Hoxa9 and Meis1 Are Key Targets for MLL-ENL-Mediated Cellular Immortalization (original) (raw)
Abstract
MLL fusion proteins are oncogenic transcription factors that are associated with aggressive lymphoid and myeloid leukemias. We constructed an inducible MLL fusion, MLL-ENL-ERtm, that rendered the transcriptional and transforming properties of MLL-ENL strictly dependent on the presence of 4-hydroxy-tamoxifen. MLL-ENL-ERtm-immortalized hematopoietic cells required 4-hydroxy-tamoxifen for continuous growth and differentiated terminally upon tamoxifen withdrawal. Microarray analysis performed on these conditionally transformed cells revealed Hoxa9 and Hoxa7 as well as the Hox coregulators Meis1 and Pbx3 among the targets upregulated by MLL-ENL-ERtm. Overexpression of the Hox repressor Bmi-1 inhibited the growth-transforming activity of MLL-ENL. Moreover, the enforced expression of Hoxa9 in combination with Meis1 was sufficient to substitute for MLL-ENL-ERtm function and to maintain a state of continuous proliferation and differentiation arrest. These results suggest that MLL fusion proteins impose a reversible block on myeloid differentiation through aberrant activation of a limited set of homeobox genes and Hox coregulators that are consistently expressed in MLL-associated leukemias.
Conditional oncogenes provide a powerful tool for studying early events in transformation. Here we applied this approach to dissect the mechanism of leukemic transformation by MLL fusion proteins. MLL, located at 11q23, is rearranged in a variety of human acute lymphoid and myeloid leukemias. Our goals were to determine if MLL-induced growth transformation is reversible, to characterize the effect of MLL fusion protein expression on cell proliferation, differentiation, and survival in physiologically relevant cells, and finally to identify genes regulated by MLL fusion that are important for transformation.
MLL is the mammalian homologue of Drosophila Trithorax (TRX) (10, 16, 43, 52). MLL and Trithorax are important constituents of transcriptional maintenance, a form of “cellular memory” that maintains preestablished transcription patterns through the introduction of epigenetic chromatin modifications. Full-length MLL protein (3,968 amino acids) is cleaved into two parts (MLLN and MLLC) that remain noncovalently bound in a binary complex (19, 46). The 300-kDa MLLN contains several motifs that are likely to be involved in targeting the protein to specific chromosomal sites. Two domains in particular, the AT hooks and MT or methyltransferase homology region, possess DNA binding activity (4, 51). These are followed by a region with several specialized zinc fingers (PhD fingers for plant homeodomain) that include a bromodomain. MLLC is responsible for the epigenetic effector functions through a conserved SET domain with histone H3 (K4) methylation activity (27) and by providing binding sites for the histone acetyltransferase CREB-binding protein (11) as well as for the SWI-SNF chromatin remodeling complex (33). The MLLN/C couple is embedded into a megadalton complex that serves the overall purpose of transcriptional activation of specific genes (30).
Translocations involving MLL delete sequences encoding the C-terminal portion of the protein, ultimately leading to the production of an in-frame fusion protein with transforming activity. 11q23 translocations are especially prevalent in infant cases of acute lymphoblastic leukemia, and they are associated with a very aggressive course of disease that is correlated with a dismal prognosis (3, 9, 12).
In knockout experiments the clustered homeobox (Hox) genes have been identified as targets of mammalian MLL (47, 48). Hox genes are not only major determinants of segment identity during embryonic development but also are crucial regulators of hematopoiesis. Mll knockout mice displayed a perturbed hematopoietic development in the yolk sack and fetal liver (18, 44). Importantly, Hox genes have been recognized as activated oncogenes in leukemia. In particular, Hoxa9 and Hoxa7 are frequently affected by retroviral insertion mutagenesis and the HOXA9 protein is a fusion partner of the nucleoporin protein NUP98 in a subset of AML with the translocation t(7;11) (28, 29, 31, 41, 42).
Although knock-in and retroviral transduction studies unequivocally established that the combination of MLL and the respective partner protein leads to oncogenic activation of the chimeric molecule, the underlying transforming mechanism of the MLL fusion derivatives is still unknown. Structure-function studies suggest transcriptional transactivation is the essential activity acquired through the fusion (8, 22, 25, 37, 39). This was supported by experiments from our laboratory and others showing that fusion of MLL to the strong transcriptional transactivator domain VP16 of the Herpes simplex virus was transforming (38, 49). In addition, the frequent translocation partners are nuclear proteins that are probable transcription factors that seem to work by converting the N terminus of MLL into an aberrant transcription factor, resulting in ectopic gene expression that ultimately disturbs growth control or differentiation and cooperates with further mutations to establish a leukemic state.
To gain insight into the mode of action of an exemplary MLL fusion protein, we constructed a conditional derivative of MLL-ENL that can be transduced with a single retrovirus. By this strategy it was possible to analyze the consequences of MLL-ENL expression in primary hematopoietic cells, to determine if MLL fusion protein-induced leukemogenesis is reversible, and to identify a set of MLL-ENL target genes that are potentially important for leukemogenesis. Here we demonstrate that MLL-ENL immortalizes cells mainly through inducing a reversible block on myeloid differentiation that is dependent on upregulation of Hoxa9 and Meis1 and that enforced expression of these two genes is sufficient to substitute for MLL-ENL function.
MATERIALS AND METHODS
Plasmid construction.
The cDNA coding for amino acids 281 to 599 of a mutated estrogen receptor was fused to the cDNA of the MLL-ENL derivative pΔEN2 (37), resulting in pMSCVneo-MLL-ENL-ERtm. The cDNAs for Hoxa7, Hoxa9, and Meis1a were cloned into the pMSCV puro, pMSCV hygro, or pMSCV green fluorescent protein (GFP) vector by standard techniques.
Tissue culture, transfection procedures, and growth assays.
Human embryonic kidney cells (293T) were cultured in high-glucose Dulbecco's modified Eagle medium (Life Technologies); human pre-B REH cells and mouse myeloid M1 cells were kept in RPMI 1640 (Life Technologies). For protein expression the cells were transfected by a standard Ca-phosphate method. The ecotropic packaging cell line Phoenix was obtained from Gary Nolan (Stanford, Calif.). The maintenance of Phoenix cells is outlined at http://www.stanford.edu/group/nolan/.
Transduced bone marrow cells were kept either in MethoCult (M3234) methylcellulose medium (Stem Cell Technologies, Vancouver, Canada) or in RPMI 1640. Recombinant mouse cytokines (Strathmann Biotech, Hannover, Germany) were added in the following concentrations: interleukin 3 (IL-3), IL-6, and granulocyte-macrophage colony-stimulating factor (GM-CSF), 10 ng/ml; SCF, 100 ng/ml. All liquid media were supplemented with 10% bovine fetal calf serum (FCS Gold; PAA Laboratories, Pasching, Austria) and penicillin-streptomycin. 4-Hydroxy-tamoxifen (4-OHT) was added at a 100 nM final concentration as a 1 mM stock solution in ethanol.
For luciferase reporter studies REH cells or M1 cells were electroporated in RPMI 1640 containing 5 μg of DEAE-dextran/ml at 300 V and 925 μF in 4-mm cuvettes without a pulse controller in a Bio-Rad electroporator. In total, 1 μg of DNA was transfected in a ratio of expression plasmid to reporter of 9 to 1. Luciferase activity was determined with the luciferase assay system (Promega, Madison, Wis.) according to the instructions of the manufacturer.
Western blotting, enzyme-linked immunosorbent assay, and antibodies.
