The assembly of a GTPase–kinase signalling complex by a bacterial catalytic scaffold (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 7.
Published in final edited form as: Nature. 2010 Dec 19;469(7328):107–111. doi: 10.1038/nature09593
Abstract
The fidelity and specificity of information flow within a cell is controlled by scaffolding proteins that assemble and link enzymes into signalling circuits1,2. These circuits can be inhibited by bacterial effector proteins that post-translationally modify individual pathway components3–6. However, there is emerging evidence that pathogens directly organize higher-order signalling networks through enzyme scaffolding7,8, and the identity of the effectors and their mechanisms of action are poorly understood. Here we identify the enterohaemorrhagic Escherichia coli O157:H7 type III effector EspG as a regulator of endomembrane trafficking using a functional screen, and report ADP-ribosylation factor (ARF) GTPases and p21-activated kinases (PAKs) as its relevant host substrates. The 2.5 Å crystal structure of EspG in complex with ARF6 shows how EspG blocks GTPase-activating-protein-assisted GTP hydrolysis, revealing a potent mechanism of GTPase signalling inhibition at organelle membranes. In addition, the 2.8 Å crystal structure of EspG in complex with the autoinhibitory Iα3-helix of PAK2 defines a previously unknown catalytic site in EspG and provides an allosteric mechanism of kinase activation by a bacterial effector. Unexpectedly, ARF and PAKs are organized on adjacent surfaces of EspG, indicating its role as a ‘catalytic scaffold’ that effectively reprograms cellular events through the functional assembly of GTPase-kinase signalling complex.
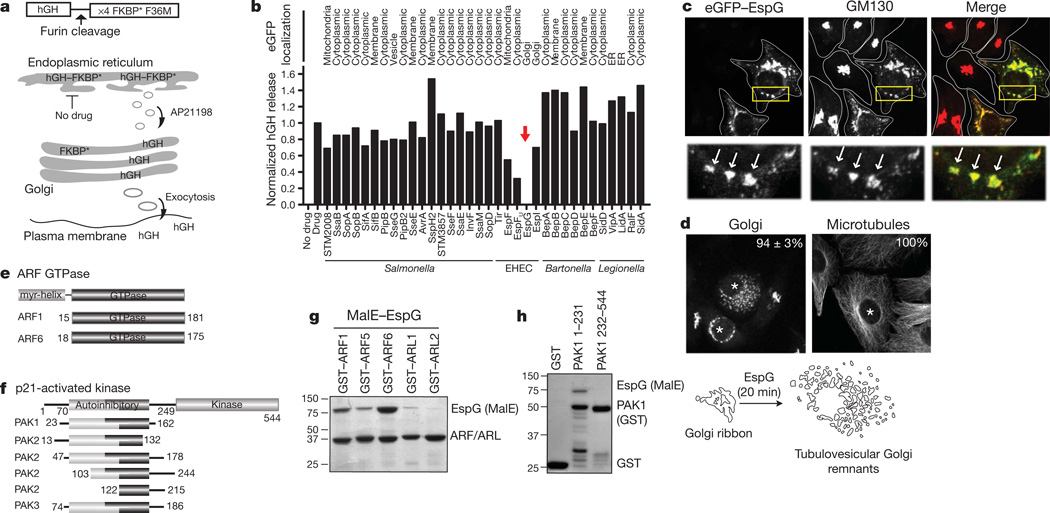
To identify new signalling pathways targeted by bacterial pathogens, we used a human growth hormone (hGH) secretion assay9 to measure the ability of type III and type IV effector proteins to regulate vesicle trafficking through the general secretory pathway (Fig. 1a, b). Consecutively, each bacterial effector was tagged with enhanced green fluorescent protein (eGFP) and assessed for localization at host organelles (Fig. 1b). We noted that several type III effectors encoded by the extracellular pathogen enterohaemorrhagic E. coli (EHEC) O157:H7 inhibited host trafficking events, whereas effectors secreted by Salmonella Typhimurium, Legionella pneumophila and Bartonella henslae showed little inhibitory functions, consistent with their intracellular life cycles (Fig. 1b). In particular, the EHEC type III effector EspG blocked exocytosis of hGH through an unknown molecular mechanism (Fig. 1b). eGFP-tagged EspG localized to the _cis_-Golgi apparatus, where it induced severe fragmentation of the organelle (Fig. 1c, d and Supplementary Fig. 1a, b). The Golgi disruption phenotype was observed when 10 nM recombinant EspG protein was microinjected into cells to mimic the protein concentration delivered by E. coli through the type III secretion apparatus10 (Fig. 1d). In addition, EspG disrupted the recycling endosome compartment in both transfection (Supplementary Fig. 1c, d) and microinjection experiments (data not shown). Previous genetic studies have implicated EspG11,12 and related Shigella family members13 in microtubule depolymerization. However, microtubules were intact in EspG microinjected cells (Fig. 1d), consistent with previous reports showing that these effectors do not disrupt cytoskeletal architectures14,15. Thus, EspG represents a new class of bacterial signalling effector that functionally regulates cargo trafficking from membrane organelles.
