Fresh Approaches to Anti-Infective Therapies (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 16.
Abstract
If discovery of new antibiotics continues to falter while resistance to drugs in clinical use continues to spread, society's medicine chest will soon lack effective treatments for many infections. Heritable antibiotic resistance emerges in bacteria from nonheritable resistance, also called phenotypic tolerance. This widespread phenomenon is closely linked to nonproliferative states in ways that scientists are just beginning to understand. A deeper understanding of the mechanisms of phenotypic tolerance may reveal new drug targets in the infecting organisms. At the same time, researchers must investigate ways to target the host in order to influence host-pathogen relationships. Government must reform the regulatory process for approval of new antibiotics. The private sector, government, and academia must undertake multiple, organized, multidisciplinary, parallel efforts to improve the ways in which antibiotics are discovered, tested, approved, and conserved, or it will be difficult to sustain the modern practice of medicine.
INTRODUCTION
Throughout most of human history, infectious diseases have been a leading cause of death. However, by the late 20th century, infectious diseases caused by bacteria fell off the public's radar in wealthier regions as society and medicine erected an effective four-walled fort: sanitation, nutrition, immunization, and antibacterial drugs (herein, “antibiotics” for simplicity). Recent years have brought a concerted effort to enlarge the protective zone globally, because infectious diseases remain the leading cause of loss of disability-adjusted life years (1). Tragically, however, the fourth wall of the fort is crumbling as antibiotic resistance allows microbial pathogens to surge over it. Much of the pharmaceutical industry has withdrawn from its effort to rebuild the wall, whereas the food industry is inadvertently helping to tear it down, using more than half of our antibiotic output to promote growth in healthy animals and plants, hastening the spread of resistance. This Review discusses the linked challenges of antibiotic resistance and discovery and recommends new policies and efforts to rebuild and maintain a supply of effective treatments for bacterial infection.
Why is the problem urgent? And why has there been so little response? Our shrinking ability to cure bacterial infections threatens to impair much of modern medical practice, undermines global economic growth, compromises national security, and drives up mortality rates for individuals in all stages and stations of life. The complacency with which much of society has met this onrushing calamity may stem from two factors. First, everyone shares the risk. Second, in contrast to people infected by HIV (the cause of AIDS), those who go on to suffer from untreatable bacterial infections are rarely acquainted before infection and, once infected, may die quickly. Their families rarely can identify each other because medical histories are confidential. Thus, no subgroup of concerned citizens has emerged or is likely to emerge to organize and advocate as a counterweight to inertia and to business interests that resist change. Elected leaders will have to act, not in response to donors or opponents, but out of informed civic concern.
The goal is not to return to a golden age when antibiotics held sway over bacterial diseases. After antibiotic use became widespread in the United States and life expectancy rose, the U.S. Surgeon General was said to have testified to Congress in 1969 that the time had come to “close the books on infectious diseases.” However, the vision that a handful of broad-spectrum agents would provide a permanently effective defense against serious infections was a mirage. The control these agents afforded was a high point in the history of medicine, but the protection they afforded was brief and incomplete.
To regain, maintain, and extend substantial control over bacterial infections will require continuous development and application of fresh approaches based on new knowledge, practices, and policies. We need to learn more about how antibiotics work, how bacteria resist them, and how to discover, test, approve, and conserve them. We need to learn how to advantage the host side of the host-pathogen relationship, both to make better use of host immunity and to deny bacteria access to dispensable host factors on which pathogenesis depends. Finally, we need to figure out better ways for industry, government, and academia to work together to enable these advances.
DEFINING PATHOGENS
To discover antibiotics and use them effectively, we need to understand the traits that define a given bacterial species as the cause of a disease. However, nearly 130 years after the introduction of Koch's postulates provided the definitive guide, we are coming to recognize their limitations.
Koch maintained that the causative microbe is one that (i) is abundant in all individuals with the disease, but not in healthy individuals; (ii) can be isolated from diseased individuals and grown in pure culture; (iii) when taken from the culture can cause the disease in healthy subjects; and (iv) can be isolated again from the experimentally infected hosts. However, most bacteria cannot currently be cultured. Indeed, a number of diseases once considered to be noninfectious in origin, such as gastric and duodenal ulcers and Whipple's disease—a disorder of absorption in the small intestine—were reclassified as infectious only when it became possible relatively recently to culture causative bacterial agents. Other diseases such as sarcoidosis and rheumatoid arthritis invite speculation as to possible infectious causes, but none has been established. One or more species among the intestinal microbiota in mice can act as co-causes in diverse diseases that are not considered infectious, such as demyelination (2), arthritis (3), and atherosclerosis (4).
Another limit to Koch's postulates is that certain diseases, such as Crohn's disease, ulcerative colitis, and environmental enteropathy, may be triggered in part by bacteria, but not necessarily by one pathogen or invasive pathogens so much as by the particular composition of the polymicrobial microbiota in an individual of a given genetic background (5).
Moreover, virulence is not an intrinsic property of a microbe but is context-dependent. A particular microbe may colonize the host harmlessly or cause disease, depending on the status of the host's immune system and epithelia (the tissues exposed to the external environment, including the skin and the lining of the airways, gut, bladder, and genital tract), the other microbes present (6), and whether the microbe is growing in planktonic form (in suspension) or in a biofilm (embedded in an extracellular matrix produced by itself or other microbes) (7). Sometimes, virulence emerges only when antibiotics reduce the numbers of other bacteria, both because this alters microbe-microbe interactions and because reduction in bacterial burden can lead to the waning of innate immune mechanisms (8, 9).
Even when Koch's postulates have placed a single bacterial species in the crosshairs as the causative agent in a particular disease, we may struggle to define the species well enough to know how best to attack it. Bacteria in a given species can exhibit extensive genomic diversity, such that a species may resemble a swarm (10). A species can diversify its genome by mutation and by horizontal gene transfer through transformation, conjugation, and viral transduction. In the middle ear, the lung, sinuses, teeth, intravenous lines, and urinary catheters, and on heart valves, artificial joints, and implanted devices, bacteria aggregate in antibiotic-tolerant biofilms that favor horizontal gene transfer because of their high population density, prolonged cell-cell contact, and high content of free DNA trapped in the matrix upon release from cells (7). Antibiotic treatment itself promotes genetic diversity by stimulating mutagenesis (11).
Even when their set of genes is fixed, bacterial pathogens from a single species express a changing ensemble of mRNAs—the transcriptome—under different circumstances. For example, _Mycobacterium tuberculosis_—the single leading cause of death from bacterial infection worldwide—remodels its transcriptome extensively as it moves from growth in standard broth culture to residence in host macrophages (12). The genes whose expression is essential for M. tuberculosis to survive in vitro overlap only partially with those whose expression is required for the bacterium to survive in the host. Classically, antibiotics are sought using populations of bacteria that express the genes essential for growth in vitro. However, only genes essential for bacteria to survive in and cause disease in the host encode targets for clinically useful antibiotics.
