Cryo-EM structure of a 3D DNA-origami object (original) (raw)
Abstract
A key goal for nanotechnology is to design synthetic objects that may ultimately achieve functionalities known today only from natural macromolecular complexes. Molecular self-assembly with DNA has shown potential for creating user-defined 3D scaffolds, but the level of attainable positional accuracy has been unclear. Here we report the cryo-EM structure and a full pseudoatomic model of a discrete DNA object that is almost twice the size of a prokaryotic ribosome. The structure provides a variety of stable, previously undescribed DNA topologies for future use in nanotechnology and experimental evidence that discrete 3D DNA scaffolds allow the positioning of user-defined structural motifs with an accuracy that is similar to that observed in natural macromolecules. Thereby, our results indicate an attractive route to fabricate nanoscale devices that achieve complex functionalities by DNA-templated design steered by structural feedback.
Natural macromolecular machines have complex 3D shapes with subnanometer-precise structural features that enable executing tasks such as signal transduction, molecular transport, and enzymatic catalysis (1). A key goal for nanotechnology is to fabricate synthetic objects with similarly precise features to ultimately control tasks known today only from natural “nano-machines.” Molecular self-assembly with DNA is considered a candidate route to achieve this goal (2–13). Densely packed 3D DNA-origami objects (14–18) seem particularly suited for use as rigid scaffolds to position reactive groups at target locations in space, and to implement features known from natural macromolecular complexes, such as shape complementarity and controlled domain movement. Designing such objects to meet precise structural specifications will benefit strongly from detailed 3D structural feedback. Here we present a cryo-EM map of an asymmetric, densely packed DNA object that comprises 15,238 nt in 164 chains. Combined with prior knowledge about the topology of chain connectivity, this map provided sufficient detail to construct a full pseudoatomic model for this particle. Thereby, our results provide structural feedback on DNA-templated design that will prove valuable for the design of complex functionalities.
Results and Discussion
DNA-Templated Design and Synthesis.
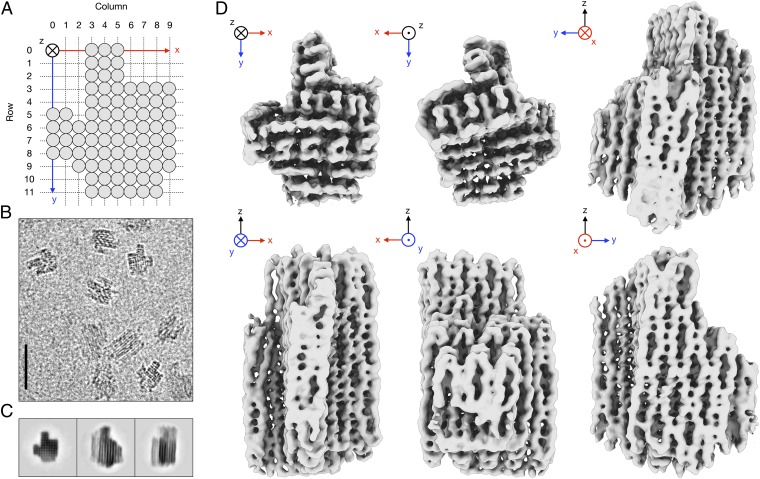
Our DNA object was designed to be suitable for structure determination by cryo-EM single-particle analysis, because the choice of a distinctly asymmetric shape facilitated the recognition of different particle orientations in electron micrographs (Fig. 1_A_). The object was designed to assemble as 82 parallel dsDNA helices of varying length in a square lattice (17) of 10 columns and 12 rows and comprising a total of 15,238 nt with a molecular mass of 4.8 MDa (Fig. S1). A total of 740 nt were distributed among flexible loops at the ends of the helices to prevent aggregation by blunt-end stacking (19). Synthesis of the object was based on templated molecular self-assembly (3) with a 7,249-nt-long “scaffold” DNA strand derived from M13 bacteriophage and 163 shorter “staple” strands (Table S1). The object formed overnight in a one-pot reaction with high yield and few byproducts, such as higher-molecular-weight aggregates. It was purified from excess staple strands by molecular-weight cutoff filtration, which yielded particle solutions free from nucleic acid stains and gel matrix residues (Fig. S2).
Fig. 1.
Cryo-EM reconstruction of a designed, densely packed DNA object. (A) Schematic representation of the designed rectangular lattice comprising 82 parallel dsDNA helices (gray circles). (B) Representative part of an electron micrograph. (Scale bar, 50 nm.) (C) Reference-free 2D class averages. (D) Six orthogonal views of the 3D reconstruction, shown as iso-density surface at density level 0.1.