Fiure 1. EspG inhibits endomembrane trafficking and disrupts Golgi architecture.
a, hGH trafficking assay showing how the hGH–FKBP* (Phe 36 Met mutant) aggregates in the endoplasmic reticulum until drug application (AP21998), whereby hGH enters the general secretory pathway and is secreted into the culture medium. b, hGH release assay showing the effects of type III and type IV effector proteins on trafficking through the general secretory pathway (Methods). hGH was quantified by enzyme-linked immunosorbent assay and normalized to GFP control (Drug) experiments. The subcellular localization of eGFP-tagged effectors is indicated. ER, endoplasmic reticulum. c, Co-localization of eGFP–EspG (green) with _cis_-Golgi matrix protein GM130 (red). The Golgi in untransfected cells appears as tightly associated cisternae. d, Golgi and microtubule phenotypes induced by EspG protein microinjection (asterisk). The percentage of microinjected cells exhibiting each phenotype is indicated (n = 3, from >40 cells per experiment). e, f, ARF GTPase (e) and PAK isoforms (f) that interact with EspG by yeast two-hybrid. g, h, Glutathione pull-down of GST–ARF isoforms (g) and GST–PAK1 fragments (h) with recombinant MalE-tagged EspG.
We used a yeast two-hybrid screen to identify host enzymes targeted by EspG. The screen resulted in 26 positive interactions with multiple overlapping complementary DNA clones expressing two ARF GTPase family isoforms (ARF1 and ARF6) and three p21-activated kinase family members (PAK1, PAK2 and PAK3) (Fig. 1e, f). ARF GTPases function within a broad range of organelle systems, where they organize vesicle transport machinery, phospholipids and signalling molecules at membrane microdomains16,17, whereas the PAK family of serine/threonine kinases transduce Cdc42 and Rac1 GTPase signals that establish intracellular polarity18. Direct interactions between EspG and the GTPase domain of ARF family members (Fig. 1g) and the autoinhibitory domain (AID) of PAK kinases (Fig. 1h) were shown by in vitro binding studies using purified recombinant proteins. These findings establish two EspG substrates that are consistent with its regulatory function in host protein trafficking identified here and in bacterial infection studies conducted in vivo19,20.