DEFINING DEATH
Assuming we know which microbe causes a disease and which of its gene products are required to sicken the host, how can we tell that an appropriately targeted antibiotic has killed it? Nothing should be less ambiguous. However, there is no one operational definition of microbial death that applies to all bactericidal antibiotics and scales from single bacterial cells to bacterial populations in culture to bacteria in the infected host.
Death of a single bacterium under direct observation can be defined by its lysis, as documented by time-lapse microscopy (13, 14). However, not all clinically useful antibiotics are bacteriolytic, nor do bacteriolytic antibiotics kill all members of a genetically homogeneous bacterial population (13, 14), as will be discussed below.
Death is more commonly assessed at the level of bacterial populations by numerical reduction in colony-forming units (CFUs) of bacteria growing in or on agar, a method introduced by Koch. However, for unknown reasons, many antibiotics are markedly less effective when tested against dense bacterial cultures than against dilute cultures. This “inoculum effect” can be seen with tiny numbers of bacteria, provided they are confined in a small enough volume of culture medium, for example, 150 organisms in 2 pl (15). Such an inoculum effect might confer resistance on the bacterial clumps that commonly form in infected tissues. Thus, a conventionally performed drug susceptibility test may yield a single minimum inhibitory concentration (MIC; the concentration of the drug required to reduce growth of the overall population in vitro by >99%), but the concentration of the antibiotic that is effective in vivo may vary from one site to another.
Finally, death of bacterial pathogens can be defined in terms of the host's clinical response, as will be discussed later. First, though, it is necessary to disentangle death of bacteria from their nonreplication.
DISSECTING NONREPLICATION
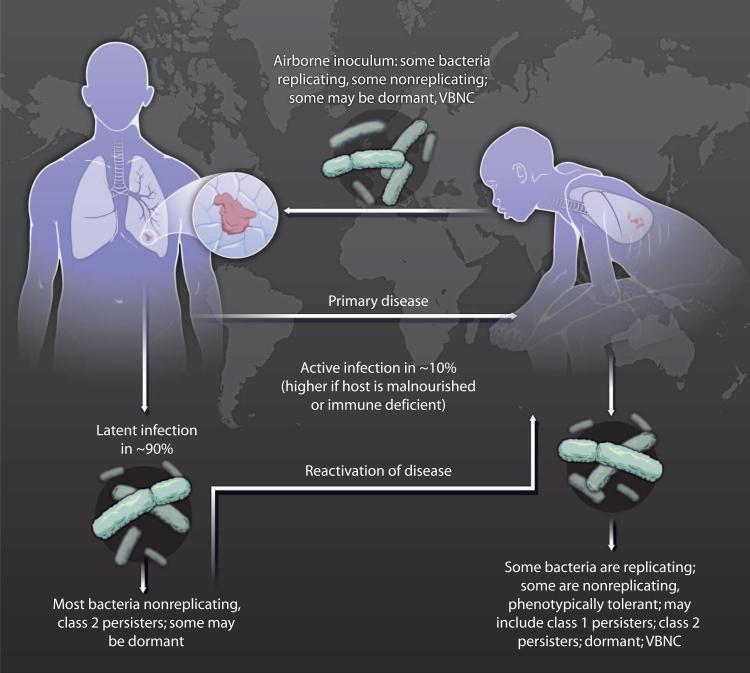
Making bacterial death particularly hard to define are diverse states in which bacteria can refrain from replicating for varying periods, yet retain the potential to resume replication and cause disease (Figs. 1 and 2). By the CFU criterion, some such bacteria may be scored as dead. States of nonreplication are often also states of relative resistance to antibiotics that were developed on the basis of their ability to kill replicating bacteria (16, 17). Our limited understanding of the mechanisms of entry into, maintenance of, and exit from nonreplicating states and of the relationship between their manifestations in vitro and in vivo leaves the distinctions among them imprecise, their clinical relevance speculative, and our ability to control them marginal. The following discussion compares and contrasts five states of bacterial nonreplication: stationary phase, viable but nonculturable (VBNC), persistent, dormant, and latent. Frequent reference is made to M. tuberculosis in deference to the burden of disease for which it is responsible, but these states can be manifest by many bacterial pathogens.
Fig. 1. Diverse bacterial states.
Many infectious diseases are complicated by the ability of bacterial pathogens to adopt diverse functional states that reduce the organisms’ susceptibility to antibiotics. This phenomenon is illustrated for tuberculosis, the leading cause of death from a single bacterial infection. VBNC, viable but nonculturable.
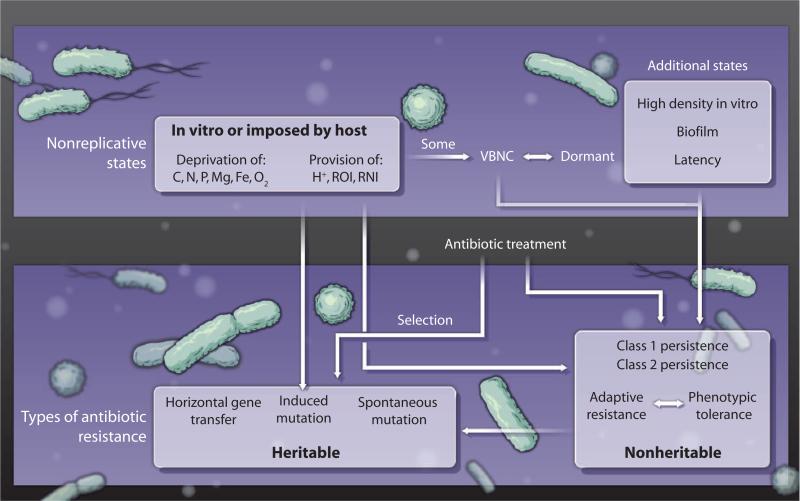
Fig. 2. Types of antibiotic resistance.
Various bacterial states are associated with antibiotic resistance. See text for definitions and discussion.
Stationary phase
In vitro, bacteria stop replicating—that is, enter stationary phase—when they exhaust sources of carbon, nitrogen, phosphorus, magnesium, iron, or other macro- or micronutrients. Even if nutrients are not limiting, cultured bacteria stop replicating when exposed to certain concentrations of acid or reactive oxygen or nitrogen intermediates. Aerobes stop replicating if deprived of oxygen, and anaerobes if exposed to oxygen. In vivo, the host can restrict particular sources of carbon, deprive the pathogen of iron or oxygen, acidify the microenvironment, and impose oxidative and nitrosative stress. Likewise, residence in biofilms can impose at least some of these conditions, such as oxygen deprivation. In such situations, nonreplicating bacteria may up-regulate the transcription of stress-response genes, down-regulate genes that encode enzymes that catalyze central carbon metabolism, and become relatively resistant to antibiotics that kill the organism when it is replicating.