Cryo-EM Single-Particle Analysis and Model Generation.
Imaging of the sample by cryo-EM revealed monodisperse particles with the expected dimensions and shape (Fig. S3). The particles showed suitable contrast in ice and adopted multiple orientations on the supporting continuous carbon film (Figs. 1 B and C). This allowed us to calculate a 3D reconstruction (20) from 28,502 individual particles with an overall resolution of 11.5 Å (Fig. S4). The resolution of the map varies with position, with an estimated range of 9.7 Å at the core to 14 Å at the periphery (Fig. 1_D_; Materials and Methods). With the exception of a region comprising helices in row 11 at the periphery of the object and a single cross-over in row 10, all helices and connecting cross-overs are well resolved in the map. The global shape of the reconstructed density is in agreement with the designed topology of the object. The overall dimensions of the object and the extent of global twist that results from a mismatch of B-form DNA helicity and the imposed square-lattice packing also agree with a prediction (21, 22) when using 2.6-nm effective helix diameter and 10.44 bp/turn reciprocal twist density (17) (Fig. S5).
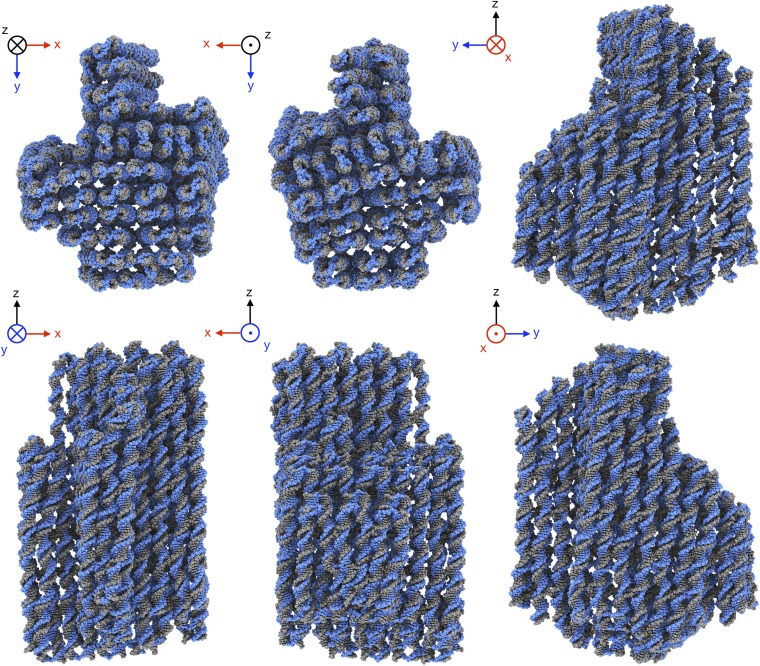
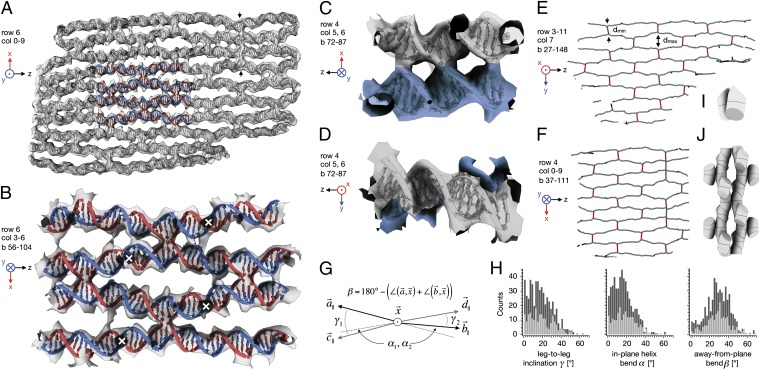
Aided by knowledge of the designed topology of the 164 strands in the object, we constructed an initial atomic model that consisted of canonical B-form DNA segments (23) for all expected dsDNA helices and cross-overs. Flexible fitting of this model inside the cryo-EM density (24) yielded a pseudoatomic model for the entire object (Fig. 2 and Movie S1), except for the single-stranded loops at the boundaries, for which no density was observed. Throughout the object, the fitted model is in good agreement with the cryo-EM density, and nearly all residues form expected base pairs. Note that in the EM density, connections between neighboring helices are often visible at positions that do not coincide with covalent strand cross-over (Fig. 3 A–C), suggesting additional contacts beyond the two covalent strand cross-overs. However, low-pass filtering of our atomic model to the same resolution as the EM map shows similar contacts (Fig. S6), indicating that the additional connections are due to the limited resolution of our map. The quality of the pseudoatomic model is highest at the core of the object, where it reproduces the observed major and minor grooves resolved in the map (Fig. 3_B_). According to the frequency-dependent decay of the power in our EM reconstruction (25), the rmsd of the atoms at the core of the object was estimated to be in the range of 2–3 Å (SI Text), thus providing experimental evidence that structural order that approaches that of natural nanomachines may be attained in discrete DNA objects.