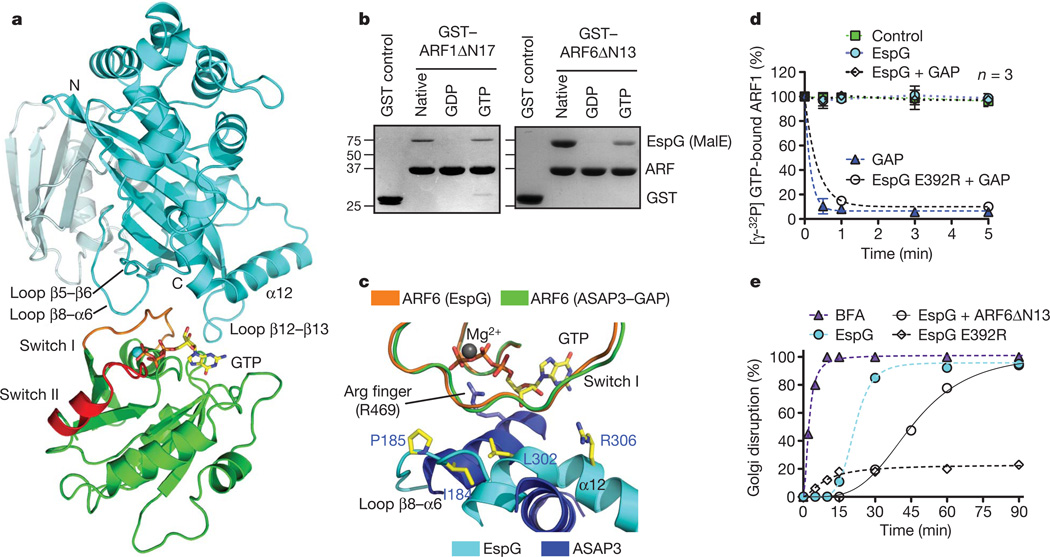
Next we crystallized EspG (residues 42–398) in complex with the GTPase domain of ARF6 (residues 13–175) and solved the structure to a resolution of 2.5 Å (Supplementary Table 1). EspG buries 602 Å2 of ARF6 surface area and the complex interface is mediated by a collaboration of EspG loops (loops connecting β5 and β6, β8 and α6, and β12 and β13) that specifically engage the switch I loop of ARF6 and several residues lining the guanine-nucleotide-binding pocket (Fig. 2a and Supplementary Fig. 2a, b). The conformational state and amino-acid sequence of switch I are highly conserved between ARF family members, indicating that the EspG–ARF6 structure illustrates the nature of EspG’s interaction with several ARF isoforms (Supplementary Fig. 3a, b). The importance of conserved switch I residues for binding EspG were confirmed by mutational analysis on ARF6 (Supplementary Fig. 3c).
Figure 2. The structure of EspG in complex with GTP-bound ARF6.
a, The overall structure of EspG–ARF6GTP complex. EspG is shown in cyan and ARF6 in green. Switch I and switch II on ARF6 are coloured orange and red, respectively. b, EspG selectively binds the GTP-loaded ARF1 and ARF6 (GST tagged) in glutathione pull-down assays. The native lane represents ARF GTPases purified from bacteria without removing or loading specific nucleotides. c, Structural overlay of EspG–ARF6GTP and ASAP3(GAP)–ARF6GDP·AlFX (Protein Data Bank ID, 3LVQ) showing how EspG sterically hinders ARF binding to ASAP3–GAP. The catalytic Arg finger of ASAP3 is labelled. d, GTP hydrolysis assay showing that EspG inhibits GAP-assisted GTP hydrolysis on ARF1. The rate of γ32P[GTP] hydrolysis was measured as the percentage of γ32P[GTP] remaining on ARF1 over time. Intrinsic ARF1 GTPase activity (control, green), GAP-stimulated activity (GAP, blue triangle), and EspG inhibition of GAP activity (EspG + GAP, open diamond) or mutant EspG Glu 392 Arg (open circle) are shown. e, Time course of the Golgi disruption phenotype presented as the percentage of microinjected cells with altered Golgi morphology as shown in Fig. 1d. At least 45 microinjected cells were scored in each trial for a Golgi disruption phenotype, and the data are representative of three experimental trials. BFA, brefeldin A.
ARF6 is GTP bound in the crystal and adopts an active-state conformation nearly identical to that of ARF6GTPγS (ref. 21; Supplementary Fig. 4a). Further structural analyses revealed that switch I is inaccessible to EspG when ARF6 adopts the GDP-bound conformation (Supplementary Fig. 4a). EspG selectively bound the GTP-loaded forms of ARF1 and ARF6 but did not recognize GDP–ARF complexes (Fig. 2b). Moreover, EspG interacted with ARF6GTP in its full-length myristoylated form, which was isolated from membrane fractions (Supplementary Fig. 4b). Thus, EspG preferentially targets the active ARFGTP signalling molecule.