Viable but nonculturable
After bacteria remain in stationary phase for a period that varies greatly among species, the number of CFUs measured in vitro falls. Thus, variable proportions of the bacteria are considered to be dead. However, some of the bacteria may resume replication in alternative assays. Bacteria that do not form CFUs and whose replication is only evident in other kinds of assays are imprecisely termed “viable but nonculturable” (VBNC). For example, late stationary-phase bacteria that do not form CFUs may grow after dilution in liquid medium to the point that only one bacterium is present in each of several replicate copies of the culture, a method called “dilution to extinction.” In this case, a statistical approach called “the most probable number method” is used to back-calculate the number of viable bacteria in the original sample (18). Why this method of culture is sometimes restorative is not understood. Other assays that may reveal VBNC bacteria include culture with “resuscitation-promoting factors” (19), injection into an experimental animal, or immunosuppression of an animal serving as the host, as is discussed further below.
It was recently reported that the vast majority of M. tuberculosis in sputum from untreated people with tuberculosis were VBNC (19). If confirmed, this observation has major implications for the diagnosis of tuberculosis and the monitoring of clinical trials of drugs for that disease. Drug trials in tuberculosis currently depend on quantitative reductions in bacterial numbers in the sputum early in the course of treatment as a biomarker for efficacy.
Persistence
“Persisters,” first so designated nearly 60 years ago in a study of the actions of penicillin on staphylococci (20), are the minority of bacteria in an antibiotic-sensitive population that are not killed by that antibiotic but, when allowed to grow in its absence, give rise to populations that can be killed by the antibiotic in the same proportion as before. Time-lapse microscopy of Escherichia coli maintained in a microfluidic device allowed an analysis of antibiotic action at the level of individually identifiable bacterial cells. This study revealed that the persisters in a population, most of which succumbed to ampicillin, were the individual bacilli that had not been dividing before the ampicillin was applied (13). Such an observation suggested that stationary-phase bacteria and persisters might both be antibiotic-tolerant for the same reason and that the reason has something to do with nonreplication (21). In contrast, however, a single-cell analysis revealed that persisters in a population of Mycobacterium smegmatis challenged with isoniazid had been replicating at the time of antibiotic exposure (14).
In vivo, antibiotics tend to kill most of a genetically susceptible bacterial population quickly but a small proportion of the population slowly or not at all, giving rise to a biphasic “hockey-stick” kill curve. The survivors, when expanded in vitro, remain as sensitive to the antibiotic as the starting population and are therefore described as “phenotypically tolerant” (22). They are also called persisters, but it is not known how persisters in vivo are related to persisters in vitro. Persisters in vivo are particularly prominent in biofilms.
To avoid using the same term for distinct phenomena, researchers may find it useful to distinguish between two classes of persisters. Class I persisters correspond to those originally designated by Bigger (20): They are a small minority in a given population before antibiotic exposure. Most of the population may be replicating. The mechanisms of class I persistence may differ among species, within a given species for different antibiotics, and for the same bacterium-antibiotic combination when cultured in vitro or as found in vivo. Class I persistence might arise when a few members of a bacterial population engage stochastically in any process that can impart heritable antibiotic resistance, but do so through changes in gene expression or posttranslational modification in the absence of mutation.
In contrast, class II persisters are the majority of bacteria in a population, most of which have become nonreplicating and antibiotic-tolerant—for example, during stationary phase. The mechanisms of antibiotic tolerance in class II persistence may differ from those in class I. One mechanism for class II persistence in E. coli involves some of the bacteria secreting the organic compound indole. Indole acts on most of the bacteria in the population to induce genes that protect against oxidative and other stresses and that promote antibiotic tolerance by unknown mechanisms (23). Given that the conditions imposing nonreplication can create metabolic dysfunction in bacteria that leads to excess generation of oxidants, it is possible that low concentrations of oxidants serve as a signal to induce class II persistence. In contrast, higher concentrations of oxidants can kill bacteria or sensitize them to other agents that do so.
It is commonly held that chronic infections take a long time to treat because of persisters. Until recently, it has been difficult to distinguish this premise from circular reasoning. Now, several reports—such as one with Candida albicans, a fungal pathogen, and another with _Pseudomonas aeruginosa_—show that the proportion of antibiotic-tolerant pathogens in a microbial population recovered from patients and tested ex vivo rose during long-term antibiotic treatment without a rise in the MIC (24, 25). That is, the vast majority of the microbes recovered late in the course of treatment remained as drug-sensitive as those recovered before treatment or early in treatment, but the small minority that were resistant increased proportionately. When this small subpopulation was expanded in vitro, the population to which it gave rise remained as antibiotic-sensitive as the original population. These observations lend credence to the close relationship between class I persistence in vitro and in vivo.
Dozens of genes in E. coli, P. aeruginosa, Streptococcus pneumoniae, and M. tuberculosis have been implicated in setting the level of class I persisters in a given population cultured in vitro, without affecting the MIC of the same antibiotic (26). Some of these genes encode transcriptional regulatory proteins that induce stress responses; some specify components of toxins or antitoxins. One such toxin is HipA, a kinase that phosphorylates the bacterial EF-Tu translation factor (27) and presumably suppresses translation of bacterial proteins. Other toxins implicated in persistence can degrade bacterial mRNAs (21). Mechanisms that execute class I persistence—and thereby allow a small subpopulation of bacteria to resist being killed by a conventional antibiotic—might be targets for new anti-infectives that on their own would kill very few bacteria but, in combination with conventional antibiotics, could shorten treatment times and enhance efficacy.
Recently discovered features of the biology of nonreplicative states may suggest new antibiotic targets in class II persistence. Stationary-phase bacteria accumulate irreversibly oxidized proteins (28). Upon resumption of replication, some bacteria distribute these proteins asymmetrically, so that one of the progeny is endowed with newly synthesized copies and the other is burdened with the damaged versions. Only the former progeny can sustain a maximum replicative rate (28). Some stationary-phase bacteria release d-amino acids and the polyamine norspermidine. These metabolites reduce the synthesis of peptidoglycans, alter their composition, and/or promote the disassembly of biofilms (29–31).
Dormancy
A “dormant” bacterial population is nonreplicating, contains a preponderance of VBNC members, and is presumed to have very low metabolic activity. Dormant bacteria are substantially resistant to most antibiotics. Persistent bacteria are also sometimes called dormant. However, both class I and class II persisters in vitro can be detected by CFU assays and by this convention are called “culturable.” Persisters in vivo are often culturable as well, as noted above (24, 25). Changes in antibiotic treatment can dictate whether bacteria exhibit class I persistence or become VBNC and, therefore, potentially dormant. For example, when mice are treated for tuberculosis, monotherapy often leads to or selects for persisters, whereas combination chemotherapy can lead to or select for VBNC organisms (32, 33).
Latency
“Latency” refers to the clinical state of an infection that has not caused disease but has the potential to do so. For example, an estimated one-third of the human population is latently infected with M. tuberculosis. The mycobacterial population in latent tuberculosis exhibits phenotypic tolerance to current antibiotics. Presumably as a result, latent tuberculosis takes many months to cure, as defined by markedly reducing the risk of later emergence of the disease, despite a very small bacterial burden. M. tuberculosis collected from tissues of people with latent infection can be difficult or impossible to culture, and there is little information about its metabolic state.