For the detection of MLL-ENL and MLL-ENL-ERtm fusion proteins nuclear extracts were prepared from transfected 293T cells in a high-salt elution buffer (500 mM NaCl, 20 mM HEPES [pH 7.5], 0.5 mM EDTA, 0.1% Triton X-100, 0.5 mM sodium vanadate, 2 mM NaF, 2 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 20 μg of leupeptin/ml, 40 μg of pepstatin A/ml). After separation on standard sodium dodecyl sulfate (SDS)-polyacrylamide gels the proteins were blotted onto nitrocellulose in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid-0.01% SDS-1% methanol. The detection was done with monoclonal antibodies against MLL (5).
Antibodies for fluorescence-activated cell sorter (FACS) analysis (isotype control, c-kit, Gr-1, Mac-1/CD11b) were purchased from BD Biosciences and used according to the recommendations of the manufacturer.
Retroviral transduction of mouse primary hematopoietic cells.
High-titer retrovirus supernatants were produced by transient transfection of the packaging cell line Phoenix-E by a standard Ca-phosphate precipitation method. Viral titers usually reached approximately 5 × 106 CFU/ml. The retroviral transduction of primary hematopoietic cells was done according to a method described previously (34).
RT-PCR, quantitative RT-PCR, Southern blotting, and cDNA array analysis.
Total RNA was isolated by ion-exchange chromatography with kits from Qiagen (Hilden, Germany) according to the recommended protocols. MLL fusion-specific RNA and Bmi-1 RNA were detected by reverse transcription-PCR (RT-PCR) after reverse transcription of total RNA with a random hexamer primer. Southern blotting was done on DNA isolated by phenol-chloroform extraction and blotted under alkaline conditions according to standard procedures.
Real-time PCR was performed at least in triplicate using Taqman probes and the ABI prism 7700 sequence detection system. Samples were quantified relative to a cDNA dilution series and normalized to β-actin as outlined in ABI user bulletin no. 2. Probes used were as follows: Hoxa9, Taqman probe, CCCCATCGATCCCAATAACCCAGC, forward primer, GAATGAGAGCGGCGGAGAC, reverse primer, GAGCGAGCATGTAGCCAGTTG; Meis1, Taqman probe, ACCGGTCCACCACCTGAACCACG, forward primer, GCATGCAGCCAGGTCCAT, reverse primer, TAAAGCGTCATTGACCGAGGA; Hoxa7, Taqman probe, CGCAGTTCAGGACCCGACAGGAA, forward primer, CGGCCGAAGCCAGTTTC, reverse primer, GCGCCGCGTCAGGTAG; Pbx1, Taqman probe, CAGTGACGGACTCGCAGCCAGTCA, forward primer, TGTTATCAGCCAGACAGGAGGAT, reverse primer, TGCCCTGCGGACTGTACA; Pbx3, Taqman probe, CCATGCAGGCTCTCATCATCGTTCTCA, forward primer, TAAGCTGAACCCTTGCGGATT, reverse primer, TCCAAGCCCATCCGTGAT.
Target preparation and hybridization of microarrays was conducted as described in the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, total RNA was converted to first-strand cDNA, using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 min, and hybridized for 16 h at 45°C to the microarrays. The microarrays were then washed at low (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}]) and high (100 mM morpholineethanesulfonic acid, 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect fluorescence signal at 3-μm resolution after excitation at 570 nm. The average signal from two sequential scans was calculated for each microarray feature.
Affymetrix Microarray Suite 5.0 was used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (tau = 0.015) was used to assign a call of absent, present, or marginal for each assayed gene, and a P value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated using the one-step Tukey's biweight estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays: after exclusion of the highest and lowest 2%, the average total chip signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor.
RESULTS
Construction of a conditional MLL-ENL derivative.
We adapted the available in vitro transformation assay for MLL-ENL activity, which relies on retroviral transduction of primary hematopoietic cells, by fusing the ligand-binding domain of the estrogen receptor modified by a single point mutation (ERtm) to MLL. The use of this modified receptor avoids interference with endogenous estrogens. This altered domain binds specifically to the synthetic estrogen derivative 4-OHT or its precursor substance tamoxifen, which is converted to 4-OHT by cellular enzymes. Proteins fused to ERtm are presumably retained in a complex with heat shock proteins. Upon binding to 4-OHT the fusion protein is released and commences its normal function (24).
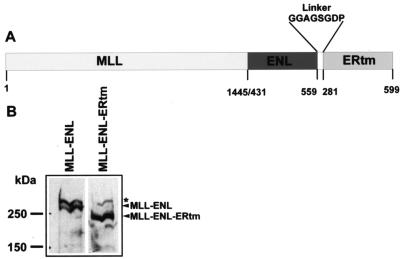
Due to the length restraints for retrovirally packaged DNA, a shorter deletion variant of MLL-ENL was chosen for this experiment. In this molecule only the last 128 amino acids of ENL are fused to the MLL N terminus. This ENL portion was shown previously to encompass the minimal essential transactivation domain that is necessary to convey transforming properties to MLL (37). The ERtm domain (amino acids 281 to 599 of the estrogen receptor) (24) was fused to the C terminus of MLL-ENL separated by an eight-amino-acid glycine-rich linker. The assembled cDNA was inserted into the retroviral backbone pMSCVneo, yielding pMSCV-MLL-ENL-ERtm. A schematic representation of the construct is depicted in Fig. 1A.
FIG. 1.
Construction of an inducible derivative of MLL-ENL. A) Schematic representation of the MLL-ENL-ERtm construct. Numbers denote the respective amino acid positions of the individual constituents. The amino acid sequence of the linker is given in the one-letter code. (B) Western blot analysis of MLL-ENL-ERtm expression. Phoenix-E retroviral packaging cells were transfected with the retroviral vectors pMSCV-MLL-ENL, coding for native MLL-ENL, and pMSCV-MLL-ENL-ERtm. Nuclear lysates were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with an anti-MLL monoclonal antibody recognizing an N-terminal epitope. The star denotes the N-terminal cleavage product of endogenous MLL.
The correct expression of the MLL-ENL-ERtm assembly was tested by transfection of the Phoenix-E packaging cell line with pMLL-ENL-ERtm and, as a control, with a similar retrovirus encoding a “full-length” MLL-ENL protein. Subsequent immunoblotting with an antibody specific for the MLL N terminus revealed comparable amounts of the respective proteins (Fig. 1B). The MLL-ENL-ERtm protein was detected in the nucleus of the transfected cells regardless of the presence or absence of 4-OHT. Therefore 4-OHT does not regulate the subcellular localization. This has been reported previously also for the ER fusion proteins JunD-ER and ER-E1A (14, 32).
4-OHT dependent functions of MLL-ENL-ERtm.