COPI coat, vesicle complex adaptors and signalling enzymes primarily associate with switch 2 and the β2/3 interswitch of ARFGTP (refs 22–26; Supplementary Fig. 5a). Given the frequent occurrence of this binding mode, we were surprised to find that EspG is rotated away from these common binding elements and is positioned directly over the guanine-nucleotide-binding pocket (Fig. 2a, c). Surprisingly, however, EspG does not function as a guanine nucleotide exchange factor (Supplementary Fig. 5b, c) or a GTPase-activating protein (GAP) (Fig. 2d, cyan circles). Rather, EspG is appropriately positioned to hinder binding of ARF–GAP and its catalytic access to the γ-phosphate of GTP (Fig. 2c). EspG completely abolished the GAP-stimulated GTPase hydrolysis on lipid-anchored ARF1 (Fig. 2d, diamonds), in comparison with the fast ARF–GAP reaction (Fig. 2d, blue triangles). The inhibition of GAP by EspG relied on a direct interaction between EspG and ARF because the binding-deficient mutant EspG Glu 392 Arg (characterized in Supplementary Fig. 6) had no affect on GAP-stimulated hydrolysis (Fig. 2d, open circles).
GTP hydrolysis and exchange on ARF is required for proper membrane transport functions, suggesting that EspG inhibits Golgi trafficking by blocking its guanine nucleotide cycle17,26. Several lines of evidence support this idea. First, EspG disrupted the Golgi complex with rapid inhibitory kinetics (Fig. 2e) and a phenotype similar to the fungal toxin brefeldin A (Supplementary Fig. 7a), a potent ARF1 GTPase inhibitor that also interferes with the guanine nucleotide cycle27. Second, microinjection of dominant-negative ARF protein (ARFΔN13) caused a significant delay in Golgi disruption induced by EspG (Fig. 2e, open circles). Third, EspG Glu 392 Arg, a mutant that does not interact with host substrates (Supplementary Fig. 6), had no affect on Golgi morphology or trafficking function (Fig. 2e and Supplementary Fig. 7b, c). Finally, EspG co-localized with the ARF1 effector β-COP (ref. 26) on Golgi membranes (Supplementary Fig. 1b). These combined structure and cellular studies provide a mechanism for bacterial regulation of membrane trafficking: EspG prevents vesicle transport by directly inhibiting ARF guanine nucleotide turnover on host membranes.
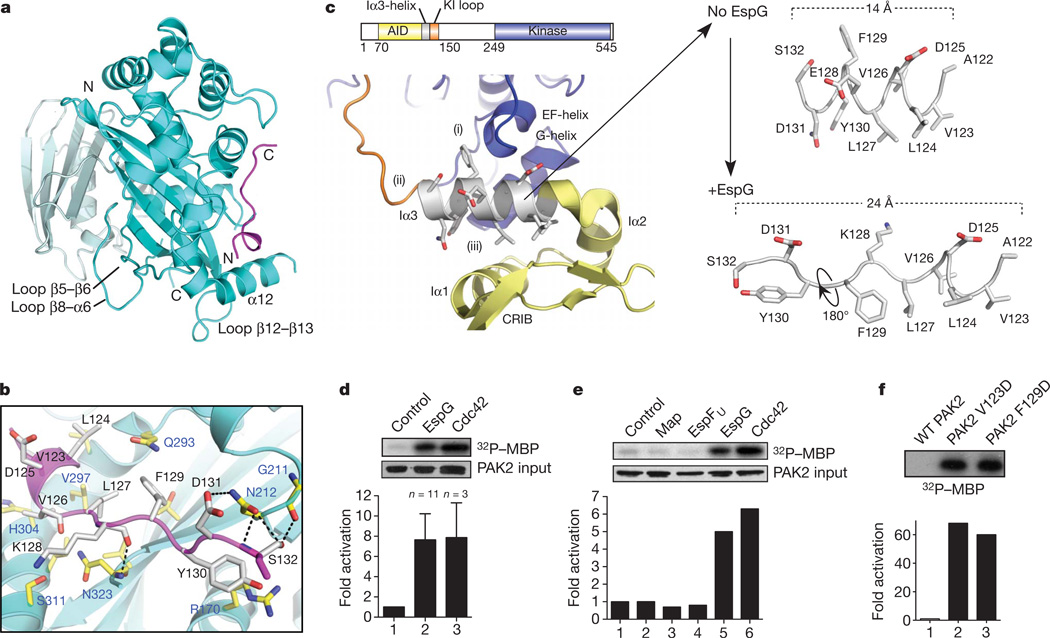
Having established the mechanism of ARF GTPase regulation, we next explored a second possible function of EspG: regulation of PAK family kinases. The EspG-binding site on PAK2 was defined to residues 121–136, a highly conserved sequence that encodes the Iα3-helix within the kinase AID (Supplementary Fig. 8). We crystallized EspG in complex with the PAK2 Iα3-helix fragment and solved the structure to a resolution of 2.8 Å (Supplementary Table 1). EspG recognized the initial turn of the