Fig. 2.
Pseudoatomic model. Six orthogonal views of the pseudoatomic model that was fitted into the EM density map.
Fig. 3.
Analysis of internal geometry. (A) Central slice through the object at row 6, showing the cryo-EM density map (transparent gray) and the fitted pseudoatomic model as ribbon/slab model. For part of the model, the scaffold is shown in blue and the staples in red. Vertical arrows indicate a vertical stack of three cross-overs. (B) Close-up of colored area in A, but rotated around the z axis by 180°. White crosses indicate out-of-plane cross-over positions. (C) Side view of a single cross-over. (D) Top view of the same cross-over as in C. (E and F) Schematic representation of the 3D chickenwire-like pattern found in the structure, depicting dsDNA helical stretches in gray and cross-overs in red. The pattern was computed using the coordinates of base pair midpoints in the pseudoatomic model. The midpoints of neighboring dsDNA helices move on average from a minimum distance <dmin> = 18.5 Å at the cross-over to a maximum distance of <dmax> = 36 Å away from each other. Cross-overs marked in blue indicate scaffold-based strand cross-overs. (G) Definition of the angles enclosed by the four helical legs of a cross-over. Vectors are computed using the coordinates of base pair midpoints at the cross-over position and 2 bp away from the cross-over in each leg. The cross-over vector x is computed from the coordinates of the midpoints between the two base pairs in each of the two helices at the cross-over position and is normal to what we call the cross-over plane. The subscript “||” indicates vectorial projections into the cross-over plane. The angle β is also computed as indicated for vectors C and D. (H) Observed distribution of angles between the legs for cross-overs with (light gray) and without (dark gray) nearby nicks for 377 cross-overs in the structure, where each cross-over contributed two γ, α, and β angle values each. (I) Schematic representation of a revised cross-over model for DNA nanostructure design. (J) 3D chickenwire-like pattern formed by multiple instances of the revised cross-over model when rotated and translated according to the square-lattice packing connectivity scheme.
Analysis of Structural Features.
The pseudoatomic model enables the type of geometrical analysis required for designing objects with precise structural specifications. For example, in agreement with default B-form DNA geometry, we find that the average distance from base pair to base pair midpoints is 3.35 Å as derived from the helical distance between consecutive cross-overs that are resolved in the cryo-EM density. The design of the object assumes parallel double helices packed on a square lattice. Although the cross-sectional square lattice is indeed realized, we find that the helices are not parallel: on either side of the majority of cross-overs they enclose a nonzero angle within, but also out of the plane normal to the cross-over direction (Figs. 1_D_ and 3 A–D). Globally, this results in a 3D chickenwire-like pattern whereby individual dsDNA helices form diamond-shaped cavities in between cross-overs (Fig. 3 E and F). The 3D chickenwire pattern is also correlated with an unusual geometry of the 377 Holliday junctions that connect the helices in the object (Fig. 3 G and H). As established by FRET (26, 27) and atomic force microscopy (AFM) measurements (28), Holliday junctions adopt stacked conformations in the presence of cations where the two opposite helices are at an angle γ of ∼60°. In crystal structures, Holliday junctions show similar features (29–31). However, both the in-plane and the out-of-plane bending that we find in our structure is absent in the crystal structures and was also not resolved in the FRET and AFM measurements. Moreover, the junctions in our object exhibit a left-handed, rather than a right-handed, interhelical conformation (Fig. S7). These large conformational differences suggest that Holliday junctions are easily deformed by any ordering scheme, be it through crystal contacts or scaffolding interactions inside a densely packed DNA object. In the light of these results, future design of DNA objects to more precise structural specifications will benefit from the consideration of more appropriate cross-over models (Fig. 3 I and J).
