Proteomic and Bioinformatic Analysis of mSWI/SNF (BAF) Complexes Reveals Extensive Roles in Human Malignancy (original) (raw)
. Author manuscript; available in PMC: 2013 May 30.
Published in final edited form as: Nat Genet. 2013 May 5;45(6):592–601. doi: 10.1038/ng.2628
Abstract
Subunits of mammalian SWI/SNF (mSWI/SNF, also called BAF) complexes have recently been implicated as tumor suppressors in a number of human malignancies. To understand the full extent of their involvement, we conducted a proteomic analysis of purified endogenous mSWI/SNF complexes. Our studies revealed several new dedicated, stable subunits not found in the yeast SWI/SNF complex including Bcl7a, b and c, Bcl11a and b, Brd9 and SS18. Incorporating these novel members, we determined the frequency of mSWI/SNF subunit mutations in recent exome- and whole-genome sequencing studies of primary human tumors. Surprisingly, mSWI/SNF subunits are mutated in 19.6% of all human tumors reported in 44 exome sequencing studies. Our analysis suggests that specific subunits protect against cancer in specific tissues. In addition, we find that mutations to more than one subunit, which we define as a type of compound heterozygosity, are prevalent in certain cancers. Our studies demonstrate that mSWI/SNF is the most frequently mutated chromatin-regulatory complex (CRC) in human cancer and that in contrast to other known tumor suppressors and oncogenes surveyed, mSWI/SNF is broadly mutated, similar to TP53. Thus, proper functioning of these polymorphic chromatin regulatory complexes may constitute a major mechanism of human tumor suppression.
Recent genome-wide sequencing efforts have provided unprecedented views of gene networks and are increasingly used to identify biological pathways underlying complex diseases1. In particular, exome and whole genome sequencing studies of human cancer have provided an opportunity to revisit the contributions of oncogenic genetic circuitries and networks and have repeatedly identified mutations to subunits of polymorphic mSWI/SNF complexes. In mammals mSWI/SNF complexes are polymorphic assemblies of at least 13 subunits encoded by 26 genes, generating an extensive diversity of complexes with specialized function in specific tissues2–4. Early studies indicated that the catalytic subunits SMARCA4 (BRG1) or SMARCA2 (BRM) are frequently inactivated in established cell lines. Reintroduction of BRG1 into cell lines lacking BRG1/BRM produced Rb-dependent cell-cycle arrest leading to the speculation that they were tumor suppressors5,6. Clear evidence that at least one mSWI/SNF subunit is indeed a tumor suppressor emerged when SMARCB1 (hSNF5) was found to be biallelically inactivated and to undergo loss of heterozygosity (LOH) in malignant rhabdoid tumors, an aggressive set of pediatric malignancies7–11. In addition, several studies reported frequent inactivation of SMARCA4, SMARCC1, SMARCD3, PBRM1, DPF2, and ARID1A in several tumor types, including breast, lung and colon carcinomas12–19.
We report a proteomic analysis in parallel with biochemical investigations to determine the composition of endogenous mSWI/SNF complexes in various cell types. We uncovered several additional mSWI/SNF subunits that do not have homologs in the yeast SWI/SNF complex. These data enabled us to perform a comprehensive meta-analysis of 44 exome and genome sequencing studies to determine the contribution of mSWI/SNF mutations compared to other chromatin regulators and established tumor suppressors and oncogenes. Our analysis demonstrates that mSWI/SNF complexes represent the most frequently mutated chromatin regulator in human cancer, and hence substantiates and extends earlier evidence from genetic studies.
Results
BCL7A/B/C, BCL11A/B, SS18, and BRD9 are novel mSWI/SNF complex subunits
We performed an affinity purification/mass spectrometry-based analysis of endogenous mSWI/SNF complexes in several primary cell types including mouse ES cells, fibroblasts, neural progenitors, and neurons to better define the combinatorial assemblies of mSWI/SNF complexes and their associated proteins. This study was performed using an antibody specific to the C-terminus of the Brg1 ATPase subunit, thereby allowing for rapid purification of the complexes after a single biochemical step4,20. We recovered several novel proteins including Bcl7a, Bcl7b, and Bcl7c, Bcl11a and Bcl11b, as well as SS18 and Brd9. Peptide numbers and percent coverage for these proteins were comparable to those of established mSWI/SNF complex subunits (Fig. 1a). To determine the extent of dedication of these putative subunits to mSWI/SNF complexes, we performed density sedimentation analyses using a human T-cell line. BCL7A/B/C, BCL11A/B, and BRD9 co-sedimented exclusively with BRG1 and other mSWI/SNF complex subunits in glycerol gradients; these proteins were not associated with lower molecular-weight complexes such as polycomb repressive complexes PRC1 or PRC2 (denoted by immunoblots for Bmi1 or Ezh2, respectively), nor did they migrate as free monomers (Fig. 1b). In separate experiments, we noted that PBAF components BAF200 and BAF180 migrate with a much larger complex of about 4–5 MDa3 and that only about 10% of these proteins are incorporated into the 2 MDa complexes (fractions 13–15) shown in Figure 1b. Hence BCL7A/B/C, BCL11A/B and BRD9 are subunits of mSWI/SNF complexes rather than PBAF complexes. This is consistent with the specialized role of PBAF defined in vitro.21
Figure 1. Identification of novel, dedicated subunits of mSWI/SNF-like BAF complexes.