INFERRING DEATH
A third operational definition of the death of bacterial pathogens is at the level, not of the single bacterium or populations of bacteria, but of the infected host. If treatment with an antibiotic returns the host to a state of wellness comparable to that which he or she enjoyed before the infection was perceived, we say the host has been “cured” and the pathogen has been “eradicated.” However, an agent can cure the host without having the ability to kill the pathogen in vitro by either of the other two definitions discussed above, that is, without being able to lyse individual bacteria or reduce the number of CFUs in the bacterial population. The first example of this phenomenon dates back 80 years when Avery and Dubos cured mice infected with S. pneumoniae by administering an enzyme that degrades the bacterial capsule (34). The enzyme was neither bacteriostatic nor bactericidal, but it allowed antibody to opsonize the bacteria so that they could be ingested and killed by cells of the host's immune system (34). In a contemporary example, an antibiotic (aureomycin) inhibited the production of an antioxidant pigment (staphyloxanthin) by Staphylococcus aureus. In vitro, the compound had no apparent effect on the bacteria, but in the infected host, it allowed the immune system to control the infection (35).
Moreover, antibiotics that do kill the pathogen in vitro can cure a host without eradicating the bacteria in the host. This clinically important phenomenon becomes apparent when the host becomes immunocompromised and cure is reversed. That is, the bacteria appear to be eliminated, but only as long as the host's immune system continues to repress them. The key role of the immune system in enabling antibiotics to cure an infected host was first demonstrated in the “Cornell model” of experimental tuberculosis developed by McDermott and colleagues about 50 years ago (32, 33). Antibiotics cured mice infected with M. tuberculosis as demonstrated by (i) the histologic absence of tuberculosis in about two-thirds of the mice at the end of their natural life span and (ii) the absence of any M. tuberculosis CFUs when some of the cohort were euthanized and their lungs, livers, and spleens were cultured in their entirety. However, about one-third of the mice eventually relapsed, and nearly all of them did so if they were treated with immunosuppressive corticosteroids (32, 33). Chemotherapy that appeared to be sterilizing was not; it only protected the mice from death if aided by the immune system.
Thus, when we confine the search for antibiotics to screens against bacterial populations growing in standard culture conditions, we are trying to kill the bacteria in a state that differs importantly from that which some of them adopt in the host. We are using a restricted definition of bacterial death as a criterion for success that ignores critical contributions of the host to effective therapy.
WAYS OF KILLING
Just as there are several ways of defining bacterial death, there is growing appreciation of the diversity of ways in which antibiotics kill bacteria.
Abortive synthesis
It has long been thought that antibiotics that kill bacteria in pure culture do so mostly by interfering with synthesis of macromolecules, such that an opposing or incomplete process leads to suicide as bacteria break down their own peptidoglycan, DNA, or protein. Alternatively, antibiotics corrupt the synthesis of proteins to produce misfolded and, therefore, toxic translation products (36). For example, β-lactams, d-cycloserine, and vancomycin promote cell wall breakdown by interfering with peptidoglycan synthesis, whereas remodeling enzymes that autolyze peptidoglycans continue to function (37). Fluoroquinolones promote autolysis of DNA by stalling DNA gyrase–topoisomerase in its progression along the chromosome while leaving its endonuclease at work. Small molecules called acyl depsipeptides that activate a bacterial protease (Clp) are thought to kill by promoting uncontrolled autolysis of intrabacterial proteins (38).
Redox suicide
A new theory holds that many antibiotics kill chiefly by promoting bacterial suicide through oxidation (11, 39). Antibiotics trigger a bacterial stress response. This leads in an undefined way to a dysregulated form of metabolic reprogramming in the pathogen that is superficially similar to the metabolic adaptation in stationary phase. The critical result is an accumulation of reducing equivalents, such as NADH (reduced form of nicotinamide adenine dinucleotide). These electron donors autoxidize, generating reactive oxygen intermediates (ROIs) that oxidize diverse targets, among them guanosine, such that life-sustaining processes are interrupted (11, 39, 40). Lending support to this view, some bacteria maintain relative resistance to antibiotics by producing nitric oxide (NO) and H2S, which synergize to induce antioxidant defenses (41).
Either mechanism—abortive synthesis followed by autotoxicity, or promotion of endogenous stress followed by autoxidation—might account for the action of antibiotics that sensitize the pathogen to exogenous stresses imposed by the host. As mentioned, S. aureus is sensitized to host-imposed oxidative stress by an antibiotic that inhibits the synthesis of an endogenous antioxidant, staphyloxanthin (35). Inhibition of the proteasome in M. tuberculosis is hypothesized to lead to death of the bacterium from the accumulation of proteins that undergo misfolding in response to stresses imposed by the host, including oxidation, nitrosation (42), and starvation (43). It is not clear whether corrupted synthesis or stress-induced autoxidation accounts for bacterial death caused by antibiotics that disrupt the cell membrane, such as daptomycin (44), or block adenosine triphosphate synthase, such as TMC207 (45). In any event, the notion that there may be one or more final common pathways of antibiotic action does not diminish, but may add to, an enlarged view of potential antibiotic targets (46).
SURVIVAL MECHANISMS
The foregoing discussion of the diverse genomic contents, transcriptional profiles, replicative states, and ways of killing bacterial pathogens sets the stage for taking stock of what we know about how bacteria evade being killed by antibiotics (Fig. 2).
Heritable resistance
Until recently, research has focused mostly on mechanisms of heritable antibiotic resistance, a phenomenon that is preexistent in nature, even against synthetic antibiotics that have never been clinically deployed (47). For clinically deployed antibiotics, resistance has been identified in organisms that could never have encountered them (48).
“Baseline creep” is an important and widespread form of heritable antibiotic resistance in which average MIC values of clinical isolates rise over time on the order of years (49). Sensitive competition experiments demonstrate that exposure to minute concentrations of antibiotics, orders of magnitude below the MIC, can select for resistant mutants (50). Thus, it should not be surprising that resistance has been detected in clinical isolates soon after the introduction of each antibiotic into clinical practice (51).
Mechanisms of heritable resistance include the acquisition or mutation of genes that lead to (i) the ability to keep out or pump out the antibiotic, (ii) the inability to activate it, (iii) the ability to inactivate it, (iv) an alteration in the structure of the antibiotic's target so as to reduce inhibition by the drug, (v) increased expression of the drug target, or (vi) expression of pathways that compensate for inhibition of the target.
Mutations that lead to heritable antibiotic resistance can be acquired by lateral gene transfer or can arise de novo. De novo mutations can accrue at random at a low frequency related to the rates and fidelities of DNA replication and repair. However, in a process called “induction,” the frequency of de novo mutations that confer antibiotic resistance can be increased under many of the conditions that are associated with interruption of replication and expression of nonheritable antibiotic resistance (52). Interruption of replication can leave some DNA single-stranded and more vulnerable to mutation, and the repair process itself can introduce mutations (52). Some of these mutations may confer resistance to a given antibiotic. Exposure of the bacterial population to that antibiotic will select for outgrowth of the resistant mutants.