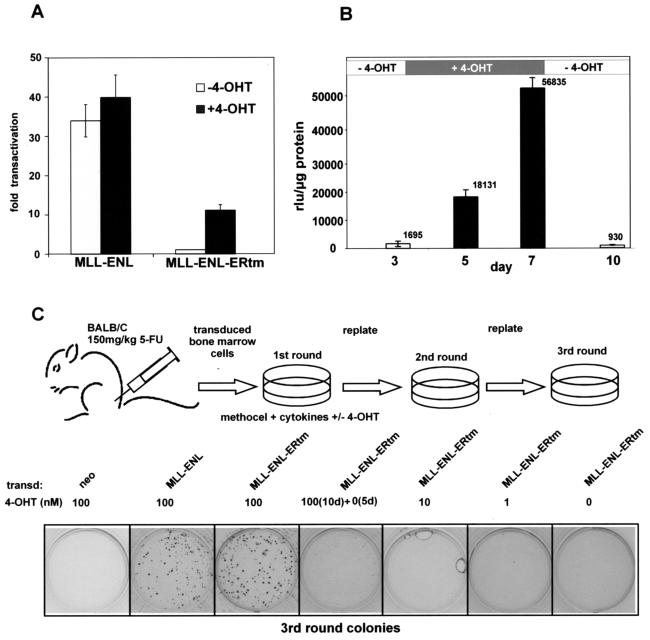
The transformation capability of native MLL-ENL correlates closely with its function as a transcriptional transactivator (35, 49). Therefore the transactivation properties of MLL-ENL and MLL-ENL-ERtm were measured in the presence or absence of 4-OHT to confirm that the inducible construct retained this key activity. To this end, 0.9 μg of the respective pMSCV constructs were coelectroporated into the pre-B-cell line REH together with 0.1 μg of a luciferase reporter driven by the murine Hoxa7 promoter (pGL3-Hoxa7). This reporter-cell line combination has been previously demonstrated to give a reliable transcriptional readout that is proportional to the transformation potency of MLL fusion proteins (35, 49). After electroporation the cells were either cultivated for 24 h in the presence of 100 nM 4-OHT or they were mock treated with solvent only. As expected the addition of 4-OHT did not significantly affect the transactivation capability of MLL-ENL. Regardless of the presence of inductor, MLL-ENL achieved an approximately 30- to 40-fold increase of luciferase activity over basal levels (Fig. 2A). In contrast, the MLL-ENL-ERtm molecule was completely inactive in the absence of 4-OHT but acquired a transactivation potential after 4-OHT addition that was capable of increasing luciferase levels about 10-fold.
FIG. 2.
Characterization of MLL-ENL-ERtm properties. (A) Transcriptional transactivator capability of MLL-ENL versus that of MLL-ENL-ERtm. The respective retroviral plasmids were coelectroporated with a luciferase reporter driven by the murine Hoxa7 promoter into the pre-B-cell line REH. The transfected cells were cultivated for 24 h in the presence or absence of 100 nM 4-OHT and lysed, and luciferase levels were determined normalized to protein content. Transactivation is given as the fold increase compared to the background levels obtained when an empty retroviral vector was coelectroporated. Shown are means and standard deviations of triplicate experiments. (B) Reversibility of 4-OHT-induced transactivation by MLL-ENL-ERtm. Mouse M1 cells stably expressing MLL-ENL-ERtm were cultivated for 3 days, and then 100 nM 4-OHT was added before the cells were depleted of inductor again at day 7 and kept for 3 more days in unsupplemented medium. At the indicated time points cell samples were drawn and electroporated with the _Hoxa7_-luciferase reporter, and luciferase levels were determined as relative luciferase units per microgram of protein. Shown are the means and standard deviations of triplicate experiments. (C) Outline and results of a bone marrow replating assay to assess the transforming properties of MLL-ENL-ERtm. Primary bone marrow cells enriched in hematopoietic precursor cells by 5-fluorouracil (5-FU) treatment of the donors were transduced with retroviruses capable of expression of MLL-ENL, MLL-ENL-ERtm, or neomycin only. The infected cells were plated under drug selection in cytokine-supplemented methocel medium. Ten thousand cells from colonies arising after the first selection round were replated in cytokine-supplemented medium, and this procedure was repeated once more. Shown are stained third-round colonies transduced with the indicated retroviruses and cultivated with the given amounts of 4-OHT. This is one representative example of five independent transduction experiments.
We next asked whether the 4-OHT induction is fully reversible, as this would suggest that the transforming activity of the protein is also completely reversible upon tamoxifen withdrawal. We established a cell line (M1-1) that stably expressed MLL-ENL-ERtm by transfection of mouse myeloid M1 cells with a linearized pMSCV-MLL-ENL-ERtm plasmid, followed by neomycin selection and subcloning. The M1-1 cell line was electroporated with the Hoxa7 promoter-luciferase reporter before 4-OHT addition, during cultivation in 4-OHT and after the cells were returned to 4-OHT-free medium. As Fig. 2B shows, luciferase levels were about 10-fold higher compared to the background 2 days after incubation with 4-OHT and increased further during culture in 4-OHT medium. Despite this strong induction, the activity returned to basal levels within 3 days after 4-OHT withdrawal, indicating a complete reinactivation of MLL-ENL-ERtm.
The transforming capacity of MLL-ENL-ERtm was assessed in retroviral transduction/replating assays, which test the ability of MLL fusion proteins to increase the self-renewing potential and block differentiation of hematopoietic cells. Untransduced bone marrow cells rapidly differentiate and die. In contrast progenitor cells that express oncogenic MLL fusions will continuously form colonies in semisolid cytokine-supplemented media even after repeated replating. A schematic depiction of the experimental outline is shown in Fig. 2C. The viral constructs coding for MLL-ENL-ERtm, native MLL-ENL, and an empty pMSCV vector were packaged in Phoenix-E cells and used to transduce bone marrow cells from BALB/c mice primed 5 days earlier with an injection of 150 mg of 5-fluorouracil/kg. Ten thousand cells were plated in duplicate per assay with neomycin selection in 1 ml of methocel medium supplemented with cytokines (IL-3, IL-6, SCF, and GM-CSF) and various concentrations of 4-OHT. After 5 days the cells were reisolated and then replated every 5 to 6 days under equal conditions in methocel without neomycin. In total five independent transduction series were performed. Figure 2C shows one representative example. As expected, neor-transduced cells did not form colonies in the third replating whereas MLL-ENL-expressing cells formed numerous colonies similar in appearance to those formed by MLL-ENL-ERtm-expressing cells in the presence of 100 nM 4-OHT. MLL-ENL-ERtm- transduced colonies formed only if the cells were kept in the continuous presence of 100 nM 4-OHT. Lower concentrations or the withdrawal of 4-OHT after two plating rounds precluded colony formation, indicating a loss of transformation potential.
Characterization of MLL-ENL-ERtm-transformed cells.
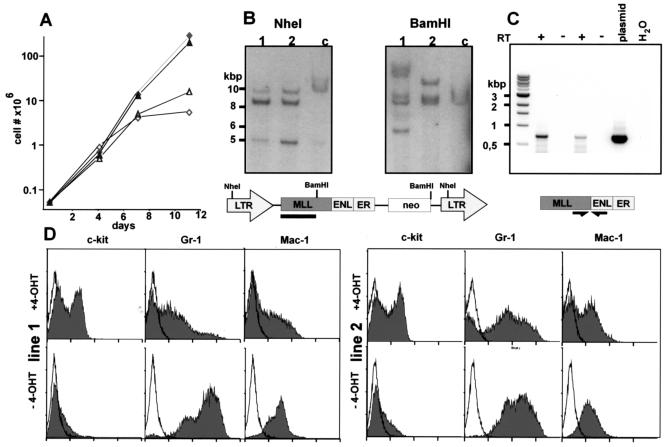
To gain material for more detailed investigations, third-round colonies of two independent transduction experiments were explanted from methylcellulose and cultivated in liquid medium either with or without 4-OHT. In contrast to cells transduced with native MLL-ENL (23), MLL-ENL-ERtm cells underwent rapid apoptosis when cultured in IL-3 alone (data not shown). Therefore for all further experiments the media for the MLL-ENL-ERtm cells were supplemented with the complete set of cytokines that was originally present in the methocel medium (IL-3, IL-6, SCF, and GM-CSF). Cells seeded without 4-OHT underwent complete growth arrest within approximately 2 weeks, whereas cells cultivated in 4-OHT-supplemented medium proliferated rapidly and established cell lines (Fig. 3A). The presence of intact provirus and the expression of MLL-ENL-ERtm RNA were confirmed in these lines by Southern blotting and RT-PCR, respectively (Fig. 3B and 3C). Additionally, the proviral integration pattern is different for line 1 and line 2 (Fig. 3B, right panel), thus confirming their independent origin.