(a) Composition of purified SWI/SNF complexes as determined by mass spectrometric analysis. Confidence (protein probability statistic) is 1.0 for all peptides. Protein identification was completed using SEQUEST (University of Washington) as described4. n.d.=not detected. (b) Glycerol gradient sedimentation analysis of CCRF-CEM human T cell nuclear extracts demonstrates that Bcl7 and Bcl11 family proteins, as well as Brd9, co-sediment with mSWI/SNF complex core subunits. Fractions of 0.5ml of the 10ml 10–30% glycerol gradient were collected and subjected to western blot analysis for various mSWI/SNF proteins. Red arrows indicate fractions with prominent BRG1 peaks. (c) Partial urea denaturation ranging from 0.25M to 5M urea prior to anti-Brg1 immunoprecipitation reveals that Bcl7 and Bcl11 family proteins, and Brd9, are mSWI/SNF complex components and must be denatured to dissociate from mSWI/SNF complexes. The co-precipitated proteins were analyzed by immunoblot with antibodies specific for established and putative subunits of mSWI/SNF complexes. (d) Reciprocal IP studies using novel anti-Bcl7a/b/c antibodies and anti-Bcl11a/b, Brd 9 also reveal BAF complex subunits. (e) Updated model of mSWI/SNF (BAF) complexes incorporating novel subunits.
The observation that subunits do not dissociate during the 16 hour centrifugation necessary to perform the glycerol gradient analysis indicates that the complexes are stable and undergo relatively little subunit exchange. To independently assess the stability of the new subunits within the complexes, we used urea denaturation as a strategy that was employed in early studies to define ribosomal subunits. We subjected nuclear extracts to a range of urea concentrations prior to anti-BRG1 immunoprecipitation (IP). Remarkably, BCL7A/B/C, BCL11A/B, and BRD9 were bound to the mSWI/SNF complex with stabilities greater than or equal to most established subunits including BAF47, BAF155 and BAF170 (Fig. 1c), reflecting associations comparable to those of ribosomal subunits22,23. In addition, reciprocal IP's using antibodies to novel complex members revealed known mSWI/SNF components (Fig. 1d). These results establish BCL7A/B/C, BCL11A/B, SS18, and BRD9 as dedicated, non-exchangeable subunits of the mSWI/SNF complex, and not simply associated proteins as was previously suggested by ourselves and others3,4,24,25 (Fig. 1e). Thus, the function of these proteins must be thought of as a critical aspect of the function of mSWI/SNF complexes.
Mammalian SWI/SNF complexes represent the most highly mutated chromatin regulator in human cancer
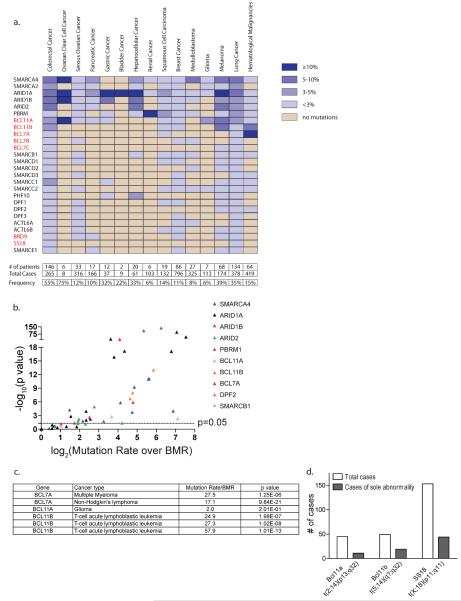
We performed an analysis of 44 published genome/exome sequencing studies26–72 to compare the mutational frequency of the mSWI/SNF subunits to that of other CRC families and established tumor suppressors and oncogenes. We determined the absolute number and overall frequency of missense, nonsense, or insertion/deletion mutations in each established or novel mSWI/SNF subunit described above (Supplementary Table 3). We limited our analysis to studies that had sequenced primary human tumors, excluding those performed on cell lines. Studies from Jones et al. (pancreas)27, Parsons et al. (glioblastoma)43, and Wei et al. (melanoma)51, in which some tumor samples were expanded as xenografts or for a limited number of passages in culture, were included (Supplementary Table 2). The mutation frequency for each gene was calculated to be the number of mutations divided by the total number of cases evaluated in one or more studies. Frequencies are displayed as a graded color scale indicating the relative contribution of each mSWI/SNF complex gene to each cancer type (Fig. 2a). Where applicable, we show the frequency calculated from the validation study (Fig. 2a, Supplementary Table 4). Finally, to calculate the frequency of mutation to the mSWI/SNF complex collectively, we divided the number of patients bearing a mutation in any mSWI/SNF subunit over the total number of cases in each cancer type (Fig. 2a, bottom). By averaging these frequencies, we find that mSWI/SNF is mutated in 19.6% of all cancer types analyzed.
Figure 2. mSWI/SNF complex subunits are mutated in human cancers with high frequency.
(a) The frequency of patients harboring a mutation in an mSWI/SNF subunit is presented according to a graded color scale. Studies on the same tissue/cancer type were summed to obtain the individual frequency of each protein. Novel subunits are highlighted in red. Bottom: the total number of patients affected by any mSWI/SNF subunit mutation was summed to calculate the overall frequency of patients with mutations in the mSWI/SNF complex. (b) A volcano plot representing the Mutation rate/BMR and p value for the most frequently mutated mSWI/SNF subunit in each of the exome sequencing studies evaluated. (c) A list of the Mutation rate/BMR and p value for all the studies in which the novel subunits BCL7A, BCL11A, and BCL11B are mutated above the BMR. (d) Translocations to mSWI/SNF subunits often represent sole genomic abnormalities (Mittelman Database).