We designed the termini of the 163 linear strands in the object to be mainly located 2 or 3 base pairs away from cross-overs. The termini result in covalent phosphodiester backbone nicks in dsDNA helices. We hypothesized that Holliday junction cross-overs with such nearby nicks might not fully form or that nicked helices might differ in shape from nonnicked helices. Our map reveals that this is not the case. Cross-overs with nearby nicks do not differ in geometry from nonnicked cross-overs (Fig. 3_H_). We also did not observe systematic differences when comparing nicked and nonnicked dsDNA helical stretches in the map. In addition, we investigated whether the nature of the local sequences at DNA cross-over positions leads to discernible differences in cross-over shape but could find no evidence for this.
Previously Unobserved DNA Topologies.
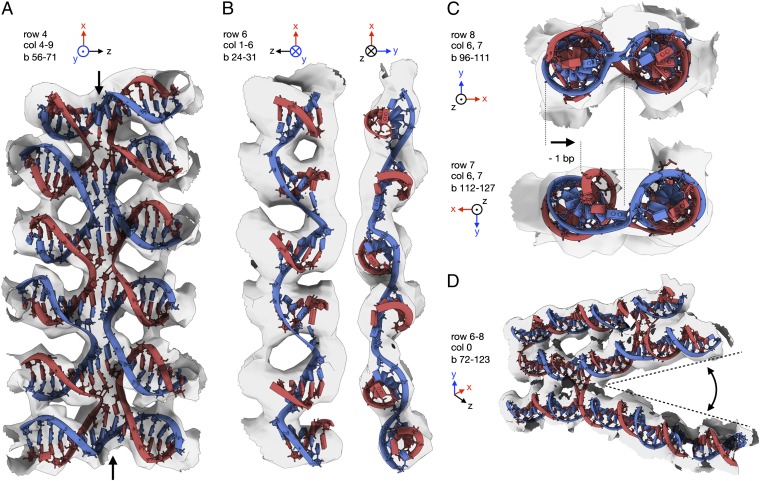
The cryo-EM map also provides structural insights into a library of highly unusual DNA topologies, some of which can only exist because of the scaffolding provided by the densely packed design. In row 4, columns 4–9 of the object, we created a vertical stack of five Holliday junctions (Fig. 4_A_; Fig. 3_A_ shows a vertical stack of three Holliday junctions). This motif is stabilized only by the stacking of base pairs at the junctions in addition to two covalent phosphodiester bonds at either end of the junction stack. In our map this motif is well resolved, thus pointing at a surprisingly high degree of structural order, contrary to what one might have intuited. We speculate that the systematic use of many of these junction stacks in future designs will lead to compaction and confer increased mechanical rigidity. In row 6, columns 1–6, we created a motif (Fig. 4_B_) that may be considered as a synthetic pseudohelix in which one strand follows a left-handed helical path with base pairs pointing along the helical direction, rather than orthogonally to the helical axis as in B-form dsDNA. The left-handed pseudohelix is stabilized by six short counter-strands of only 5- or 6-nt length. Again, the motif is well resolved in the EM density, albeit with less resolution because this motif occurs at the more flexible periphery of the object. In row 7, column 6, we omitted a single DNA base pair, which leads to a mismatch in backbone orientation at a cross-over to a neighboring helix in column 7. Previously, the introduction of many such omissions was used to design globally curved structures (15). In our map, a single base pair omission already results in a well-resolved bending deformation of both helices and a cross-over direction rotation (Fig. 4_C_), confirming that fine-tuning at the level of individual base pairs may afford precise control over orientation and location of structural features. Finally, column 0 is illustrative for how custom crevices may be created by rationally distributing cross-overs, because dsDNA helices tend to splay away from each other in the absence of stabilizing cross-overs between them (Fig. 4_D_).
Fig. 4.
Motifs beyond B-form DNA for nanotechnology. (A) A vertical stack of five Holliday junctions. (B) A pseudohelical structure that runs along the direction of the base pairs. (C) A bent helix and distorted cross-over due to an omitted base pair (Lower) and a typical cross-over without omission (Upper). (D) Crevices formed by splayed-out helices due to the absence of stabilizing cross-overs.