Iα3-helix whereas the remainder of the peptide adopted an extended strand conformation that lies orthogonal to the EspG six-stranded β-sheet (Fig. 3a, b). EspG buries 684 Å2 of the PAK2 surface area and the binding is primarily supported by a large hydrophobic interface and hydrogen bonding by residues Asn 212 and Asn 323 of EspG (Fig. 3b). This structural interface was confirmed by a series of in vitro binding studies and kinase assays using PAK2 and EspG mutant proteins (Supplementary Fig. 9).
Figure 3. The structure of EspG in complex with PAK2 Iα3 peptide.
a, The overall EspG–PAK2Iα3 complex with EspG oriented and coloured as in Fig. 2a. The PAK2 Iα3 peptide (residues 123–134) are shown in magenta. b, Detailed interactions between EspG and PAK2Iα3. Key binding residues from EspG (blue labels) and PAK2 (black labels) are shown. c, Close-up view of autoinhibited PAK1 homodimer (Protein Data Bank ID, 1F3M) focused on chain B (kinase domain, blue) and chain D (autoinhibitory domain, yellow). The Iα3-helix inhibitory functions are labelled (i)–(iii) corresponding with those outlined in the results section. The Iα3-helix extracted from the PAK1 structure (numbering corresponds to PAK2 for ease of comparison) is shown at the upper right. The corresponding PAK2 Iα3-helix extracted from the EspG structure (lower right) is oriented by the amino-terminal helical residues 123–127. KI loop, kinase inhibitory loop. CRIB, Cdc42/Rac1 interacting binding. d, e, PAK2 kinase assays comparing 2 µM EspG with equimolar GTPγS-loaded Cdc42 (d) and the indicated EHEC type III effectors (e). Phosphorylation of myelin basic protein (MBP) substrate, input levels of PAK2 and quantification of each experiment are shown. f, PAK2 kinase assays comparing autoinhibited wild-type (WT) PAK2 with PAK2 mutants Val 123 Asp and Phe 129 Asp. Data are presented as in d.
To determine how EspG may regulate the kinase, we compared the peptide structure from EspG–PAK2 with the structure of Iα3-helix in the autoinhibited PAK1 homodimer28 (Fig. 3c). In autoinhibited PAKs, the Iα3-helix is sandwiched between the kinase domain and the AID, where it has three autoinhibitory functions: (i) it folds onto the EF- and G-helices to block substrate binding; (ii) it positions the ‘kinase inhibitory’ loop across the enzyme catalytic cleft; and (iii) it stabilizes the AID three-helix bundle maintaining PAK homodimerization (Fig. 3c). Hence, EspG binding to the Iα3-helix suggested a mechanism for PAK activation. EspG induced a (7.6 ± 2.5)-fold (n = 11) increase in PAK2 activity, a profile that is comparable to PAK stimulation by GTPγS-loaded Cdc42 ((7.8 ± 3.4)-fold, n = 3) (Fig. 3d). Notably, EHEC type III effectors Map and EspFU showed no PAK stimulatory activity, demonstrating the signalling specificity of EspG (Fig. 3e).