ARID1A is the most frequently mutated among mSWI/SNF subunits, primarily in solid tumors. ARID1A mutations occur in 45.2% of endometrioid and clear-cell ovarian, 18.7% of gastric, 18.6% of bladder, 13.7% of hepatocellular, 9.4% of colorectal, 11.5% of melanoma, 8.2% of lung, 3.6% of pancreatic, and 2.5% of breast cancers (Fig. 2a, Supplementary Table 4). To address whether mutation to an mSWI/SNF subunit confers a selective advantage to a particular cancer, we compared the mutation frequencies of each subunit to the background mutation rate (BMR), accounting for gene length (Fig. 2b, Supplementary Table 5, Supplementary Text). We used the overall, non-synonymous BMR reported by each study, or the number of non-synonymous mutations divided by the total number of bases sequenced if the BMR was not reported Our statistical analysis is therefore based on the BMR for each tumor type, and is corrected for gene length (Supplementary Table 2). We find that SMARCA4, ARID1A, ARID1B, ARID2, and PBRM1 are each significantly mutated above the BMR in studies from two or more cancer types (SMARCA4, p ≤ 1.6e–02; ARID1A, p ≤ 1.0e–02; ARID1B, p ≤ 4.4e–02; ARID2, p ≤ 4.7e–02; PBRM1, p ≤ 1.9e–02), which suggests that mutations to these genes are `driver' rather than `passenger' genetic lesions (Fig. 2b, Supplementary Table 5). The significance of these mutation rates is particularly striking since ARID1A, ARID1B, ARID2, and PBRM1 had not been considered to be important contributors to oncogenesis prior to the advent of high-throughput sequencing. Several core mSWI/SNF subunits are infrequently mutated, including SMARCD1/D2/D3, SMARCE1, PHF10/DPF1/DPF2/DPF3, and ACTL6A/B, consistent with previous observations. In addition, subunits previously thought to play a role in tumorigenesis such as SMARCC1 and SMARCC215,73 are rarely mutated in primary tumors. The novel subunits BCL11B and BCL7A are frequently mutated in hematologic malignancies (Fig. 2c). BCL11B is mutated in 5.7–12.7% of T-cell acute lymphoblastic leukemia (T-ALL), and BCL7A is mutated in 19.7% of non-Hodgkin lymphoma (NHL) and 21.7% of multiple myeloma (Fig. 2a, Supplementary Table 4). BCL11B and BCL7A are significantly mutated above the BMR in these cancers (BCL11B, p ≤ 2.0e–07; BCL7A, p ≤ 1.3e–06), suggesting that mutation of these genes and subsequent loss of protein function may drive oncogenesis (Fig. 2c, Supplementary Table 5).
In addition to significant mutation frequencies in hematologic malignancies, several of the novel subunits revealed in our biochemical studies are involved in chromosomal translocations, which are thought to be driving mutations in specific cancers. Translocations involving the SS18 subunit represent the hallmark of synovial sarcoma (SS18-SSX) and are observed in >95% of cases74–76. In addition, the t(5;14)(q35;q32.2) translocation involving BCL11B is present in 20–25% of pediatric and up to 5% of adult T-ALL cases and defines a molecular subset of this malignancy62,77. BCL11A can also be a translocation partner78. To determine the significance of translocation to these novel mSWI/SNF subunits to disease, we calculated the frequency with which the translocation of interest represented the sole genomic abnormality, and thus a driving event (Fig. 2d). We analyzed BCL11A translocations t(2;14)(p13;q32) and t(2;14)(p16;q32), the BCL11B translocation t(5;14)(q35;q32.2), and SS18 t(X;18) translocations and found that they represented the sole genomic abnormality in 24.4%, 38.7%, and 28.7% of cases, respectively. These data indicate that translocations to mSWI/SNF components, particularly to three of the proteins demonstrated to be novel subunits in this analysis, contribute significantly to the impact of mSWI/SNF subunit alterations in human cancer.
Frequency of Chromatin Regulatory Mutations Among Human Cancers
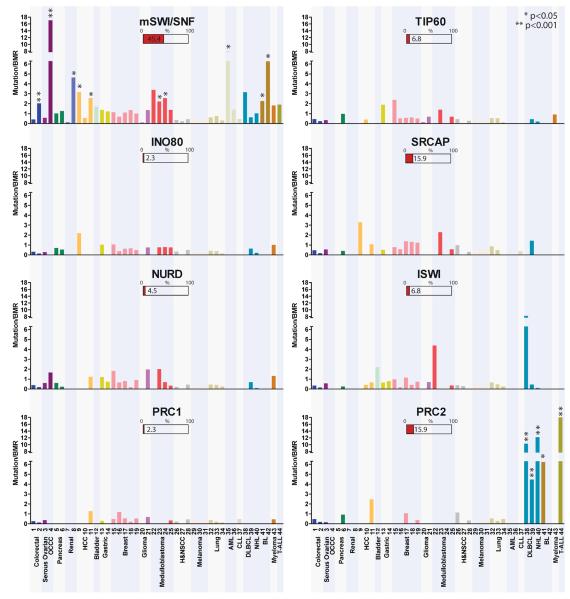
Because mSWI/SNF subunits are dedicated to the mSWI/SNF complex and are thought to function collectively as a single multi-subunit complex, we captured the propensity for mutation of the complex as a whole by considering the joint mutation frequency of mSWI/SNF components, including genes encoding novel subunits. We present the mutation frequency of the complex as the maximum-likelihood value of the mutation frequency relative to the BMR, based on the joint likelihood of the individual subunits (Fig. 3, Supplementary Text). The overall mutation frequencies are greater than the BMR in 26 of 44 studies (Mutation Rate/BMR = 1.0–17.1), including clear-cell ovarian, pancreatic, renal cell, hepatocellular, bladder, gastric, breast, glioma, medulloblastoma, and hematologic malignancies (Fig. 3). mSWI/SNF is significantly mutated in 10 studies from this group [colorectal (p = 1.4e–04), clear-cell ovarian (p = 3.6e–08), renal cell (p = 2.7e–03), hepatocellular (p < 0.05), medulloblastoma (p < 0.05), acute myeloid leukemia (p = 2.8e–02), Burkitt lymphoma (BL) (p ≤ 1.45e–02)], highlighting a potentially protective role of mSWI/SNF in the prevention of these cancers. Mammalian SWI/SNF complexes are mutated at or below the BMR in colorectal carcinoma (1 of 2 studies), squamous cell carcinoma of the head and neck, lung cancer, and melanoma (Fig. 3). Although these cancers have a high frequency of mSWI/SNF subunit mutations (Fig. 2a), several cases were considered hypermutated and therefore, high BMRs may inaccurately obscure the role of mSWI/SNF in the progression of these tumor types32,48–52,67–70 (Supplementary Fig. 1).
Figure 3. mSWI/SNF complexes are more frequently mutated in human cancer than other chromatin modifying complexes.