FIG. 3.
Establishment of MLL-ENL-ERtm-transduced cell lines. (A) 4-OHT-dependent growth of MLL-ENL-ERtm-transduced cells. Cells from two independent transduction experiments were explanted from third-round methocel cultures and incubated in cytokine-supplemented media in the presence or absence of 100 nM 4-OHT. Cell numbers were determined by direct counting. Diamonds, experiment 1; triangles, experiment 2; filled symbols, 100 nM 4-OHT; open symbols, without inductor. (B) Southern blots for detection of integrated MLL-ENL-ERtm copies. Genomic DNA was isolated from the cell lines used for panel A, digested with either _Nhe_I or _Bam_HI, blotted, and hybridized against an MLL-specific probe. The localization of the restriction site and the hybridization probe (black bar) is indicated by the graphics. Normal mouse genomic DNA was included as a control (lane c). (C) RT-PCR analysis of MLL-ENL-ERtm expression. RNA was isolated from lines 1 and 2 as for panels A and B, reverse transcribed, and subjected to PCR amplification with primers spanning the MLL-ENL breakpoint. RT-minus controls are labeled −. (D) Immunophenotype of MLL-ENL-ERtm cells. After 12 days of cultivation either in the presence or the absence of 4-OHT (as indicated) the cells used for panel A were stained with fluorophore-coupled antibodies against the surface markers c-kit, Gr-1, and Mac-1 and analyzed by FACS. The open line corresponds to the background staining with an isotype-matched control antibody.
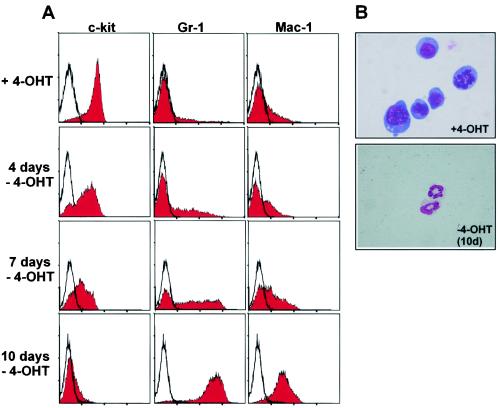
In contrast to the cells cultured without 4-OHT, which underwent myeloid differentiation, (c-kit− Gr-1+ Mac-1+), early-passage line 1 and line 2 cells consisted of a mixture of both early myeloid progenitors (c-kit+ Gr-1− Mac-1−) and more-differentiated myeloid cells (c-kit− Gr-1+ Mac-1+) (Fig. 3D). Upon further cultivation, eventually the c-kit+ precursor population became dominant. After withdrawal of the inducer, the cells ceased proliferation and underwent terminal differentiation to neutrophils characterized by downregulation of c-kit and upregulation of Gr-1 and Mac-1 (Fig. 4 and data not shown). In contrast, both cell lines proliferated indefinitely and retained myeloblast morphology in 4-OHT-supplemented medium.
FIG. 4.
Reversion of cellular immortalization after inactivation of MLL-ENL-ERtm. (A) Terminal differentiation of MLL-ENL-ERtm cells after inductor withdrawal. The cell lines generated from MLL-ENL-ERtm-transduced cells were cultivated in cytokine-supplemented media in the absence of 4-OHT. Surface marker composition was monitored by FACS staining for 10 days. Only the results for line 1 are shown, as line 2 gave virtually indistinguishable results. (B) Morphological analysis of MLL-ENL-ERtm cells grown in the presence of 4-OHT and after 10 days without inductor. May-Grünwald-Giemsa stain of cytospin slides is shown. Original magnification, ×63.
Genetic program governed by MLL-ENL-ERtm.
Microarray studies were carried out to identify genes that are regulated by MLL-ENL-ERtm. RNA isolated from line 1 at 0 and 72 h after 4-OHT withdrawal in triplicate experiments was labeled and applied to Affymetrix MG-U74Av2 microarrays, interrogating 12,000 genes of the mouse genome, including all sequences contained in the mouse UniGene cluster. Pairwise comparisons of the samples with and without 4-OHT were used to identify differentially expressed genes. Only genes that showed a concordant change in all three samples and for which the difference in numerical expression values within a pair exceeded the significance limit in at least two of three data sets were selected for further evaluation. Identified genes (excluding expressed sequence tags) that were downregulated after 4-OHT withdrawal, i.e., genes that are upregulated by MLL-ENL-ERtm, are listed in Table 1. Genes induced after inactivation of MLL-ENL-ERtm are shown in Table 2. For each gene the average fold change across the three sample sets together with the calculated standard deviation is given and the complete raw data are available as supplementary material upon request. In total, 27 genes met these criteria for targets activated by MLL-ENL-ERtm (i.e., downregulated after withdrawal of inductor).
TABLE 1.
Genes downregulated after 4-OHT withdrawala
| Accession no., gene descriptionb | Change | SD |
|---|---|---|
| Interferon-regulated genes | ||
| AF022371, Interferon-activated gene 203, leucine zipper | 3.42 | 0.85 |
| U19119, Lrg-47, G-protein like | 1.95 | 0.17 |
| M27134, H2k, histocompatibility 2, K region locus 2 | 1.86 | 0.42 |
| M31419, Interferon-activated gene 204 | 1.59 | 0.32 |
| Cytokine and chemokine signaling | ||
| X62940, _Tgf_-β-induced transcript 4, leucine zipper | 1.95 | 0.37 |
| X59398, Flt-3, receptor tyrosine kinase | 1.72 | 0.22 |
| U16985, _Tnf_-β, cytokine | 1.40 | 0.23 |
| U88327, Socs-2, inhibitor of IGF signaling | 1.37 | 0.09 |
| L40172, Jak-3, tyrosine kinase | 1.29 | 0.19 |
| Signaling, transcription regulation, and cell cycle | ||
| U33629, Meis1, transcription factor | 2.83 | 0.38 |
| L38444, Tgtp, T-cell-specific GTP-binding protein | 2.04 | 0.84 |
| AF020200, Pbx3b, transcription factor | 2.01 | 0.25 |
| J03776, _Rpt_-1, T-cell-specific suppressor of IL-2-activated transcription | 2.01 | 1.08 |
| M28845, Egr-1, transcription factor | 1.77 | 0.06 |
| X93328, F4/80, macrophage surface marker, hormone receptor-like | 1.63 | 0.12 |
| M12731, _N_-myc | 1.62 | 0.07 |
| AF080215, Par-4, megakaryocyte thrombin receptor | 1.54 | 0.16 |
| AB005458, Hoxa9, transcription factor | 1.47 | 0.21 |
| X97268, Lpap, Ptprc, B220 receptor tyrosine phosphatase associated | 1.38 | 0.02 |
| M64360, Lmo2, Rbtn2, Lim domain transcription factor | 1.32 | 0.01 |
| Enzymes and transporters | ||
| U96746, Aoe372, antioxidant enzyme | 1.73 | 0.35 |
| U75215, mAsct1, neutral amino acid transporter | 1.69 | 0.30 |
| AF011379, Adam 10, tumor necrosis factor alpha convertase | 1.40 | 0.14 |
| Others | ||
| Z11974, Mmr-1, macrophage mannose receptor | 1.88 | 0.40 |
| M36579, S100a4, calcium-binding protein | 1.87 | 0.28 |
| X98471, Emp-1, epithelial membrane protein 1 | 1.74 | 0.20 |
| AB015598, mTim1, murine timeless homolog | 1.48 | 0.52 |
TABLE 2.