To gain further insight into the mechanism of kinase activation, we first examined the details of the EspG–PAK2 interface. EspG residue Asn 212 probably initiates the kinase reaction because this residue engages the surface-accessible residues Asp 131 and Ser 132 in the autoinhibited PAK homodimer (Fig. 3b and Supplementary Fig. 10). On initial recognition, EspG displaces the Iα3-helix by reorganizing its secondary structure and by displacing side chains that normally contact the autoinhibitory interface between the kinase domain and the AID (Fig. 3c). These data suggest a novel allosteric mechanism for PAK activation. To further confirm that PAK is stimulated by local perturbations in the environment surrounding Iα3, we mutated the hydrophobic PAK2 residues Val 123 and Phe 129 that stabilize the AID and the kinase domain, respectively. Both Val 123 Asp and Phe 129 Asp resulted in a constitutively active kinase with more than 60-fold enhancement of substrate phosphorylation (Fig. 3f). We note that the mechanism of EspG binding to PAK is structurally distinct from that of Cdc42 binding28, indicating that the catalytic machinery of EspG is a unique bacterial invention.
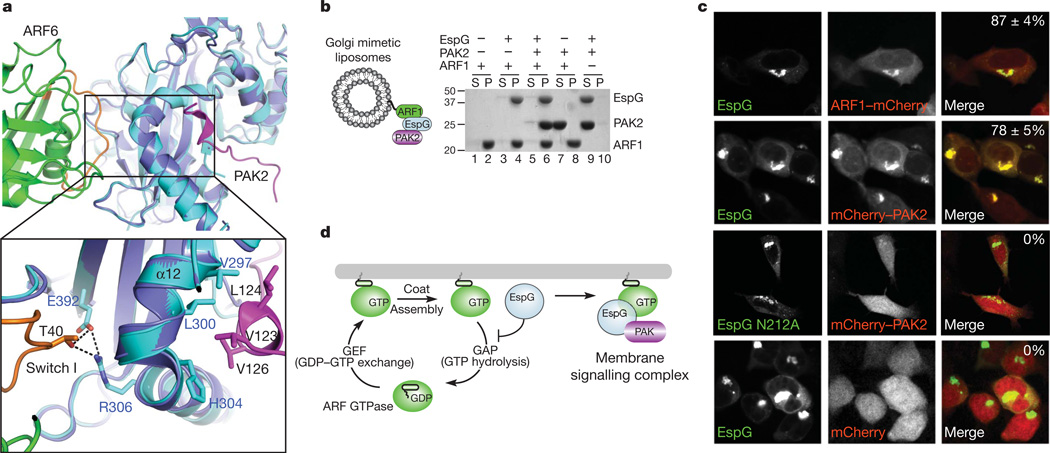
The two EspG structures reported here are nearly identical, with a root mean squared deviation of 0.612 Å over 349 Cα atoms. As shown in Fig. 4a, ARF6 and PAK2 occupy distinct, non-overlapping binding sites on adjacent surfaces of EspG. Consistent with this view, EspG nucleates a trimeric complex between the kinase and GTPase in solution (Supplementary Fig. 11). This complex could also be reconstituted on Golgi mimetic liposomes (Fig. 4b). ARF1GTP recruited EspG to the artificial membrane surface (Fig. 4b, lane 4), which in turn localized PAK2 to these sites (Fig. 4b, lane 6). Notably, PAK2 localization was strictly dependent on formation of the EspG–ARF1GTP complex (Fig. 4b, lanes 6–8) and ARF1 tethering to the membrane (Fig. 4b, lanes 9 and 10). As predicted by these findings, EspG co-localized with ARF1at the Golgi (Fig. 4c). We further speculated that PAK would also be recruited to these sites. To test this hypothesis, an in vivo ‘activity’ probe was engineered by fusing the PAK2 Iα3-helix sequence (residues 121–136) to the carboxy terminus of the mCherry fluorophore. The PAK2 probe recognized cellular EspG and was targeted to the Golgi complex in 78 ± 5% of EspG-transfected cells (Fig. 4c). By comparison, mutant EspG Asn 212 Ala that lacked all kinase stimulatory activity (Supplementary Fig. 9) localized to the Golgi complex but did not recruit PAK2 to these sites (Fig. 4c). Together, our studies support the function of EspG as a catalytic scaffold that links GTPase inhibition with kinase signal transduction pathways at membrane organelles (Fig. 4d).