The Mutation rate/BMR for 37 exome sequencing studies for the mSWI/SNF, TIP60, SRCAP, INO80, ISWI, NURD, PRC1, and PRC2 complexes is graphed. The study number refers to the publications listed in Supplementary Table 2. Statistically significant mutation rates are indicated with asterisks. The percent bar graphs under each complex name indicate the percent of studies for which that complex was the most highly mutated complex of the eight examined.
Using a similar statistical approach, we compared the mutation rates of mSWI/SNF complexes to those of several other CRCs, including TIP60, INO80, SRCAP, NURD, ISWI, PRC1, and PRC2 (Fig. 3, Supplementary Text). Mammalian SWI/SNF was the most frequently mutated of the CRCs analyzed in 45.4% of the studies. PRC2 had the highest mutation rate in 15.9% of studies, which was significant in the hematologic malignancies NHL, DLBCL, BL, and T-ALL, in agreement with previous studies79–82. The SWI/SNF-like ATRX/DAXX complex has been shown to be highly mutated in pediatric glioblastoma multiforme83,84 and pancreatic neuroendocrine tumors85, however, we found it was infrequently mutated in the 44 studies. Moreover, mSWI/SNF subunits were not mutated in these rare tumors, suggesting that ATRX/DAXX mutations are isolated and mutually exclusive of mSWI/SNF mutations (data not shown). Our analysis indicates mSWI/SNF complexes possess significantly higher rates of mutation than other CRCs in human cancers pointing to a specific mechanism underlying their role in tumor suppression.
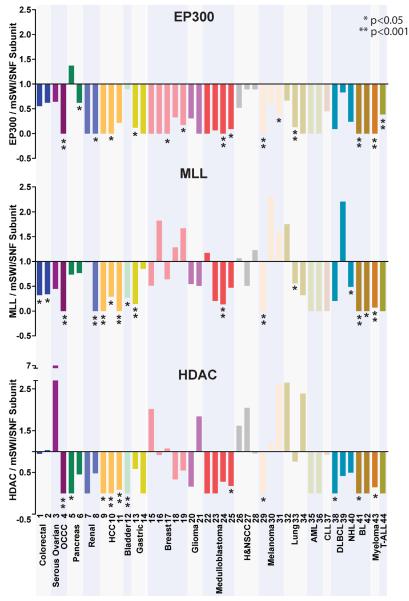
Chromatin modifiers such as the HDAC and MLL families and EP300 have also been strongly implicated in cancer86. We thus compared the mutation rates of the most frequently mutated mSWI/SNF subunit in each study to the mutation rates of the most frequently mutated of the MLL genes (MLL1-MLL5), the most frequently mutated of the HDAC genes (HDAC1-HDAC11), or to EP300 (Fig. 4, Supplementary Table 6). We represent the ratio of the mutation frequency of each chromatin modifier relative to the mSWI/SNF subunit such that ratios <1 indicate that the peak mutation frequency of mSWI/SNF genes is higher than the chromatin modifier; ratios >1 indicate that the mutation frequency of the chromatin modifier is higher (Fig. 4). With few exceptions, the mSWI/SNF subunits are more frequently mutated compared to other chromatin modifiers suggested to play important roles in tumorigenesis. The increased mutational prevalence of mSWI/SNF was statistically significant for 36.3%, 25%, and 34.1% of the studies as compared to MLL (p ≤ 4.0e–02), HDAC (p ≤ 3.8e–02), and EP300 (p ≤ 1.76e–02) respectively, indicating a major role for mSWI/SNF perturbation in the development of human cancers.
Figure 4. mSWI/SNF subunits are more frequently mutated than EP300, MLL, and HDAC family proteins.
The ratio of the mutation frequency of EP300, the highest mutated MLL family member, or the highest mutated HDAC member, was plotted relative to the most frequently mutated mSWI/SNF subunit for each study. Ratios of <1 indicate that the peak mutation frequency of mSWI/SNF genes is higher than the chromatin modifier; values of this ratio >1 indicate that mutation frequency of the chromatin modifier is higher. Statistically significant ratios favoring mSWI/SNF (ratios of less than 1) are indicated.
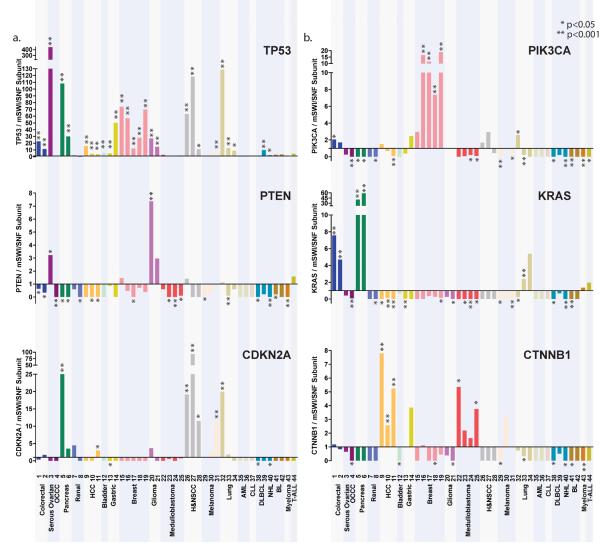
Comparison of mSWI/SNF mutational frequencies and patterns with known tumor suppressors and oncogenes
To further estimate the relative impact of potential tumor suppressive functions for mSWI/SNF complexes, we compared the mutation rates of established tumor suppressors and oncogenes to the most frequently mutated mSWI/SNF subunit from each study (Fig. 5a, b). We selected TP53, PTEN, CDKN2A, PIK3CA, KRAS, and CTNNB1 for comparison based on well-established evidence of their tumor suppressive and oncogenic functions. We limited our analysis to non-synonymous coding mutations, and did not include deletions or amplifications. Remarkably, mSWI/SNF subunits are more broadly mutated, spanning multiple malignancies of varying tissue origins, than any other tumor suppressor or oncogene analyzed with the exception of TP53. Tumor suppressors and oncogenes analyzed here are mutated with high frequencies in specific cancers rather than across several tumor types. For example, PTEN is more highly mutated than the most frequently mutated mSWI/SNF subunit in serous ovarian cancer and glioma, CDKN2A is more highly mutated in melanoma, pancreatic, lung, and squamous cell carcinoma, PIK3CA in colorectal, breast, and head and neck squamous cell cancer, KRAS in colorectal, lung, and pancreatic cancer, and CTNNB1 in hepatocellular and medulloblastoma cancers (Fig. 5a, b). These profiles highlight the specificity of these tumor suppressors and oncogenes in driving particular cancers; mSWI/SNF subunit genes by contrast are significantly mutated in a range of cancers (Fig. 3), suggesting an extensive role of tumor suppression that protects fundamental cellular functions.