Genes upregulated after 4-OHT withdrawala
| Accession no., gene description | Change | SD |
|---|---|---|
| Genes associated with myeloid immune function | ||
| X94353, Mclp, cathelin-like protein cysteine protease inhibitor | 7.61 | 5.53 |
| L37297, Clp, cathelin-like, myeloid secondary granule protein | 5.98 | 1.64 |
| AF076482, Pgrp, peptidoglycan recognition peptide | 1.86 | 1.06 |
| X51547, Lzp-s, lysozyme P | 1.45 | 0.27 |
| M21050, LysM, lysozyme M | 1.44 | 0.11 |
| U04962, Ela2, neutrophil elastase | 1.38 | 0.25 |
| Surface antigens | ||
| X70920, Ly-6G.1, Gr-1 surface marker | 3.77 | 1.70 |
| AF039663, Ac133, Prom, surface marker | 1.75 | 0.84 |
| M65027, Gp49a, glycoprotein 49 A, mast cell progenitor surface antigen | 1.74 | 0.64 |
| U05265, Gp49b, glycoprotein 49 B | 1.59 | 0.32 |
| Cytokine and chemokine signaling | ||
| X54542, Il-6, interleukin 6 | 1.81 | 0.24 |
| Z80112, Cxcr4, lcr-1, chemokine receptor | 1.55 | 0.27 |
| Signaling, transcription regulation, and cell cycle | ||
| Z11664, Sos2, son of sevenless homolog 2 | 1.91 | 0.23 |
| M26391, Rb, retinoblastoma 1 | 1.63 | 0.59 |
| M62362, Cebpa, cebp-alpha, myeloid transcription factor | 1.60 | 0.08 |
| Enzymes transporters | ||
| J03298, Ltf, lactotransferrin | 7.35 | 7.90 |
| J05118, Cpa3, mast cell carboxypeptidase | 2.80 | 0.69 |
| M32599, Gapdh, glyceraldehyde-3-phosphate dehydrogenase | 2.21 | 1.28 |
| AF052453, Sk2, ATP sulfurylase | 2.07 | 0.44 |
| M63245, _Alas-_h, amino levulinate synthase | 1.80 | 0.04 |
| M75135, Glut3, glucose transporter | 1.76 | 0.69 |
| X83202, Hsdrla, 11beta-hydroxysteroid dehydrogenase/carbonyl reductase | 1.62 | 0.13 |
| AF078752, Dgat, diacylglycerol acyltransferase | 1.60 | 0.17 |
| L10244, Ssat, spermidine/spermine N1-acetyl transferase | 1.51 | 0.31 |
| U51014, Pep4, peptidase 4 | 1.49 | 0.20 |
| M63848, Lta4, leukotriene A-4 hydrolase | 1.36 | 0.06 |
| Others | ||
| M94584, Ym-1, macrophage lectin | 4.11 | 3.67 |
| M83218, Mrp8, myeloid-associated calcium-binding protein | 2.09 | 0.31 |
| M83219, Mrp14, myeloid-associated calcium-binding protein | 2.07 | 0.26 |
| AJ001633, Anx3, annexin 3, lipocortin 3 | 1.69 | 0.14 |
| AB025406, Sid23, actin-depolymerizing factor | 1.59 | 0.11 |
| M69260, Lpc1, lipocortin 1, annexin 1, phospholipase 2 inhibitor | 1.57 | 0.16 |
| U59807, CstB, cystatin B, protease inhibitor | 1.55 | 0.40 |
| M12481, Actb, cytoplasmic β-actin | 1.52 | 0.23 |
| Z19543, Cnn2, calponin 2, cell adhesion molecule | 1.51 | 0.22 |
| Z16410, Btg1, B-cell translocated gene 1, antiproliferative factor | 1.44 | 0.21 |
Seven of the putative target genes upregulated by MLL-ENL-ERtm (Flt-3, Meis-1, Hoxa9, Ptprc, Lmo2, Adam10, and S100a4) have been previously found to be specifically overexpressed in human leukemias with MLL rearrangements compared to those lacking MLL rearrangements (2, 45). Of these, Hoxa9 and Meis1 were of particular interest because the respective proteins form trimeric complexes with Pbx proteins in myeloid cells and most importantly Hoxa9 and Meis1 strongly cooperate to induce myeloid leukemias in mice (6). Two other genes implicated in malignant transformation, Lmo2 and N-myc, were also upregulated by MLL-ENL-ERtm and are likely to contribute to transformation.
Thirty-six genes were upregulated after MLL-ENL-ERtm inactivation. Many of these were proteins involved in myeloid differentiation and/or myeloid cell function. For example, the gene encoding the mature myeloid marker Gr-1 was strongly upregulated on the arrays after 4-OHT withdrawal, closely correlating with the onset of Gr-1 protein expression as detected by FACS analysis (Fig. 4). In addition, secondary granule proteins and enzymes involved in immune defense (e.g., the lysozyme family) were also upregulated. The Glut3 glucose transporter and glyceraldehyde-3-phosphate dehydrogenase genes were upregulated, consistent with a shift toward anaerobic metabolism. Interestingly, the RNA of the retinoblastoma tumor suppressor gene also increased after inductor withdrawal, preceding the proliferation arrest occurring during more advanced differentiation. The changes in gene expression levels for targets that were activated by MLL-ENL-ERtm appeared to be closely coordinated across the individual cell populations, as indicated by small standard deviations. In contrast, the genetic program accompanying differentiation that commenced after MLL-ENL-ERtm inactivation was less synchronized between cultures, in particular for genes with the highest changes in expression levels, resulting in large standard deviations.
Hox and TALE genes are the crucial downstream targets of MLL-ENL for immortalization.