Conclusions
Our results demonstrate that designed, densely packed DNA objects are amenable to detailed structural characterization and may provide near atomic-level positional control. The range of topological motifs in our structure illustrates the versatility of DNA as a material for nanotechnology and highlights the existence of many unexplored design options for creating richer, more complex, but also more precise objects. Thereby, our results support a perspective in which chemical motifs may be arranged with precise structural specifications through an iterative strategy of DNA-templated design and 3D structural feedback. By using chemical groups attached to DNA strands or even reactive motifs formed by DNA itself, this strategy offers an attractive route to achieving complex functionalities known today only from natural nanomachines.
Materials and Methods
DNA-Templated Design and Synthesis.
Our object was designed in iterative cycles of using caDNAno v0.2 (16) for strand routing and CanDo (21, 22) for estimation of rigidity and global twist. DNA scaffold strands were prepared as previously described (32). DNA staple oligonucleotide strands (Table S1) were prepared by solid-phase chemical synthesis (Eurofins MWG) with Eurofins MWG high purity salt free purification grade. The objects were synthesized in a one-pot mixture containing 20 nM 7,249-bases-long M13mp18 phage DNA, 200 nM oligonucleotide staple in a pH = 8 buffer that included 5 mM Tris·base, 1 mM EDTA, 20 mM MgCl2, and 5 mM NaCl. The mixture was incubated at 65 °C for 15 min, then annealed from 56 °C to 44 °C over the course of 12 h, and then stored at 4 °C. Analysis of the reaction products by agarose gel electrophoresis (Fig. S2) showed that the object assembled with high yield with negligible formation of byproducts. Purification from excess staple strands could therefore be performed using simple molecular weight cutoff filtration using 100 kDa Amicon filters (Millipore).
Electron Cryo-Microscopy.
The purified sample at a concentration of ∼10 nM in 10 mM Tris buffer (pH 7.6) with 20 mM MgCl2, 5 mM NaCl, and 1 mM EDTA was heated in a water bath at 37 °C for 1 h before preparing the grids to resolve multimers formed by blunt-end stacking. Aliquots of 3 μL were incubated for 3 min on glow-discharged holey carbon grids with an ultrathin carbon film on top (Agar Scientific, catalog no. S187-4), blotted, and plunge-frozen in liquid ethane using a Vitrobot (FEI Company). Grids were transferred to an FEI Polara G2 microscope that was operated at 300 kV. Images were recorded on a back-thinned FEI Falcon detector at a calibrated magnification of 42,277× (yielding a pixel size of 3.55 Å). The defocus was varied from 1 to 4 µm, using a dose of ∼10 or 20 e−/Å2. Electron micrographs were evaluated for astigmatism and drift, and 456 micrographs were selected for further analysis.
Image Processing.
Contrast transfer function parameters were estimated using CTFFIND3 (33), and 28,502 particles were selected manually using the boxer program in the EMAN package (34). Reference-free 2D class averages and 3D reconstructions were calculated using RELION (20). The initial model for 3D refinement was calculated using CanDo (21). To avoid model bias against false high-resolution features, the initial model was low-pass filtered to 60 Å before refinement. The data set was split into two separate halves at the outset of refinement, and two independent models were refined simultaneously. This procedure allowed us to prevent overfitting and thereby obtain a reliable estimate of the resolution based on so-called “gold-standard” Fourier shell correlation (FSC) (35). The overall resolution of the final model was estimated to be 11.5 Å, based on the FSC = 0.143 criterion (35) (Fig. S4_A_). Low-pass filtering of the atomic model to the same resolution yielded a density map with similar features as the ones observed in our experimental map.
Inspection revealed that the density at the core of the object showed more high-resolution features than at the periphery. FSC calculations that used soft masks at different positions throughout the object indicated that the resolution varied from 9.7 Å at the core to ∼14 Å at the periphery. To optimally represent the information content throughout the object, we calculated a composite map by low-pass filtering a sharpened map (with a B-factor of −2,000 Å2) at different resolution cut-offs in the range of 10–14 Å and combining these maps, depending on the estimated resolution at each position.
We confirmed the overall correctness of our structure by performing tilt-pair validation (36) (Fig. S4_B_). Using similar grids and microscope settings as the ones described above, 50 pairs of images were recorded at tilt angles of 0° and 10°, from which 342 particle pairs were selected manually. Alignment of the particles against the final composite map and analysis of the tilt-pair transformations were performed in the XMIPP package (37). Apart from validating our map, this analysis also indicated that the individual particles may be aligned with an accuracy of ∼2°.
Model Building.