Figure 5. Mutations to mSWI/SNF subunits occur in a broad spectrum of cancer types.
The ratio of the mutation frequency of the (a) tumor suppressors TP53, PTEN, and CDKN2A, and (b) oncogenes PIK3CA, KRAS, and CTNNB1 was plotted relative to the most frequently mutated mSWI/SNF subunit. Ratios of <1 indicate that the peak mutation frequency of mSWI/SNF genes is higher than the tumor suppressor/oncogene; values of this ratio >1 indicate that mutation frequency of the tumor suppressor/oncogene is higher. Statistically significant ratios are indicated.
Compound heterozygosity of mSWI/SNF subunits occurs in some cancer types
While prototypical tumor suppressors often undergo LOH, many mSWI/SNF subunit mutations analyzed from primary tumors were found to be heterozygous28,63. We hypothesized that heterozygous mutations in more than one mSWI/SNF subunit may result in a compound heterozygous phenotype due to the dedicated nature of mSWI/SNF subunits within the complex and may explain why many of the tumors do not have homozygous inactivation of a single subunit. We analyzed patients bearing >1 mutation or deletion of any mSWI/SNF subunit and found that up to 50% (mean = 14.3%) of patients exhibit compound heterozygosity in certain cancer types (Fig. 6a). We observed a particularly high occurrence of this phenomenon in squamous cell carcinoma (50%), renal cell carcinoma (28%), and clear cell ovarian carcinoma (25%). Limited number of data points precluded us from being able to define a single pairing which occurs with higher frequency than others, although greater than a third (36.8%) of the compound heterozygous patients were noted to have one or more mutations in ARID1A (Fig. 6a). The prevalence of patients with multiple mSWI/SNF subunit mutations suggests that compound heterozygosity may have significant functional repercussions, which could contribute to human malignancy. In addition, because mutations that were present in the adjacent normal tissue were often not reported in these studies, our figures may underestimate the frequency of compound heterozygosity.
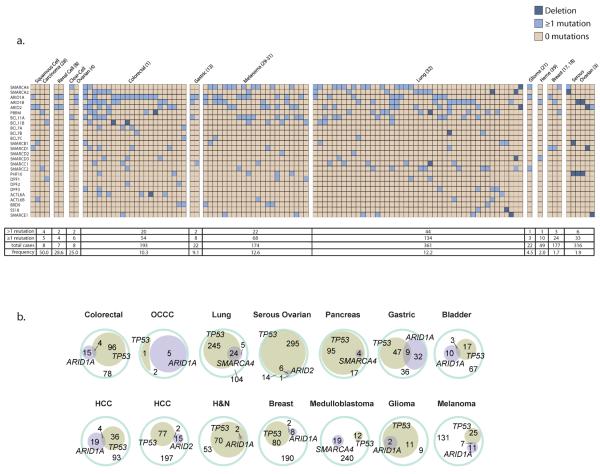
Figure 6. Co-occurrence of mSWI/SNF mutations in human cancers.
(a) The patient profiles of all the patients harboring a mutation or deletion in more than 1 mSWI/SNF subunit are represented from the studies that provided this information (study number is listed after cancer type). Hypermutated patients are marked with a # sign. The number of patients with <1 mutation in mSWI/SNF subunits, 1 mutation, the total number of cases, and the frequency of compound heterozygous patients is displayed. (b) The cooccurrence of mSWI/SNF mutations and TP53 mutations for all the studies that had more than 1 tumor with a TP53 mutation are displayed in a Euler diagram against the total number of cases.
Mutations to mSWI/SNF subunits are frequently mutually exclusive of TP53 mutations
We assessed the frequency with which mSWI/SNF mutations co-occur with mutations in other known oncogenic or tumor suppressor pathways to determine what pathways are sufficient to cause transformation in the setting of an mSWI/SNF subunit mutation. For example, ARID1A mutations were shown to be mutually exclusive of TP53 mutations, but coincident with PIK3CA and CTNNB1 mutations in clear-cell ovarian carcinoma28,63. We calculated the mutual exclusivity of TP53 and the most highly mutated mSWI/SNF subunits, namely ARID1A, SMARCA4, or ARID2, among the solid tumors (Fig. 6b). Although mutations to mSWI/SNF subunits and TP53 are not strictly mutually exclusive, there is a tendency toward mutual exclusivity in colorectal (odds ratio, OR=0.217), clear-cell ovarian (OR=0.077), gastric (OR=0.215), hepatocellular (OR=0.544 for ARID1A, OR=0.341 for ARID2), medulloblastoma (OR=0.493), and breast (OR=0.594) cancers, which is statistically significant for colorectal (OR=0.217, 95% CI=0.07–0.68, _p_=0.0043) and gastric (OR=0.215, 95% CI=0.091–0.51, _p_=0.0002) cancers. In contrast, all patients with an ARID1A mutation in serous ovarian, glioma, and squamous cell carcinoma, or a SMARCA4 mutation in pancreatic cancer also had a mutation in TP53, perhaps reflecting a requirement for TP53 loss-of-function in these cancers (Fig. 6b). This dichotomy might also reflect the fact that ARID1A and mSWI/SNF are not significantly mutated in these cancers, and suggest that these mutations are instead merely passenger mutations to a TP53 driver mutation (Supplementary Table 5, Fig. 3). Despite a limited cohort of patients, we similarly observed that mutations to mSWI/SNF subunits and PTEN in colorectal and serous ovarian carcinoma tend to be mutually exclusive, which was significant for ARID1A and PTEN in colorectal cancer (OR=0.303, 95% CI=0.110–0.838, _p_=0.0121) (Supplementary Fig. 2). In contrast, mSWI/SNF subunit mutations appear to co-occur with mutations in some oncogenes. ARID1A and CTNNBI mutations co-occur in colorectal (OR=4.545, 95% CI=1.326–15.58, _p_=0.0283) and hepatocellular carcinoma (OR=2.667, 95% CI=1.075–6.618, _p_=0.0306), and SMARCA4 and CTNNBI mutations co-occur in medulloblastoma (OR=8.819, 95% CI=3.431–22.67, p<0.0001), in keeping with observations that SMARCA4 mutations are present in CTNNB1- and MYC-, but not SHH-driven medulloblastomas (Supplementary Fig. 2)46,87. Similarly, mutations to ARID1A and PIK3CA pair in clear-cell ovarian cancer as noted previously (OR=7.000, 95% CI=1.590–30.81, _p_=0.0106)28, and exhibit overlap in colorectal and breast carcinoma (Supplementary Fig. 2). Thus, mSWI/SNF complex mutations represent a significant contribution to oncogenesis, which we demonstrate is independent of the loss of tumor suppressors TP53 and PTEN, and potentially cooperative with oncogenes such as CTNNB1 and PIK3CA.