For example, emergence of rifampin resistance during treatment of experimental tuberculosis was shown to depend strongly on expression of the DNA repair enzyme DnaE2 (53), suggesting that the DNA repair process introduced most of resistance-conferring mutations. Alternatively, a partial deficiency in DNA repair can foster a hypermutator phenotype, because DNA damage itself, if not repaired, constitutes mutation. Of course, if DNA damage is extensive and repair is lacking, a bacterium will die without help from antibiotics. Earlier, it was noted that lethal antibiotic treatment leads to and depends in part on oxidative stress (39). Sublethal antibiotic treatment can lead to lower levels of oxidative stress that, although not sufficient to kill, are nonetheless mutagenic. Some of the resulting mutations can confer resistance to the same and other antibiotics (11). Biofilms appear to harbor hypermutator bacteria and antibiotics select more frequently for resistant organisms in biofilms than in other settings (52).
Nonheritable resistance
In recent years, researchers have begun to recognize that heritable resistance is the tip of an iceberg, the base of which is nonheritable antibiotic resistance, or phenotypic tolerance (22). Phenotypic tolerance is a property of a bacterial population that is susceptible to a given antibiotic under some conditions (typically, conditions supporting exponential replication of planktonic forms) but has become relatively and reversibly resistant to the antibiotic under other conditions, without mutation of its genes or acquisition of new ones from resistant strains. Conditions associated with phenotypic tolerance include residence in biofilms plus all of the nonreplicative or slowly replicating states discussed above, including those that promote “induced mutation” (52). Such states include nutritional deprivation, oxygen deprivation, Mg deprivation, Fe deprivation, acidification, oxidative or nitrosative stress, viability without culturability, persistence, dormancy, latency, and culture at high density, as seen in the inoculum effect.
The association of phenotypic tolerance with nonreplicative states is sometimes attributed to the fact that most antibiotics interfere with biosynthetic processes needed to build biomass, whereas nonreplicating organisms have less need to build biomass than replicating organisms. However, recent evidence supports an alternative view, that antibiotic tolerance is achieved through active responses to growth-limiting conditions. Such adaptations include the starvation-signaling stringent response in P. aeruginosa (54) or the ability of growth-limiting conditions of hypoxia, iron limitation, or acidification to redirect central carbon metabolism away from the tricarboxylic acid cycle toward triglyceride synthesis in M. tuberculosis (16). The stringent response enhances antioxidant defense (54), consistent with the theory that antibiotics kill by eliciting oxidative injury (39). Similarly, diversion of carbon metabolism away from substrate oxidation toward storage (16) might be linked to antibiotic tolerance by diminishing the production of reactive oxygen species. In contrast, the ability of sub-inhibitory concentrations of diverse antibiotics to induce cross-tolerance to other antibiotics in M. tuberculosis appeared to depend on an increase in the reductive state in the bacterial cytosol, which in turn led to induction of the whiB7 transcriptional regulator (55). WhiB7 induces genes whose products pump out some antibiotics or modify the ribosome to reduce binding of others (56).
“Adaptive resistance” is a term introduced to describe phenotypic tolerance with an emphasis on three features: (i) resistance is inducible in response to specific signals that may or may not affect replication; (ii) an important additional class of signals is sublethal exposure to antibiotics; and (iii) upon removal of such signals, antibiotic susceptibility may not be fully restored (49). Speculative mechanisms for adaptive resistance are epigenetic modifications or transcriptional responses that bring about any of the same effects responsible for heritable resistance, except mutation of the target (49). However, without genome resequencing, it is difficult to know if adaptive resistance that does not fully revert on removal of the stimulus is different from the induced mutation response that has been documented in response to many of the same stimuli. Moreover, it is striking that most of the signals that can trigger adaptive resistance and induced mutation can also impose a nonreplicative state on the bacteria. The chief exception is sublethal levels of antibiotics that induce adaptive resistance without inducing nonreplication. Given the heterogeneity in responses to antibiotics by individual bacteria, it may be misleading to evaluate the issue at the population level. Researchers should consider whether the individual cells in which sublethal antibiotic treatment promotes nonreverting adaptive resistance with or without mutation are those whose replication is impaired by the antibiotic at that concentration. Without such studies, the relationship will likely remain obscure between nonreplication, induced mutation, and adaptive resistance.
Many of the conditions that trigger adaptive resistance to antibiotics, such as nutritional deprivation, are likely to exist in niches occupied by microbial pathogens in nature when they are not infecting a host. Is there an evolutionary advantage to a bacterium that becomes antibiotic-tolerant in response to conditions such as starvation, acid, hypoxia, or oxidative stress? In considering this question, it may be helpful to reflect that it is common in biology for one organism to compete with another by imposing maladaptive signaling—such as forcing a signal to come at the wrong time, at too high a level, or for too long a time (57).
For example, two of the major sets of antimicrobial molecules used by the host—ROIs and reactive nitrogen intermediates (RNIs)—are physiologic mediators of intra- and intercellular signaling, particularly when produced for short periods at low concentrations. Most antibiotics in use today are based on microbial products or act on the same targets as microbial products. Natural product antibiotics can be seen as that subset of intra- or intermicrobial signaling molecules that we have learned to apply at such high and sustained concentrations that they become inhibitory or lethal to bacteria (58). Presumably, when intermicrobial signaling molecules in the wild reach concentrations that inhibit bacterial replication, this serves as a signal to display adaptive resistance. Likewise, conditions such as carbon starvation, acid, and hypoxia may serve as advance, if imprecise, warnings that competitors in the microenvironment are numerous and may soon release antibiotics at concentrations that need to be resisted. If so, bacteria may have evolved ways to use nonreplication to trigger adaptive resistance.
It is striking that many of the states that lead to phenotypic tolerance on the part of the pathogen can be imposed by the host. Thus, the host immune system can provide an indispensable assist to antibiotics in achieving cure and can also render the same antibiotics unable to cure on their own.
TARGETING RESISTANCE
How fast and well we improve our understanding of antibiotic resistance and organize to restrict its spread will play a major role in shaping the course of medicine in the years ahead. Important fronts in the campaign include much-improved husbandry of the antibiotics we have and a much wider list of targets for antibiotics we seek (Fig. 3). This section focuses on one set of new targets: those involving resistance itself.