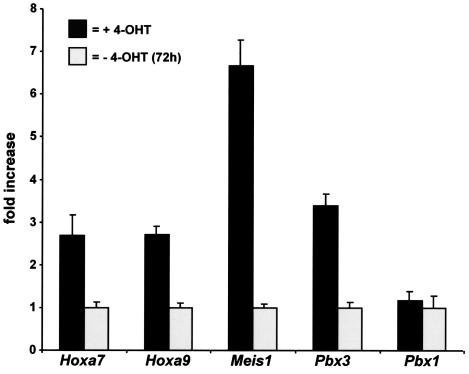
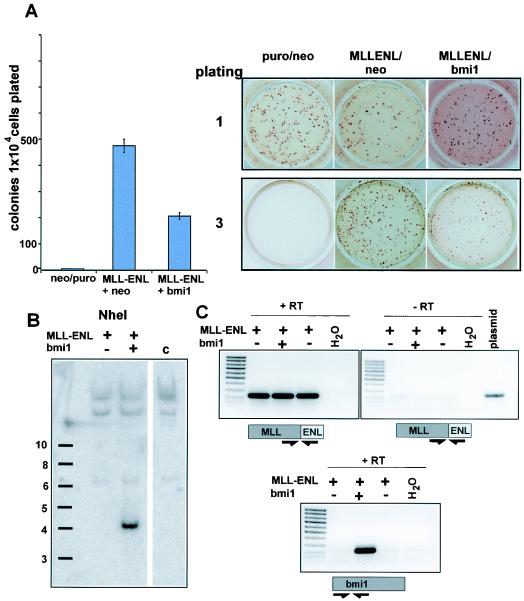
One of the most striking results of the microarray analysis was that Hox genes, including Hoxa9, as well as the Hox cofactors Meis1 and Pbx3, are coordinately upregulated. These results were confirmed by quantitative RT-PCR, which yielded results in close agreement with the microarray data. In addition we determined that Hoxa7 was also upregulated by the MLL fusion protein (Fig. 5). This is potentially a key mechanism of transformation by MLL fusion proteins because both Hoxa7 and Hoxa9 in combination with Meis1 have been shown to cooperatively transform myeloid progenitors to produce aggressive leukemias in experimental models (6, 21). Importantly, these genes are also consistently upregulated in human leukemias (20). Hoxa7 and Hoxa9 have been recognized as target genes of unaltered MLL (48). Therefore, we tested whether MLL and the MLL fusion proteins regulate a largely overlapping set of genes and whether overexpression of these originally MLL-regulated genes is at the basis of the cellular immortalization process. If this hypothesis holds true it should be possible to abrogate MLL-ENL-mediated transformation by repression of MLL-regulated genes. In mice the direct biological counterpart of Mll is the Bmi-1 repressor oncoprotein. The haploinsufficieny phenotype of an Mll heterozygote animal is completely reverted if crossed into a Bmi−/− background, demonstrating that both proteins have opposite effects on a mostly identical set of targets (17). If MLL-activated genes play a decisive role in MLL fusion protein-mediated transformation, the overexpression of Bmi-1 should counteract the oncogenic transformation process. To test this supposition, the MLL-ENL cDNA was subcloned into a pMSCV vector, conferring puromycin resistance, and cotransduced with either a Neor Bmi-1 retrovirus (pMSCVneo-Bmi1) or a neomycin control virus into hematopoietic precursor cells. Transduction with two empty viruses served as an additional control. The cells were plated under double puromycin-neomycin selection in the standard methocel assay. As the results of triplicate experiments show in Fig. 6A, Bmi-1 was able to significantly reduce the formation of third-round colonies, indicating a weakened transformation potency of MLL-ENL in the presence this protein. This was not due to a simple toxic effect, as the growth properties and the number of colonies obtained in the first round of plating were comparable between different transductions. Figure 6B shows results of a representative example out of three experiments. The presence of the Bmi-1 virus was verified in the cell populations by southern blotting (Fig. 6B), and the coexpression of MLL-ENL and Bmi-1 RNA could be demonstrated by RT-PCR (Fig. 6C).
FIG. 5.
Quantitative real-time PCR analysis. Expression of Hoxa7, Hoxa9, Meis1, Pbx3, and Pbx1 as indicated was quantitated by real-time PCR in RNA samples isolated from cells grown in the presence of 4-OHT (dark columns) or 72 h after 4-OHT withdrawal (light columns). Means and standard deviations of triplicate experiments are shown.
FIG. 6.
Coexpression of the Bmi-1 repressor gene inhibits MLL-ENL. (A, left) Colony count in the third round of a methocel assay with cotransduction of MLL-ENL and Bmi-1. Hematopoietic progenitor cells were coinfected with retroviruses encoding MLL-ENL and Bmi-1 and subjected to a methocel replating assay. Third-round colonies obtained from 10,000 cells were counted. Shown are the means and standard deviations of three cotransduction experiments. (A, right) Colonies obtained in the first and the third round of plating during the cotransduction test. Results of one of three conducted experiments are shown. (B) Southern blot performed with DNA isolated from cells either transduced with the indicated constructs or control cells (lane c) and hybridized with a Bmi-1 specific probe. (C) Expression of MLL-ENL and Bmi-1 specific RNA in cotransduced cells as detected by RT-PCR.
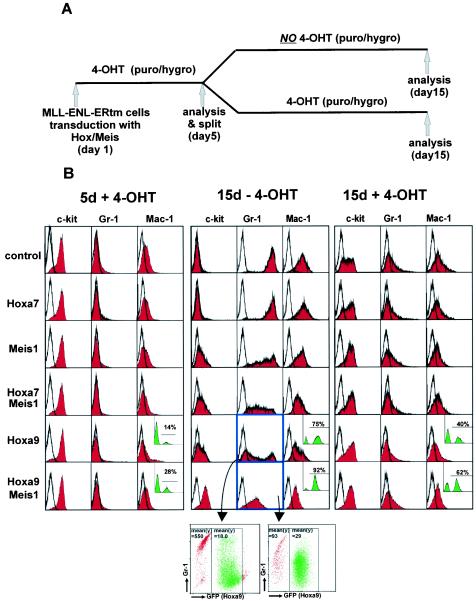
Our data show that MLL fusion proteins impose a reversible block on myeloid differentiation and that this involves changes in a limited set of target genes. One powerful feature of this model is that it allows for testing the role of individual target genes in the immortalization process. Given their strong association with leukemias with MLL rearrangements we focused our initial studies on the role of Hox and TALE domain cofactors. To explore the role of these cells transformed by MLL-ENL-ERtm, they were further transduced with retroviruses containing the cDNAs for Hoxa7, Hoxa9, and Meis1 either singly or in combination. Homogeneous populations were obtained by employing MSCV vectors conferring hygromycin or puromycin resistance (used for Hoxa7 and Meis1) followed by antibiotic selection or an MSCV plasmid that coexpressed GFP together with Hoxa9. Empty viruses served as a control. The rationale behind this strategy was that the enforced expression of crucial MLL-ENL-ERtm targets should substitute for MLL-ENL-ERtm and maintain the immortalized phenotype and the differentiation arrest in the absence of 4-OHT. MLL-ENL-ERtm cells transduced with the various Hox and Meis expression vectors were kept for 5 days under selection (where appropriate) in the presence of 4-OHT (for an outline of the experimental strategy, see Fig. 7A). The surface marker profile (c-kit, Gr-1, and Mac-1) at this time point was identical for all transduced cell populations (Fig. 7B, left panel). Subsequently, the cultures were split into two subpopulations that were grown for a further 10 days either with or without 4-OHT (Fig. 7B, middle and right panels, respectively). When analyzed 10 days after 4-OHT depletion puromycin control cells had ceased proliferation completely. As expected, c-kit expression was lost and Gr-1 and Mac-1 levels were greatly elevated, indicating terminal differentiation (Fig. 7B, middle panel, upper row). In the presence of 4-OHT the same control cell population grew vigorously and maintained the early myeloid surface marker pattern (Fig. 7B, right panel, upper row). Transduction of single Hox genes or Meis1 alone, or a combination of Hoxa7 and Meis1, partially blocked differentiation of the MLL-ENL-ERtm cells after 4-OHT withdrawal but ultimately did not inhibit terminal differentiation of the cell populations (Fig. 7, middle panel, and data not shown).
FIG. 7.
Hoxa9 and Meis1 are crucial downstream targets of MLL-ENL-ERtm. (A) Description of the experimental outline. puro, puromycin; hygro, hygromycin. (B) FACS analysis. MLL-ENL-ERtm cells were overtransduced as indicated with retroviral constructs coding for Hoxa7, Hoxa9, Meis1,or a combination thereof. After 5 days in selective medium supplemented with 4-OHT the surface marker distribution and the percentage of GFP-positive cells were determined by FACS (left panel). The black outline in all graphs represents a control staining with an isotype-specific antibody. After a further 10 days in selective medium but now without 4-OHT the differentiation status of the respective cells was assessed again by FACS (middle panel) and compared to a subpopulation of the same cells treated identically except that 4-OHT was present during culture (right panel). The insets in the graphs represent the distribution of GFP-positive _Hoxa9_-positive and -negative cells where appropriate. A two-dimensional analysis of GFP/Hoxa9 expression versus Gr-1 staining is depicted for two samples (highlighted in blue).