Discussion
We present a comprehensive proteomic and biochemical analysis of endogenous mSWI/SNF complexes in several non-transformed cell types, which facilitated a complete bioinformatic assessment of mutational frequencies to mSWI/SNF components in human cancers. Efforts to identify mSWI/SNF complex components have generally used overexpressed, tagged proteins in transformed cell lines, which disturb stoichiometric relationships. Our studies allowed us to accurately determine the subunit composition of endogenous mSWI/SNF complexes, revealing several new subunits as well as interacting proteins lacking the dedicated, non-exchangeable features of a subunit. We find that mSWI/SNF complexes have approximately the same stability as the ribosome22,88,89 using urea-based denaturation methods. Hence, the function of BCL7A/B/C, BCL11A/B, BRD9, and SS18, similar to established mSWI/SNF subunits, should be considered in the context of mSWI/SNF complex function. Interestingly, these subunits as well as ARID1A/1B/2, PHF10, DPF1/2/3, PBRM1 (BAF180), beta-actin and SMARCE1 (BAF57) are not present in the yeast SWI/SNF complex, though proteins homologous to PBRM1, SMARCE1, and beta-actin are found in the yeast RSC and SWR1 complexes90,91. Thus, they likely perform some function related to evolutionarily newer strategies of chromatin regulation, such as greater complexity/specificity in mSWI/SNF complex targeting, Polycomb-mediated repression, or DNA methylation. These differences compared to yeast SWI/SNF led us to refer to the complexes as BAF rather than mSWI/SNF to prevent inappropriate extrapolation92,93, however here we used mSWI/SNF in accordance with common usage.
Our proteomic studies enabled a comprehensive examination of mSWI/SNF mutation frequencies in tumors from 44 whole-genome and exome sequencing studies. Mammalian SWI/SNF complexes were mutated above the BMR in the majority of studies, which spanned a wide spectrum of solid and hematologic tumors. However, there are also tumor types in which mSWI/SNF is frequently mutated, but does not reach significance due to insufficient numbers of patients and hypermutation. Nevertheless, the breadth of mSWI/SNF involvement uncovered in this analysis is striking and resembles that of TP53, albeit at lower frequencies, suggesting that mSWI/SNF prevents tumorigenesis through basic cell biological functions.
A second point emerging from our analysis is that specific tissues are protected from malignancy by the function of specific subunits. Examples of a predominance of one subunit over its polymorphic partner suggest that certain complex assemblies carry out the tumor suppressive role of mSWI/SNF. For example, ARID1A, ARID1B, and ARID2 are mutually exclusive subunits occupying one position in the complex, and are mutated with different frequencies in different cancer types. Similarly, SMARCA4 and SMARCA2 have similar ATPase activity, but SMARCA2 is rarely mutated in cancer, while SMARCA4 appears to be the second most frequently mutated subunit (Fig. 2a). Cellular context also seems to play an important role; for example, although SMARCA4 is ubiquitously expressed, mutations to SMARCA4 occur in only specific cancer types. Likewise, BCL11B and BCL7A are mutated in hematologic malignancies, but rarely in solid tumors. Interestingly, the subunits of a specialized mSWI/SNF complex found only in post mitotic neurons (nBAF)3,94–96 are rarely mutated in any of the cancers examined. Finally, SMARCB1 (hSNF5) is biallelically inactivated in nearly all malignant rhabdoid tumors, but is rarely mutated in other cancer types97,98. This indicates that the apparent broad general role of the complexes in tumor suppression is likely related to the specific functions of different polymorphic assemblies in different tissues.
At present, there is little knowledge of how the individual subunits function within polymorphic complexes. It is generally assumed that the function of the ATPase subunits is to promote nucleosome mobility and the function of the PHD domains, bromo domains, chromodomains and DNA binding domains is to target the complex to specific genetic loci, however this has yet to be demonstrated conclusively. The novel subunits BCL11A and BCL11B are zinc finger-containing transcriptional repressors which are critical for the development of the erythroid and lymphoid lineages,99,100 while little is known of the function of SS18 or the BCL7 family members. Since mSWI/SNF is known to directly repress transcription by binding to about 15,000 islands over the genome101, one can speculate that aberrant chromatin binding, mis-targeting or loss of ATPase activity might underlie the role(s) of different mSWI/SNF subunits in human malignancy. Alternatively, Roberts et al. have shown that loss of hSNF5 results in enhanced PRC2-mediated repression of critical repressors of cell proliferation such as INK4A/ARF102. EZH2/hSNF5 double mutants are completely rescued from hSNF5-driven tumorigenesis, strongly implicating the ability of mSWI/SNF to evict or oppose PRC2. Indeed, loss of Smarca4 also results in global increases in PRC2 binding and H3K27me3 deposition at mSWI/SNF binding sites in mouse ES cells101.
