank M. Holz, E. Castillo, A. Webson for laboratory support, C. Murray for critical reading and editing of the manuscript, and A. Ploss, M. Scull, M.T. Catanese, S. You for thoughtful discussions. This work was supported in part by NIH grants AI057158 (Northeast Biodefense Center-Lipkin) to C.M.R and AI064003 to P.B. Additional funding was provided by the Greenberg Medical Research Institute, the Starr Foundation and the Ronald A. Shellow, M.D. Memorial Fund (C.M.R.). J.W.S. and C.T.J. were supported by National Research Service Awards DK082155 and DK081193, respectively, from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions J.W.S. and C.M.R. designed the project. J.W.S, S.J.W., M.P., M.Y.M, and C.T.J performed the experimental work. J.W.S, S.J.W., P.B, and C.M.R. analyzed results and wrote the manuscript. S.J.W., C.T.J., and P.B. contributed reagents and technical expertise. Microarray data was submitted to NCBI GEO database with accession GSE26817.
Author information Reprints and permissions information is available at www.nature.com/reprints. The authors declare the following conflicts of interest, which are managed under University policy: C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and the HCVcc cell culture system.
References for Print Article
- 1.de Veer MJ, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 2.Knight E, Jr., Korant BD. Fibroblast interferon induces synthesis of four proteins in human fibroblast cells. Proc Natl Acad Sci U S A. 1979;76:1824–1827. doi: 10.1073/pnas.76.4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larner AC, et al. Transcriptional induction of two genes in human cells by beta interferon. Proc Natl Acad Sci U S A. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 5.Keskinen P, et al. Impaired antiviral response in human hepatoma cells. Virology. 1999;263:364–375. doi: 10.1006/viro.1999.9983. [DOI] [PubMed] [Google Scholar]
- 6.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky LI, et al. A novel unsupervised method to identify genes important in the anti-viral response: application to interferon/ribavirin in hepatitis C patients. PLoS One. 2007;2:e584. doi: 10.1371/journal.pone.0000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He XS, et al. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology. 2006;44:352–359. doi: 10.1002/hep.21267. [DOI] [PubMed] [Google Scholar]
- 9.Hilkens CM, Schlaak JF, Kerr IM. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J Immunol. 2003;171:5255–5263. doi: 10.4049/jimmunol.171.10.5255. [DOI] [PubMed] [Google Scholar]
- 10.Hultcrantz M, et al. Interferons induce an antiviral state in human pancreatic islet cells. Virology. 2007 doi: 10.1016/j.virol.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Indraccolo S, et al. Identification of genes selectively regulated by IFNs in endothelial cells. J Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 12.Lanford RE, et al. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 13.Leaman DW, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 14.Rani MR, et al. Novel interferon-beta-induced gene expression in peripheral blood cells. J Leukoc Biol. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- 15.Sarasin-Filipowicz M, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy S, et al. Control of the interferon-induced 68-kilodalton protein kinase by the HIV-1 tat gene product. Science. 1990;247:1216–1219. doi: 10.1126/science.2180064. [DOI] [PubMed] [Google Scholar]
- 17.Neil S, Bieniasz P. Human immunodeficiency virus, restriction factors, and interferon. J Interferon Cytokine Res. 2009;29:569–580. doi: 10.1089/jir.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furtak V, et al. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS One. 2010;5:e9081. doi: 10.1371/journal.pone.0009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HC, Niikura M, Fulton JE, Cheng HH. Identification of chicken lymphocyte antigen 6 complex, locus E (LY6E, alias SCA2) as a putative Marek’s disease resistance gene via a virus-host protein interaction screen. Cytogenet Genome Res. 2003;102:304–308. doi: 10.1159/000075767. [DOI] [PubMed] [Google Scholar]
- 21.Spindler KR, et al. The major locus for mouse adenovirus susceptibility maps to genes of the hematopoietic cell surface-expressed LY6 family. J Immunol. 184:3055–3062. doi: 10.4049/jimmunol.0903363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284:29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 2009;37:1073–1085. doi: 10.1093/nar/gkn1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 26.Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 28.Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 30.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Supplemental Information
- 31.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGee CE, et al. Infection, Dissemination, and Transmission of a West Nile Virus Green Fluorescent Protein Infectious Clone by Culex pipiens quinquefasciatus Mosquitoes. Vector Borne Zoonotic Dis. 2009 doi: 10.1089/vbz.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsetsarkin K, et al. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006;6:325–337. doi: 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- 34.Suthar MS, Shabman R, Madric K, Lambeth C, Heise MT. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J Virol. 2005;79:4219–4228. doi: 10.1128/JVI.79.7.4219-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 37.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrakova O, et al. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YJ, et al. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J Virol. 2002;76:6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 41.Zennou V, et al. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 42.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
4