An initial atomic model with the designed topology of parallel dsDNA helices in a rectangular lattice and with the known nucleotide sequence was calculated using custom software that relied on 3DNA (23) to calculate canonical B-DNA for all designed helical fragments. Separate rigid body fitting of each of the 82 helices in the EM density map yielded a model that was suitable for flexible fitting using MDFF (24). Because the MDFF approach was not capable of handling the entire structure (which, including hydrogens, consists of 460,641 atoms), we split the structure into multiple substructures, consisting of three neighboring columns of the rectangular lattice each. Before molecular dynamics (MD) simulation, the geometry of these models was improved by up to 5,000 steps of energy minimization, which included hydrogen bond restraints but did not include restraints toward the EM density. Subsequent MD simulations were performed at 300 K for 200,000 steps using both hydrogen bond and dihedral angle restraints and a weight on the EM density (GSCALE) of 0.3 kcal/mol. After MD, another 2,000 steps of energy minimization were performed using hydrogen bonds and dihedral restraints and a weight on the EM density of 1.0 kcal/mol. A final model for the entire object was obtained by combining the central columns of all substructures. Low-pass filtering of the atomic model density yielded a map with similar features as the ones observed in the experimental reconstruction (Fig. S6 A and B). Additionally, FSC calculations between the final model and the reconstruction showed significant correlation up to 12 Å, indicating that the quality of the atomic model reflects the quality of the reconstructed density (Fig. S6_C_).
Supplementary Material
Supporting Information
Acknowledgments
We thank Lori Passmore, Andrew Carter, and Friedrich Simmel for discussions; Jake Grimmet, Shaoxia Chen, and Monika Rusp for technical support; Wei Zhao and Carlos Olguin (Autodesk Research) for support with graphics rendering; and Do-Nyun Kim and Mark Bathe for support with CanDo structure predictions. This work was supported by the Deutsche Forschungsgemeinschaft through the Excellence Cluster Center for Integrated Protein Science, Nano Initiative Munich, the Technische Universität München Institute for Advanced Study, European Research Council Starting Grant 256270 (to H.D.), and Medical Research Council Grant MC_UP_A025_1013 (to S.H.W.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM map and the atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB accession no. 2210) and in the Protein Data Bank, www.pdb.org (PDB ID codes: 2ymf–2ymi and 2ymr).
References
- 1.Alberts B, et al. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 2.Seeman NC. Nanomaterials based on DNA. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 4.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427(6975):618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 5.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461(7260):74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han D, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332(6027):342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 7.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406(6796):605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 8.Omabegho T, Sha R, Seeman NC. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324(5923):67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wickham SF, et al. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7(3):169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, et al. DNA-directed three-dimensional protein organization. Angew Chem Int Ed Engl. 2012;51(14):3382–3385. doi: 10.1002/anie.201108710. [DOI] [PubMed] [Google Scholar]
- 11.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459(7243):73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 12.Voigt NV, et al. Single-molecule chemical reactions on DNA origami. Nat Nanotechnol. 2010;5(3):200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 13.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9(7):2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 14.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459(7245):414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325(5941):725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37(15):5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke Y, et al. Multilayer DNA origami packed on a square lattice. J Am Chem Soc. 2009;131(43):15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke Y, Voigt NV, Gothelf KV, Shih WM. Multilayer DNA origami packed on hexagonal and hybrid lattices. J Am Chem Soc. 2012;134(3):1770–1774. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo S, Rothemund PWK. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat Chem. 2011;3(8):620–627. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro CE, et al. A primer to scaffolded DNA origami. Nat Methods. 2011;8(3):221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 22.Kim DN, Kilchherr F, Dietz H, Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012;40(7):2862–2868. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu XJ, Olson WK. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31(17):5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16(5):673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson R, et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol. 2011;413(5):1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murchie AI, et al. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- 27.Eis PS, Millar DP. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochemistry. 1993;32(50):13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- 28.Mao C, Sun W, Seeman NC. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J Am Chem Soc. 1999;121:5437–5443. [Google Scholar]
- 29.Ortiz-Lombardía M, et al. Crystal structure of a DNA Holliday junction. Nat Struct Biol. 1999;6(10):913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- 30.Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme. Nat Struct Biol. 1999;6(2):151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 31.Eichman BF, Vargason JM, Mooers BH, Ho PS. The Holliday junction in an inverted repeat DNA sequence: Sequence effects on the structure of four-way junctions. Proc Natl Acad Sci USA. 2000;97(8):3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci USA. 2007;104(16):6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 35.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Scheres SHW, Núñez-Ramírez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3(6):977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information