Notably, however, MLL-ENL-ERtm cells that had been transduced with a combination of Meis1 and Hoxa9 remained proliferative (>3 months in continuous culture), retained c-kit expression, and did not upregulate Mac-1 regardless of the presence of 4-OHT. The only suggestion of maturation in these cells was a slightly elevated level of Gr-1 in the absence of 4-OHT (Fig. 7B, middle panel, lower row) compared to cultivation with 4-OHT (Fig. 7B, right panel, lower row), strongly suggesting that Hoxa9 and Meis1 coexpression essentially replaces the activity of the MLL fusion protein and that these are the critical downstream targets. In keeping with this the percentage of GFP-positive (_Hoxa9_-bearing) cells increased from 28 to 92% in the culture kept without 4-OHT, demonstrating a strict selection for Hoxa9 under these conditions (Fig. 7B, left and middle panels, lower row, inset). In addition, a high GFP fluorescence of _Hox9_-transduced MLL-ENL-ERtm cells was generally correlated with a low intensity of lineage marker staining and vice versa (Fig. 7B, middle panel, two-dimensional FACS plots, and data not shown). In the presence of 4-OHT a significantly lower number of Hoxa9/GFP-positive cells were detectable (62%) at the same time point, suggesting less selection for Hoxa9 expression in cells that also contain active MLL-ENL-ERtm (Fig. 7B, right panel, lower row, inset).
DISCUSSION
The results presented here indicate that MLL fusion proteins transform by imposing a reversible block on myeloid differentiation. In this way MLL fusion proteins resemble the core binding factor fusions (AML1 fusions) (40) and the retinoic acid alpha fusion proteins (50), which appear to act primarily by establishing a block in blood cell maturation. In the case of MLL fusion proteins this transformation block involves cooperative upregulation of Hox, Meis, and Pbx genes, which have been shown to associate as a trimeric complex in myeloid cells (36). This coordinate upregulation may be a common theme in leukemogenesis because it is strikingly similar to that seen in Nup98-Hox fusion protein-transformed cells, which also show persistent high-level expression of Hoxa7, Hoxa9, and Meis1 (7).
One of the advantages of the model we have developed is that it allows for testing of the role of individual targets of the fusion protein in the transformation process. Our experiments suggest that in isolation Hoxa7, Hoxa9, and Meis1 have similar partial effects on myeloid differentiation but that the combination of Hoxa9 and Meis1 in particular results in a complete block in differentiation that results in immortalization. Expression of these synergistic genes appears to be coordinately regulated, raising the question of whether all are direct targets of MLL. Considerable evidence suggests that Hox genes are directly regulated by MLL fusions. Expression of both Hoxa7 and Hoxa9 is dependent on wild-type MLL (17). Recently it has been shown that wild-type MLL binds to the Hoxa9 promoter and that changes in histone modifications at the promoter are MLL dependent (27, 30). In addition, using chromatin immunoprecipitation we have found that MLL fusion proteins also bind directly to a CpG-rich region in the first exon of the Hoxa9 gene in hematopoietic cells (26). These results suggest that Hox deregulation is mediated by direct fusion protein binding.
Increasing evidence supports a model for leukemogenesis in which disruption of transcription factors results in chimeric proteins that block differentiation and these act in concert with receptor tyrosine kinases that drive proliferation (15). It is noteworthy in this regard that the receptor tyrosine kinase Flt3, which is consistently expressed in MLL-associated leukemias, was also upregulated by the inducible MLL fusion protein. Thus, the MLL fusion protein might generate both the block in differentiation as a result of Hoxa7, Hoxa9, and Meis1 overexpression as well as overexpression of Flt3 to provide the “second hit” required for full transformation. Flt3 is likely to play a contributory role in MLL-induced leukemia because drugs that target the kinase activity effectively kill cells harboring MLL rearrangements (1).
Although we have shown that upregulation of Hox genes and Meis1 by MLL fusion proteins plays a pivotal role in blocking myeloid differentiation, other oncogenes upregulated by MLL-ENL-ERtm may contribute to transformation. For example N-myc expression was activated by MLL-ENL-ERtm. Given our previous results that MLL-ENL can cooperate with c-myc to establish an irreversible differentiation arrest (34), this suggests that N-myc may also contribute to blocking differentiation. Furthermore, Lmo2, which is overexpressed in some T-cell leukemias (13), also was upregulated by MLL-ENL-ERtm, suggesting that cross talk between several leukemogenic pathways may be operative in MLL-induced leukemia.
Despite the statistical significance the amplitude of the expression changes appears to be small for some of the genes listed. This is most likely because the study design allows only the sampling of one particular time point during a continuous process within a nonsynchronous cell population. Changes reflect only an average value and might be higher at other time points after 4-OHT withdrawal. In addition a small change in the expression of a master transcription factor or an initiating member of a signal transduction cascade will translate in a much larger and amplified signal output of the respective pathway. Therefore the numerical value alone is not necessarily proportional to the importance of the corresponding gene in the regulation of the differentiation process.
Finally, our results indicate that at least initially the transformed state established by MLL fusion proteins is reversible. This suggests that drugs that target the gain-of-function activity of the fusion proteins or perhaps the activity of downstream Hox genes may effectively reverse the differentiation block imposed by these oncogenes and ultimately be used as effective therapy for MLL-associated leukemias.
Acknowledgments
We are grateful to Trevor Littlewood, Keith Humphries, and Gary Nolan for the gift of reagents. Additionally, we wish to thank Renate Zimmermann for technical assistance and Georg Fey for continuous support.
This work was supported by DFG grants SL27/6-1 and SFB473/B10 to R.K.S. and by a Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society and National Institutes of Health grant CA-92251 to J.L.H. R.KS. is a recipient of a Ria Freifrau-von-Fritsch Stiftung career development award.