Figure 4. EspG functions as a catalytic scaffold at membrane organelles.
a, Structural overlay of EspG–ARF6GTP and EspG–PAK2Iα3 highlighting the close association between ARF and PAK on the surface of EspG. Colours are as in Figs 2a and 3a except that EspG from the PAK2 structure is coloured purple. b, Golgi-mimetic-liposome-binding assays showing that EspG nucleates a trimeric complex between ARF1 and PAK2 on membrane surfaces. After centrifugation, proteins remaining in the supernatant (S) or those associated with liposomes in the pellet (P) are indicated. c, HEK239A cells co-transfected with the indicated constructs showing that eGFP–EspG co-localizes with ARF1–mCherry and recruits a PAK activity probe (mCherry–PAK2121–136) to Golgi membranes. The percentage of cells exhibiting co-localized EspG with mCherry-tagged proteins (n = 3) is shown in the upper right of the merged micrographs. d, Model of the dual function of EspG as an inhibitor of membrane trafficking and as a catalytic scaffold that assembles a GTPase–kinase signalling complex at cellular membranes. GEF, guanine nucleotide exchange factor.
EspG belongs to large family of type III effectors secreted by diverse bacterial pathogens. Our studies show that EspG has structural homology with VirA (refs 14, 15) from Shigella flexneri, suggesting that it too may function as an enzyme scaffold (root mean squared deviation, 3.1 Å; _Z_-score, 5.9) (Supplementary Fig. 12). However, a detailed structural comparison indicates that VirA is unlikely to target the same signalling pathways as EspG during Shigella pathogenesis (Supplementary Fig. 12). We provide mechanistic insights and structural evidence that EspG harbours two unique pathogenic activities, ARF GTPase inhibition and PAK stimulation. Moreover, EspG targets PAK to specific membrane surfaces through its association with ARFs. Froma strategic point of view, the assembly of an artificial enzyme complex enables bacteria to precisely control signalling events with little competition between endogenous regulatory systems. Thus, it is intriguing to speculate that EspG organizes a higher-order signalling network to effectively subvert key cellular processes including cell polarity, adhesion, receptor trafficking and protein secretion.
METHODS SUMMARY
Recombinant protein preparation and cloning were done using standard methods. In immunofluorescence experiments, we transfected cells with Fugene 200 or microinjected them with 10 nM protein, where indicated. We performed kinase assays with rabbit pEGFP–PAK2, which was immunoprecipitated from 293T cell lysates and incubated with the protein of interest in the presence of MBP, 10 µM ATP and 5 µCi γ32P[ATP]. Reactions were stopped by the addition of SDS buffer, separated by SDS–polyacrylamide gel electrophoresis and kinase activity was measured as 32P counts per minute. EspG–ARF6 and EspG–PAK2–Iα3 were purified by ion exchange and gel filtration chromatography and crystallized by the hanging-drop vapour diffusion method. We collected X-ray diffraction data at the Structural Biology Center, Advanced Photon Source, Argonne National Laboratory (USA). The structure of EspG–ARF6 was phased to a resolution of 2.5 Å by the multiwavelength anomalous dispersion method using selenomethionine-labelled EspG and ARF6 proteins. The EspG–PAK2 structure was solved to a resolution of 2.8 Å by the molecular replacement method using the EspG monomer of the EspG–ARF6 structure as the initial search model. Further details can be found in Supplementary Information.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supplemental Information
02
Acknowledgements
We would like to thank our colleagues, specifically K. Orth, M. Rosen, M. Cobb, J. Seeman, C. Brautigam and T. Fox, for helpful discussions in preparation of this manuscript, and we are particularly indebted to members of the Structural Biology Lab and mass spectrometry facilities for their efforts on this project. We would also like to thank J. Goldberg and G. Bokoch for providing valuable reagents. We would particularly like to thank J. Cherfils for providing preliminary insights into this work and for key reagents. The structure shown in this report is derived from work performed on beamlines 19-BMand 19-ID at the Structural Biology Center, Advanced Photon Source, Argonne National Laboratory. Argonne National Laboratory is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. R.C.O. was supported by a NIH Molecular Microbiology training grant (5T32AI007520-12). This work was supported by the Welch Foundation (#I-1704) and a grant from the NIH (NIAID; 1RO1AI083359-01) to N.M.A.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions N.M.A. and A.S.S. had the general ideas for this manuscript. A.S.S., N.M.A. and S.M.B. crystallized the protein complexes and D.R.T. solved the complex structures. N.M.A., A.S.S., S.E.S., B.A.W., L.E.R. and R.C.O. planned, performed and interpreted the experiments. N.M.A. and A.S.S. wrote the manuscript and all authors provided editorial input.