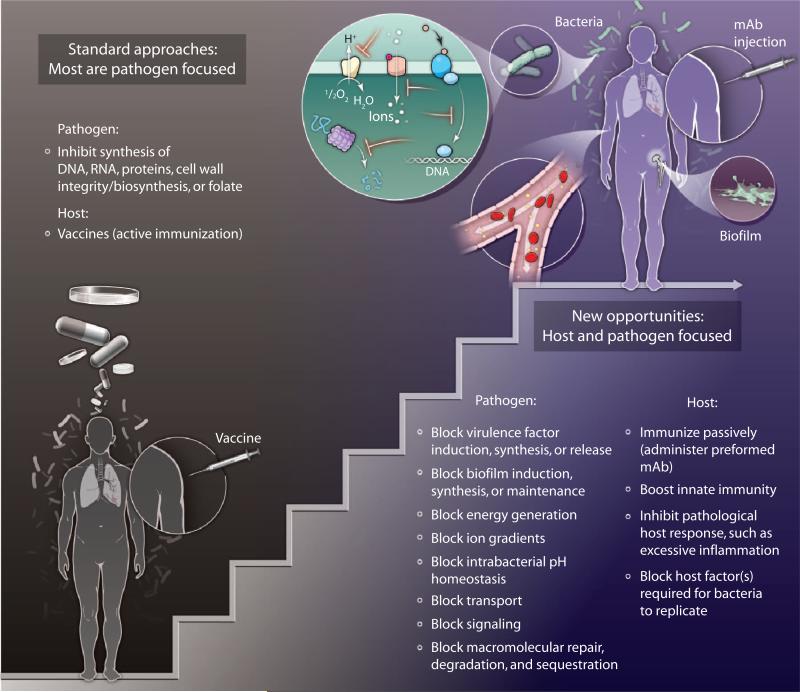
Fig. 3. Intervening in infection.
Standard approaches to the prevention or treatment of bacterial infection are pathogen-focused; either the host is immunized with an attenuated or killed pathogen or some of its components, to which the host's adaptive immune system responds, or the host is given drugs that inhibit bacterial synthesis of nucleic acids, proteins, cell walls, or folate. Researchers are beginning to explore new opportunities for the treatment of bacterial infections by targeting a wider range of pathways in the pathogen, as well as processes in the host that the pathogen relies on to cause disease. mAb, monoclonal antibody.
Resistance mechanisms
To reduce heritable antibiotic resistance, we must develop effective blockers of bacterial drug efflux pumps and learn more about mechanisms of induced resistance. To cope with nonheritable resistance, we need to learn how antibiotic tolerance arises in antibiotic persistence, phenotypic tolerance, and adaptive resistance. Some promising targets are emerging, such as the transcriptional regulator PhoU; this protein mediates a downshift in E. coli's metabolism under conditions that slow replication, which is accompanied by an increase in nonheritable antibiotic resistance (59). We need to learn more about how induced resistance and phenotypic tolerance are related to DNA damage, stress responses, and nonreplicative or slowly replicative states. Metabolomics will likely be an invaluable complement to genetics and transcriptomics in identifying critical features that are shared by these states (60). However, harnessing the power of metabolomics to study bacterial pathogens as they exist in the host may require innovations in sampling and advances in sensitivity.
Researchers should initiate screens for antibiotics that block induced resistance and phenotypic tolerance, not only in planktonic, replicating bacteria, but also under the very conditions that make pathogens tolerant to conventional antibiotics or induce resistance to them (46)—such as residence in biofilms, high inoculum density, nutrient limitation, oxygen deprivation, metal insufficiency, acidification, oxidative or nitrosative stress (61, 62), and sublethal exposure to other antibiotics (63). Scientists need to characterize and target mechanisms of bacterial entry into, maintenance of, and exit from nonreplicative states.
Biofilms
Given that biofilm infections are a major contributor to antibiotic tolerance and resistance, we need to explore ways to (i) discourage biofilm formation in tissues (64) or on implanted devices (65), (ii) incorporate into implanted foreign surfaces the catalytic means to generate anti-infective activity from endogenous host precursors (66), (iii) block bacterial synthesis of extracellular components that make biofilms resistant to penetration by liquids (such as body fluids containing antibiotics) and gases (such as oxygen) (67), and (iv) dispel biofilms (29, 30). Possible ways to achieve these goals include the interruption of bacterial signaling pathways involved in forming and maintaining biofilms (68, 69); the activation of processes that promote dispersal of biofilms into planktonic cells that reacquire antibiotic sensitivity, such as an NO-triggered decrease in intrabacterial cyclic diguanosine monophosphate (70); or the supply of specific cations, such as norspermidine, that destabilize the biofilm matrix at the site of infection (30).
Virulence mechanisms, multitargeting, and other approaches
An indirect way to avoid antibiotic resistance is to target virulence mechanisms without trying to kill the pathogen (71). Another way to minimize antibiotic resistance is to focus on, rather than shy away from, agents that can target the bacterial membrane (44), and to seek, rather than shun, antibiotics with multiple targets (72). Lewis has proposed the development of a prodrug that a bacteria-restricted enzyme would activate to a form that is reactive enough to inactivate diverse intrabacterial targets (21). Evolution has already devised two classes of anti-infectives that have multiple targets: ROIs and RNIs (73). The nitroimidazole PA824 is a drug in clinical trials for tuberculosis that fits Lewis’ description: A mycobacterial nitroreductase uses a mycobacterial flavin as a cofactor to metabolize the drug to generate RNI (74, 75). Unfortunately, resistance arises in strains that are unable to generate the cofactor. Still, the substantial success of this intrabacterial RNI generator reminds us that another way to control infection is to interfere with the ability of pathogens to catabolize host-derived ROI and RNI or repair their damage (76), particularly if key enzymes involved have essential additional roles (77).
Enzymes as drugs
Researchers should further explore bacteriolytic therapies. Bacteriophage-derived lysins are enzymes that can degrade peptidoglycans in the bacterial cell wall in a species-selective manner (78, 79). They may be effective in cases where the lysin can reach a sufficient proportion of the pathogenic species in the host (for example, in extracellular infections that do not involve biofilms) and cure the infection before the host immune response inactivates the lysin.
TARGETING THE HOST
Treatment of infection has two goals: to prevent morbidity and mortality and, in the case of infectious diseases that are contagious, to prevent transmission. Morbidity, mortality, and transmission are all manifestations of the host-pathogen relationship. Given the difficulty of finding new antibiotic targets in bacterial pathogens, scientists should devote attention to the host side of the host-pathogen relationship in designing treatments. Intervention at the level of the host could take two forms: boosting host immunity or blocking something in the host that the pathogen must exploit to cause disease (Fig. 3).
Boosting host immunity
In theory, active immunization is the ideal way to control infectious disease through medical intervention. Immunization should be extended to more bacterial diseases. However, the public is unlikely to accept numerous universal immunizations of healthy people with a finite risk of adverse effects in an effort to prevent specific infections, any one of which only a minority of the population is otherwise likely to experience.
Passive immunization by administration of monoclonal antibodies to bacterial proteins, lipopeptides, glycolipids, or carbohydrates holds promise for prophylaxis in settings where there is sufficient epidemiologic intelligence to specify which group to protect against which infection at which time. Post-infection treatment of bacterial infections with monoclonal antibodies may be effective if they are administered early in the course of the disease (80). Despite their cost to produce and stockpile, monoclonal antibodies may be an attractive adjunct or alternative to small chemical compounds in treating infections by the pathogens for which other options are running out, such as the “ESKAPE” pathogens, Enterococcus faecium, S. aureus, Klebsiella species, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species (81), and drug-resistant Neisseria gonorrhoeae (82).