Based on the dedicated nature mSWI/SNF complex subunits, we hypothesized that an otherwise passenger heterozygous mutation in one subunit paired with a heterozygous mutation in another subunit could result in loss of mSWI/SNF tumor suppressive function. We think of this as a form of compound heterozygosity. For example, we found that many tumors had an ARID1A or SMARCA4 driver mutation paired with a mutation in an infrequently mutated subunit (Fig. 6a). Using the MutationAssessor platform, we found that most of these passenger mutations are predicted to be deleterious and thus could indeed further disable the mSWI/SNF complex in the context of compound heterozygosity (Supplementary Fig. 3). These mutational pairings may confer a unique loss of function, or they may be less detrimental to cells and thus selected over homozygous inactivation of ARID1A or SMARCA4. Increased sample size will help determine whether mSWI/SNF compound heterozygosity is a driving mechanism in tumorigenesis, and define oncogenic pairings.
Our studies support the previously expressed notion that mSWI/SNF subunits are tumor suppressors rather than oncogenes5,7. Indeed, the fact that mSWI/SNF mutations are generally exclusive of TP53 and PTEN mutations suggests an analogous role for mSWI/SNF in tumor suppression (Fig. 6b, Supplementary Fig. 2). However, specific mutations or translocations to mSWI/SNF subunits may produce gain-of-function properties that result in a specific mSWI/S91NF complex becoming oncogenic. Future studies will be necessary to define the mechanism(s) that underlie the frequent mutation of mSWI/SNF subunits in human cancer. Although tumor suppressors are often considered difficult therapeutic targets, detailed mechanistic studies might elucidate unexpected avenues for treatment of this broad class of human cancers.
Materials and Methods
Affinity purification and mass spectrometry
A rabbit polyclonal antibody raised against aa1257–1338 of hBrg that recognizes both mouse Brg and Brm was used for affinity purification from nuclear extracts obtained from E14 ES cells. Immunoprecipitation of endogenous complexes was performed in 300 mM NaCl, 50mMTris-HCl (pH 8.0), 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 1 mM DTT. Purified complexes were separated further by strong cationic exchange, and fractions were analyzed on LTQ-Orbitrap (Thermo Scientific). Peptides were identified by searching acquired mass spectra using SEQUEST (University of Washington) against the Mouse IPI database version 3.34. Peptide identifications were validated statistically using PeptideProphet, and the protein inference was performed using ProteinProphet, available as a part of the TransProteomic Pipeline (2). The list of protein identifications in each analysis was filtered using a 0.95 probability threshold or as otherwise stated (estimated error rate of less than 1%). All proteins identified in the control runs and other known contaminants were subtracted from the final list.
Preparation of ES Nuclear Extracts
CCRF-CEM T-cell leukemia cells were grown under standard conditions to confluence and lysed and homogenized in Buffer A(10 mM Hepes (pH 7.6), 25 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, and protease inhibitors (complete mini tablets (Roche) supplemented with 1 mM PMSF) on ice. Nuclei were sedimented by centrifugation (1,000×g), resuspended in Buffer C (10 mM Hepes (pH 7.6), 3 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and protease inhibitors), and lysed by the addition of ammonium sulfate to a final concentration of 0.3 M. Soluble nuclear proteins were separated by insoluble chromatin fraction by ultracentrifugation (100,000×g) and precipitated with 0.3 mg/ml ammonium sulfate for 20 min on ice. Protein precipitate was isolated by ultracentrifugation (100,000×g), and resuspended in IP buffer (150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1% NonidetP-40, 0.5% deoxycholate, 1 mM DTT, 1mM PMSF with protease inhibitors) for immunoprecipitation analyses or HEMG-0 buffer (25mM HEPES (pH 7.9), 0.1mM EDTA, 12.5mM MgCl2, 100mM KCl, supplemented with fresh DTT and PMSF) for glycerol gradient analyses.
Density sedimentation analyses
800 ug of CCRM-CEM T-cell nuclear extracts from above were resuspended in 300 microliters of 0% glycerol HEMG buffer containing 25mM HEPES pH 7.9, 0.1mM EDTA, 12.5 mM MgCl2, 100mM KCl, freshly supplemented with DTT and PMSF before use and carefully overlaid on to a 10ml 10–30% glycerol (in HEMG buffer) gradient prepared in a 14×89 mm polyallomer centrifuge tube (Beckman, part# 331327). Tubes were placed in a SW-40 swing bucket rotor and centrifuged at 4 degrees for 16 hours at 40,000 RPM. 0.5 ml fractions were harvested and used in gel electrophoresis and subsequent western blotting analyses.
Collation of SWI/SNF complex mutations
Discovery screens were searched for mutations in mSWI/SNF, TIP60, SRCAP, INO80, ISWI, NURD, PRC1, and PRC2 subunits (complete term sets listed in Supplementary Table 3), TP53, MLL1–5, HDAC1–11, EP300, PIK3CA, PTEN, KRAS, CTNNB1, and CDKN2A. For each gene, the number of patients harboring an mSWI/SNF subunit mutation was summed to calculate the frequency of patients with mutations in the mSWI/SNF complex. Studies on the same tissue/cancer type were summed to obtain the individual frequency of each protein. In cases where a particular gene was further evaluated in validation screens, the frequency from these screens was calculated and is represented in the table. In cases in which one patient harbored more than 1 mutation/deletion in mSWI/SNF subunit proteins, the patient was considered a compound heterozygote.
Translocations
The number of patients in the Mitelman Database harboring a particular translocation was determined using http://cgap.nci.nih.gov/Chromosomes/AbnCytSearchForm. Of these patients, those for whom the translocation of interest was the sole abnormality were summed.