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3PCR and 3PCS.
The authors declare no competing financial interests.
References
- 1.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim WA. Designing customized cell signalling circuits. Nature Rev. Mol. Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duesbery NS, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 4.Yarbrough ML, et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt G, et al. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 6.Li H, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 7.Alto NM, et al. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J. Cell Biol. 2007;178:1265–1278. doi: 10.1083/jcb.200705021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vingadassalom D, et al. Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspFU during pedestal formation. Proc. Natl Acad. Sci. USA. 2009;106:6754–6759. doi: 10.1073/pnas.0809131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera VM, et al. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 2000;287:826–830. doi: 10.1126/science.287.5454.826. [DOI] [PubMed] [Google Scholar]
- 10.Winnen B, et al. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS ONE. 2008;3:e2178. doi: 10.1371/journal.pone.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw RK, et al. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 disrupt the microtubule network of intestinal epithelial cells. Infect. Immun. 2005;73:4385–4390. doi: 10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomson FL, et al. Enteropathogenic Escherichia coli EspG disrupts microtubules and in conjunction with Orf3 enhances perturbation of the tight junction barrier. Mol. Microbiol. 2005;56:447–464. doi: 10.1111/j.1365-2958.2005.04571.x. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida S, et al. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science. 2006;314:985–989. doi: 10.1126/science.1133174. [DOI] [PubMed] [Google Scholar]
- 14.Germane KL, Ohi R, Goldberg MB, Spiller BW. Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules. Biochemistry. 2008;47:10241–10243. doi: 10.1021/bi801533k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis J, et al. Novel fold of VirA, a type III secretion system effector protein from Shigella flexneri. Protein Sci. 2008;17:2167–2173. doi: 10.1110/ps.037978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 18.Bokoch GM. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 19.Borthakur A, et al. Enteropathogenic Escherichia coli inhibits butyrate uptake in Caco-2 cells by altering the apical membrane MCT1 level. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G30–G35. doi: 10.1152/ajpgi.00302.2005. [DOI] [PubMed] [Google Scholar]
- 20.Guttman JA, et al. Aquaporins contribute to diarrhoea caused by attaching and effacing bacterial pathogens. Cell. Microbiol. 2007;9:131–141. doi: 10.1111/j.1462-5822.2006.00773.x. [DOI] [PubMed] [Google Scholar]
- 21.Pasqualato S, Menetrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isabet T, et al. The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J. 2009;28:2835–2845. doi: 10.1038/emboj.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 25.Shiba T, et al. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nature Struct. Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, et al. Direct and GTP-dependent interaction of ADP ribosylation factor 1 with coatomer subunit beta. Proc. Natl Acad. Sci. USA. 1997;94:4418–4423. doi: 10.1073/pnas.94.9.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chardin P, McCormick F. Brefeldin A: the advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 28.Lei M, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information
02