Aside from boosting or borrowing an adaptive immune response, there is promise in boosting innate immunity (83). An advantage could be protection against diverse infections. An inherent risk is exacerbation of immunopathology. For example, in systemic inflammatory response syndrome and septic shock, the innate immune response appears to contribute more to multiple organ damage than an antibiotic-susceptible pathogen in patients who receive appropriate antibiotic treatment. It is a challenge to predict whether an individual patient would benefit from more or less of an innate immune response at any particular point in the course of the disease (84). A systems biology analysis of host responses during specific infections, interpreted in the context of coincident medical conditions, treatments, and outcomes, may help address this challenge.
Denying the pathogen a required host pathway
Pathogens often require host factors for their propagation, persistence, or pathogenicity. If such a host factor were temporarily dispensable for the host, then a drug that inhibited its expression or action might constitute an adjunctive anti-infective therapy. This approach is well advanced in the treatment of viral infections, but its application is only beginning to be explored in bacterial infections. A virtue of such an approach would be that a pathogen could not become resistant on the basis of such bacterially encoded mechanisms as impaired drug uptake or retention, reduced drug activation, increased drug inactivation, or the mutation, overexpression, or bypass of the drug target.
Numerous candidate host targets, including kinases and phosphatases, have been identified for at least one bacterial pathogen, M. tuberculosis (85–87). Deficiency of one host kinase, protein kinase R (PKR), causes little phenotype in mice on its own, yet led to sustained reductions in bacterial burden and lung pathology during tuberculosis (88). Two mechanisms appear to be at work. PKR deficiency enhanced host-protective macrophage apoptosis upon M. tuberculosis infection. Moreover, PKR played a key role in inducing the expression of interleukin-10, a macrophage-deactivating cytokine. As a result, PKR deficiency enhanced macrophage activation in response to interferon-γ, leading to higher amounts of protective RNI than those observed in wild-type mice (88). Given to mice before and after infection, the tyrosine kinase inhibitor imatinib (Gleevec) reduced the growth of M. tuberculosis over at least a month; the target or targets are unknown (89).
Intercellular signaling molecules derived from host fatty acids can also be manipulated to the benefit of the infected host. Loss of function of leukotriene A4 hydrolase increased susceptibility of zebrafish to Mycobacterium marinum. Heterozygosity for certain polymorphisms at the corresponding human locus may protect from tuberculosis and lepromatous leprosy (90). Given that the mechanism may involve the balance between pro- and anti-inflammatory eicosanoids, it may be possible to translate this discovery into an adjunctive therapy for mycobacterial diseases. Similarly, mice were shown to survive E. coli infection in higher proportions and to lower the numbers of viable E. coli and S. aureus further when antibiotic treatment was supplemented with resolvins and protectins, anti-inflammatory meta-bolites of ω-3-eicosapentaenoic or docosahexaenoic acids (91).
STEPPING UP
The Institute of Medicine [IOM, a branch of the U.S. National Academies (92, 93)], the Infectious Diseases Society of America (IDSA) (94), and ReAct—Action on Antibiotic Resistance (95) have published comprehensive recommendations for responding to the antibiotic crisis. Several particularly important steps are emphasized below. Some of these ideas are included in the published reports; others are new.
Private-public cooperation
Drug companies face economic, regulatory, and scientific obstacles to antibiotic development (96). Legislation that was rejected in 2010 by the U.S. Congress but is presently being reconsidered, the Generating Antibiotic Incentives Now Act, would add 5 years to market exclusivity for certain new antibiotics, but this is unlikely to alter the decisions of pharmaceutical firms to close down most of their antibiotic research and development (97). More fundamental solutions to the problem of inadequate financial incentives have been proposed and compared (98–100). In brief, as summarized by ReAct, it will be necessary to develop a business model for antibiotics that “delinks revenues from sales” (95). For example, governments could establish a fund from which developers of new medicines could be rewarded in proportion to their product's medical benefit, rather than through sales at monopoly-protected prices, if the developer so chose (98).
However, even if financial incentives were in place both to develop new antibiotics and to conserve them—a contradiction in terms under the current system of reward—the problem remains that new antibiotics have become very difficult to discover using the approaches on which the pharmaceutical industry has converged in the last few decades (101). How can the industry experiment with innovative approaches?
In 2004, “open access drug companies” were proposed to foster antibiotic discovery (96). These entities were visualized as sectors within one or more campuses of major drug companies where academic, government, and biotechnology scientists could match innovative approaches to pharmaceutical expertise, sharing rights in resulting intellectual property as appropriate. This vision has been realized both in its original form and in a geographically distributed version. GlaxoSmithKline has opened its Diseases of the Developing World campus at Tres Cantos, Spain, to academic, government, and biotech scientists for collaborative discovery of antibiotics for neglected infectious diseases. The independent Tres Cantos Open Lab Foundation selects projects and helps cover visitors’ expenses (http://www.openlabfoundation.org). Another version of the “Open Lab” is the Lilly TB Drug Discovery Initiative, a consortium of Eli Lilly and Company, the U.S. National Institute of Allergy and Infectious Diseases (NIAID), the not-for-profit Infectious Disease Research Institute, and Academia Sinica of Taiwan (http://www.tbdrugdiscovery.org). These groups contribute resources to TB drug development projects selected from submissions from any source, public or private. In another manifestation of the concept, Pfizer has rented space for “Centers of Therapeutic Innovation” near high concentrations of academic laboratories in San Francisco, Boston, and New York City, where company and academic scientists work together to develop peptide and protein therapeutics. As industrial drug discovery shifts to a more distributed model, numerous partnerships are being forged between pharmaceutical companies and academic institutions or individual academic laboratories.
It is time to take this movement to the next level: intercompany open labs. In this model, laboratories at one or more sites, including virtual sites, would house cooperative efforts by academic, government, and industrial scientists—the last from more than one company—to share knowledge, risk, and reward in anti-infective development. The collaborators would jointly plan to test diverse approaches while avoiding redundant efforts. An experiment in pulling together multiple academic and industrial groups in a cooperative effort is already under way in the TB Drug Accelerator program led by the Bill & Melinda Gates Foundation. Participants include Abbott Laboratories, AstraZeneca, Bayer, Weill Cornell Medical College, Eli Lilly, GlaxoSmithKline, Infectious Disease Research Institute, Merck, NIAID, Sanofi, and Texas A&M University.
One off-the-shelf way to extend the intercompany open lab concept to other neglected infectious diseases is for pharmaceutical companies to contribute to the Tres Cantos Open Lab Foundation and post some of their personnel to the Tres Cantos campus or open one of their own campuses in a similar way. However, intercompany open labs could tackle a wider range of infectious diseases. One fitting project for an intercompany open lab would be to fulfill Richard Baltz's vision for a cooperative approach to reinvigorate natural product isolation from actinomycetes (102), a group of Gram-positive bacteria that have furnished or inspired many antibiotics. Moreover, pharmaceutical firms could lend their now rarely used collections of defined natural products to an intercompany open lab, rather than shelving them, discarding them, or spinning them off to small firms or not-for-profit organizations that have little chance of exploiting them fully and effectively. A public-private consortium could mine the collective natural product resource for antibiotic discovery, with appropriate reach-through rights for the donors of the compounds in the event of product development. An important additional benefit of open labs would be to share, and thus preserve, the knowledge base about microbial physiology and antibiotic action that has been built up in industry and is now endangered by industry's extensive withdrawal from the field (96).