Statistics
Full details of statistical methods are provided in the Supplementary Text. Briefly, mutation frequencies of individual genes were compared to the BMR by assuming a uniform underlying mutation frequency. Log-likelihood ratio tests were performed by comparing the maximum-likelihood mutation frequency for a gene to the BMR. Groups of genes were compared to other groups of genes by a similar method of comparing the joint maximum-likelihood mutation frequency for the group of genes obtained based on empirical mutation frequencies.
Supplementary Material
01
Acknowledgements
This work was supported in part by NIH grants NS046789 (G.R.C.) and CA163915 (G.R.C.). G.R.C. is an Investigator of the Howard Hughes Medical Institute. C.K. is supported by the National Science Foundation (Graduate Research Fellowship Program). D.C.H. is supported by the Helen Hay Whitney Foundation Fellowship. C.H. is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Fellowship F32HD072627.
Footnotes
Contributions C.K. performed and interpreted experiments. D.C.H. and C.H. performed genomic data analysis and interpretation. L.E., L.H. and J.R. performed proteomic mass spec data collection and analysis. C.K., D.C.H., and G.R.C. conceived of and wrote the manuscript.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996;15:5370–82. [PMC free article] [PubMed] [Google Scholar]
- 3.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunaief JL, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–30. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 6.Strobeck MW, et al. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci USA. 2000;97:7748–53. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 8.Sevenet N, et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–8. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 10.Taylor MD, et al. Familial posterior fossa brain tumors of infancy secondary to germline mutation of the hSNF5 gene. Am J Hum Genet. 2000;66:1403–6. doi: 10.1086/302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–6. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reisman DN, et al. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21:1196–207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- 13.Wong AK, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–7. [PubMed] [Google Scholar]
- 14.Rodriguez-Nieto S, et al. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- 15.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–30. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Zhao YL, Li Y, Fletcher JA, Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007;46:745–50. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 17.Heeboll S, et al. SMARCC1 expression is upregulated in prostate cancer and positively correlated with tumour recurrence and dedifferentiation. Histol Histopathol. 2008;23:1069–76. doi: 10.14670/HH-23.1069. [DOI] [PubMed] [Google Scholar]
- 18.Takita J, et al. Gene expression profiling and identification of novel prognostic marker genes in neuroblastoma. Genes Chromosomes Cancer. 2004;40:120–32. doi: 10.1002/gcc.20021. [DOI] [PubMed] [Google Scholar]
- 19.Hoyal CR, et al. Genetic polymorphisms in DPF3 associated with risk of breast cancer and lymph node metastases. J Carcinog. 2005;4:13. doi: 10.1186/1477-3163-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–4. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 21.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–8. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 22.Spitnik-Elson P, Atsmon A. Detachment of ribosomal proteins by salt. I. Effect of conditions on the amount of protein detached. J Mol Biol. 1969;45:113–24. doi: 10.1016/0022-2836(69)90214-9. [DOI] [PubMed] [Google Scholar]
- 23.Roberts ME, Walker IO. Structural studies on Escherichia coli ribosomes. 3. Denaturation and sedimentation of ribosomal subunits unfolded in urea. Biochim Biophys Acta. 1970;199:184–93. [PubMed] [Google Scholar]
- 24.Middeljans E, et al. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–63. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–9. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 34.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 37.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 38.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durinck S, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–43. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 54.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 56.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hug I, et al. Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J Biol Chem. 2011;286:37887–94. doi: 10.1074/jbc.M111.287755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Keersmaecker K, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med. 2010;16:1321–7. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guan B, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–32. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richter J, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44:1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 73.DelBove J, et al. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics. 2011;6:1444–53. doi: 10.4161/epi.6.12.18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crew AJ, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. Embo J. 1995;14:2333–40. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clark J, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–8. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 76.de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–9. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- 77.Berger R, et al. t(5;14)/HOX11L2-positive T-cell acute lymphoblastic leukemia. A collaborative study of the Groupe Francais de Cytogenetique Hematologique (GFCH) Leukemia. 2003;17:1851–7. doi: 10.1038/sj.leu.2403061. [DOI] [PubMed] [Google Scholar]
- 78.Satterwhite E, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–20. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- 79.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park SW, Chung NG, Eom HS, Yoo NJ, Lee SH. Mutational analysis of EZH2 codon 641 in non-Hodgkin lymphomas and leukemias. Leuk Res. 2011;35:e6–7. doi: 10.1016/j.leukres.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 81.Bodor C, et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia. 2011;25:726–9. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 82.Ryan RJ, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6:e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kannan K, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 85.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 87.Pugh TJ, Cho Y-J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations within a broad landscape of genetic heterogeneity. Nature. 2012 doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Atsmon A, Spitnik-Elson P, Elson D. Detachment of ribosomal proteins by salt. II. Some properties of protein-deficient particles formed by the detachment of ribosomal proteins. J Mol Biol. 1969;45:125–35. doi: 10.1016/0022-2836(69)90215-0. [DOI] [PubMed] [Google Scholar]
- 89.Robinson A, Sykes J. A study of the atypical ribosomal RNA components of Rhodopseudomonas spheroides. Biochim Biophys Acta. 1971;238:99–115. doi: 10.1016/0005-2787(71)90014-1. [DOI] [PubMed] [Google Scholar]
- 90.Cairns BR, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 91.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 92.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 93.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 94.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–17. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–6. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uno K, et al. Aberrations of the hSNF5/INI1 gene are restricted to malignant rhabdoid tumors or atypical teratoid/rhabdoid tumors in pediatric solid tumors. Genes Chromosomes Cancer. 2002;34:33–41. doi: 10.1002/gcc.10052. [DOI] [PubMed] [Google Scholar]
- 98.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97:13796–800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Durum SK. Bcl11: sibling rivalry in lymphoid development. Nat Immunol. 2003;4:512–4. doi: 10.1038/ni0603-512. [DOI] [PubMed] [Google Scholar]
- 100.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–42. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 101.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–13. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–64. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01