REFERENCES
- 1.Armstrong, S. A., A. L. Kung, M. E. Mabon, L. B. Silverman, R. W. Stam, M. L. Den Boer, R. Pieters, J. H. Kersey, S. E. Sallan, J. A. Fletcher, T. R. Golub, J. D. Griffin, and S. J. Korsmeyer. 2003. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell 3**:**173-183. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. A., J. E. Staunton, L. B. Silverman, R. Pieters, M. L. den Boer, M. D. Minden, S. E. Sallan, E. S. Lander, T. R. Golub, and S. J. Korsmeyer. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30**:**41-47. [DOI] [PubMed] [Google Scholar]
- 3.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20**:**5695-5707. [DOI] [PubMed] [Google Scholar]
- 4.Birke, M., S. Schreiner, M. P. Garcia-Cuellar, K. Mahr, F. Titgemeyer, and R. K. Slany. 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 30**:**958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, L. H., R. Slany, X. Cui, M. L. Cleary, and D. Y. Mason. 1997. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 89**:**3361-3370. [PubMed] [Google Scholar]
- 6.Calvo, K. R., P. S. Knoepfler, D. B. Sykes, M. P. Pasillas, and M. P. Kamps. 2001. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc. Natl. Acad. Sci. USA 98**:**13120-13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo, K. R., D. B. Sykes, M. P. Pasillas, and M. P. Kamps. 2002. Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. Oncogene 21**:**4247-4256. [DOI] [PubMed] [Google Scholar]
- 8.DiMartino, J. F., P. M. Ayton, E. H. Chen, C. C. Naftzger, B. D. Young, and M. L. Cleary. 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99**:**3780-3785. [DOI] [PubMed] [Google Scholar]
- 9.Dimartino, J. F., and M. L. Cleary. 1999. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol. 106**:**614-626. [DOI] [PubMed] [Google Scholar]
- 10.Djabali, M., L. Selleri, P. Parry, M. Bower, B. D. Young, and G. A. Evans. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2**:**113-118. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, P., J. Wang, M. Huang, R. H. Goodman, and S. J. Korsmeyer. 2001. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell. Biol. 21**:**2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, P., J. Wang, and S. J. Korsmeyer. 2002. The role of MLL in hematopoiesis and leukemia. Curr. Opin. Hematol. 9**:**282-287. [DOI] [PubMed] [Google Scholar]
- 13.Ferrando, A. A., D. S. Neuberg, J. Staunton, M. L. Loh, C. Huard, S. C. Raimondi, F. G. Behm, C. H. Pui, J. R. Downing, D. G. Gilliland, E. S. Lander, T. R. Golub, and A. T. Look. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1**:**75-87. [DOI] [PubMed] [Google Scholar]
- 14.Francis, M. K., D. G. Phinney, and K. Ryder. 1995. Analysis of the hormone-dependent regulation of a JunD-estrogen receptor chimera. J. Biol. Chem. 270**:**11502-11513. [DOI] [PubMed] [Google Scholar]
- 15.Gilliland, D. G., and J. D. Griffin. 2002. The roles of FLT3 in hematopoiesis and leukemia. Blood 100**:**1532-1542. [DOI] [PubMed] [Google Scholar]
- 16.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71**:**701-708. [DOI] [PubMed] [Google Scholar]
- 17.Hanson, R. D., J. L. Hess, B. D. Yu, P. Ernst, M. van Lohuizen, A. Berns, N. M. van der Lugt, C. S. Shashikant, F. H. Ruddle, M. Seto, and S. J. Korsmeyer. 1999. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 96**:**14372-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess, J. L., B. D. Yu, B. Li, R. Hanson, and S. J. Korsmeyer. 1997. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90**:**1799-1806. [PubMed] [Google Scholar]
- 19.Hsieh, J. J., P. Ernst, H. Erdjument-Bromage, P. Tempst, and S. J. Korsmeyer. 2003. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 23**:**186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura, T., A. Morimoto, M. Takanashi, S. Hibi, T. Sugimoto, E. Ishii, and S. Imashuku. 2002. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br. J. Haematol. 119**:**119-121. [DOI] [PubMed] [Google Scholar]
- 21.Kroon, E., J. Krosl, U. Thorsteinsdottir, S. Baban, A. M. Buchberg, and G. Sauvageau. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17**:**3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavau, C., C. Du, M. Thirman, and N. Zeleznik-Le. 2000. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 19**:**4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavau, C., S. J. Szilvassy, R. Slany, and M. L. Cleary. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16**:**4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23**:**1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, R. T., C. Lavau, C. Du, F. Simone, P. E. Polak, S. Kawamata, and M. J. Thirman. 2001. The elongation domain of ELL is dispensable, but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol. Cell. Biol. 21**:**5678-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, M. E., T. A. Milne, S. Bloyer, K. Galoian, W. Shen, D. Gibbs, H. W. Brock, R. Slany, and J. L. Hess. 2003. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell 4**:**197-207. [DOI] [PubMed] [Google Scholar]
- 27.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10**:**1107-1117. [DOI] [PubMed] [Google Scholar]
- 28.Moskow, J. J., F. Bullrich, K. Huebner, I. O. Daar, and A. M. Buchberg. 1995. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell. Biol. 15**:**5434-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura, T., D. A. Largaespada, M. P. Lee, L. A. Johnson, K. Ohyashiki, K. Toyama, S. J. Chen, C. L. Willman, I. M. Chen, A. P. Feinberg, N. A. Jenkins, N. G. Copeland, and J. D. Shaughnessy, Jr. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12**:**154-158. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10**:**1119-1128. [DOI] [PubMed] [Google Scholar]
- 31.Owens, B. M., and R. G. Hawley. 2002. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells 20**:**364-379. [DOI] [PubMed] [Google Scholar]
- 32.Picard, D., S. J. Salser, and K. R. Yamamoto. 1988. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell 54**:**1073-1080. [DOI] [PubMed] [Google Scholar]
- 33.Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib, J. Blechman, T. Nakamura, C. M. Croce, A. Mazo, and E. Canaani. 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95**:**4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiner, S., M. Birke, M. P. Garcia-Cuellar, O. Zilles, J. Greil, and R. K. Slany. 2001. MLL-ENL Causes a Reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res. 61**:**6480-6486. [PubMed] [Google Scholar]
- 35.Schreiner, S. A., M. P. Garcia-Cuellar, G. H. Fey, and R. K. Slany. 1999. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia 13**:**1525-1533. [DOI] [PubMed] [Google Scholar]
- 36.Shen, W. F., S. Rozenfeld, A. Kwong, L. G. Kom ves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19**:**3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slany, R. K., C. Lavau, and M. L. Cleary. 1998. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol. 18**:**122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.So, C. W., and M. L. Cleary. 2003. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood 101**:**633-639. [DOI] [PubMed] [Google Scholar]
- 39.So, C. W., and M. L. Cleary. 2002. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell. Biol. 22**:**6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speck, N. A., and D. G. Gilliland. 2002. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2**:**502-513. [DOI] [PubMed] [Google Scholar]
- 41.Thorsteinsdottir, U., E. Kroon, L. Jerome, F. Blasi, and G. Sauvageau. 2001. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21**:**224-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorsteinsdottir, U., A. Mamo, E. Kroon, L. Jerome, J. Bijl, H. J. Lawrence, K. Humphries, and G. Sauvageau. 2002. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99**:**121-129. [DOI] [PubMed] [Google Scholar]
- 43.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71**:**691-700. [DOI] [PubMed] [Google Scholar]
- 44.Yagi, H., K. Deguchi, A. Aono, Y. Tani, T. Kishimoto, and T. Komori. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92**:**108-117. [PubMed] [Google Scholar]
- 45.Yeoh, E. J., M. E. Ross, S. A. Shurtleff, W. K. Williams, D. Patel, R. Mahfouz, F. G. Behm, S. C. Raimondi, M. V. Relling, A. Patel, C. Cheng, D. Campana, D. Wilkins, X. Zhou, J. Li, H. Liu, C. H. Pui, W. E. Evans, C. Naeve, L. Wong, and J. R. Downing. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1**:**133-143. [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama, A., I. Kitabayashi, P. M. Ayton, M. L. Cleary, and M. Ohki. 2002. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood 100**:**3710-3718. [DOI] [PubMed] [Google Scholar]
- 47.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95**:**10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378**:**505-508. [DOI] [PubMed] [Google Scholar]
- 49.Zeisig, B. B., S. Schreiner, M. P. Garcia-Cuellar, and R. K. Slany. 2003. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia 17**:**359-365. [DOI] [PubMed] [Google Scholar]
- 50.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J. D. Licht. 2001. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene 20**:**7186-7203. [DOI] [PubMed] [Google Scholar]
- 51.Zeleznik-Le, N. J., A. M. Harden, and J. D. Rowley. 1994. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl. Acad. Sci. USA 91**:**10610-10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziemin-van der Poel, S., N. R. McCabe, H. J. Gill, R. Espinosa III, Y. Patel, A. Harden, P. Rubinelli, S. D. Smith, M. M. LeBeau, J. D. Rowley, et al. 1991. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. USA 88**:**10735-10739. [DOI] [PMC free article] [PubMed] [Google Scholar]