National policy
The U.S. federal government should create a national interagency Infectious Disease Policy Board that reports to the president. Members should be appointed from the IOM, IDSA, NIAID, Centers for Disease Control and Prevention (CDC), U.S. Food and Drug Administration (FDA), National Security Council, President's Council of Advisors on Science and Technology, and the Departments of Health and Human Services, Defense, State, and Agriculture. The board should be tasked as follows:
- To draw guidelines for executive branch action and recommend legislation to combine regulation and tax policy to drastically curtail the use of antibiotics in healthy food animals and plants. A recent FDA guideline falls short in merely recommending that manufacturers voluntarily label antibiotics to eliminate animal growth promotion as an approved use. To counter the expected push-back from the food industry, evidence should be marshaled and publicized, for example, that (i) an estimated 99,000 people die each year in the United States from hospital-acquired infections (94); (ii) antibiotic-resistant infections in the United States are estimated to cost 21billionto21 billion to 21billionto34 billion per year; (iii) using antibiotics in healthy food animals disseminates resistance; and (iv) the recommended curtailment has been instituted stepwise in Europe beginning with Sweden's ban on antibiotic growth promoters in food animal production in 1986 and was extended to the entire European Union in 2006. There has been a predominantly favorable impact on antibiotic resistance without major adverse effects on food production. For example, Danish swine production increased 47% as antibiotic use in swine fell by 51% (103).
- To develop guidelines for legislation that provides patent extensions for new antibiotics that are distributed at cost or at affordable prices in least-developed and low-income countries and manufactured or packaged in such a way as to prevent counterfeiting and deter smuggling into profit-generating markets.
- To require FDA to modify clinical trial requirements for new antibiotics, such that approval may be based on in vitro evidence of activity against drug-resistant strains plus clinical safety and adequacy, without requiring “superiority”—that the new drugs must be more efficacious than current gold standards—and dropping the requirement that patients enrolled in such trials be shown to be infected with drug-resistant strains, yet not have received previous antibiotic therapy.
- To work with U.S. National Institutes of Health (NIH) and the U.S. Department of Defense to rationalize the distribution of their investments between counter-bioterrorism research and other infectious disease research, recognizing the significant impact of prevalent, poorly treatable or untreatable, nonweaponized infections on the military and on national security. These agencies should ensure that these precious research dollars support high-quality anti-infectives discovery and development in a way that combines the best elements of the Defense Threat Reduction Agency's emphasis on innovation with NIH's use of expert peer review.
- To redirect government-funded antibiotic discovery efforts away from the search for ultrabroad-spectrum agents, and to encourage, instead, the development of more focused agents active against pathogens for which the need is greatest (81).
- To couple patent life extension and the foregoing emphasis on non–ultrabroad-spectrum agents with the development of affordable, fast, point-of-care, pre-prescription diagnostic technologies, including for the detection of viral infections, so that antibiotics can be prescribed appropriately.
- To review and update guidelines, incentives, and penalties to ensure that health care providers and facilities practice appropriate methods of infection control.
- To educate doctors and the public on proper antibiotic use.
- To educate the public, particularly the elderly and parents of young children, on the value of vaccines.
- To work with CDC to implement better national and international systems of monitoring baseline creep and full-blown antibiotic resistance as well as outbreaks of infectious disease.
SUMMING UP
Except for self-limited disorders, surgical interventions, and replacement of missing nutrients, vitamins, metals, or hormones, almost the only diseases we can cure with drugs are bacterial and fungal infections and several malignancies. Most drugs palliate or mitigate. When their administration stops, disease remains. Thus, it would set medicine back immeasurably if society forfeits the ability to cure infectious diseases. Moreover, the impact of infectious disease on national security is not limited to bioterrorism but hovers over every case of trauma, surgery, pneumonia, or meningitis in the military and its civilian support infrastructure.
Challenges in the control of infectious disease have become so great that no single group or approach can meet them. We need efforts that are organized, multidisciplinary, multiple, and parallel and that bring the private and public sectors together in precompetitive space to address them in accord with a new set of national policies.
Acknowledgments
I thank S. Ehrt, B. Gold, K. Rhee, D. Schnappinger, and K. Shigyo for critical comments and J. MacMicking for discussions. Funding: The author's views were shaped during work supported by NIH grant AI064768, Defense Threat Reduction Agency grant HDTRA1-06-C-0039 (S. N. Cohen, principal investigator), grants OPP42672 and 1024029 from the Bill & Melinda Gates Foundation, and the Milstein Program in Chemical Biology of Infectious Disease. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
Competing interests: There are no competing financial interests. The author serves in an unpaid capacity on the Board of Governors of the Tres Cantos Open Lab Foundation, the Scientific Steering Committee of the Lilly TB Drug Discovery Initiative, and the Joint Steering Committee of the Pfizer–Weill Cornell Center for Therapeutic Innovation.
Citation: C. Nathan, Fresh approaches to anti-infective therapies. Sci. Transl. Med. 4, 140sr2 (2012).
REFERENCES AND NOTES
- 1.Mathers CD, Ezzati M, Lopez AD. Measuring the burden of neglected tropical diseases: The global burden of disease framework. PLoS Negl. Trop. Dis. 2007;1:e114. doi: 10.1371/journal.pntd.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 3.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall A, Fang FC, Pirofski LA. Microbial virulence as an emergent property: Consequences and opportunities. PLoS Pathog. 2011;7:e1002136. doi: 10.1371/journal.ppat.1002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich GD, Ahmed A, Earl J, Hiller NL, Costerton JW, Stoodley P, Post JC, DeMeo P, Hu FZ. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol. Med. Microbiol. 2010;59:269–279. doi: 10.1111/j.1574-695X.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willing BP, Russell SL, Finlay BB. Shifting the balance: Antibiotic effects on host–microbiota mutualism. Nat. Rev. Microbiol. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 10.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional adaptation of Myco-bacterium tuberculosis within macrophages: Insights into the phagosomal environment. J. Exp. Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 14.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Connell JL, Wessel AK, Parsek MR, Ellington AD, Whiteley M, Shear JB. Probing prokaryotic social behaviors with bacterial “lobster traps”. MBio. 2010;1:e00202–e00210. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011;2:e00100–e00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shleeva MO, Bagramyan K, Telkov MV, Mukamolova GV, Young M, Kell DB, Kaprelyants AS. Formation and resuscitation of ‘non-culturable’ cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology. 2002;148(Pt. 5):1581–1591. doi: 10.1099/00221287-148-5-1581. [DOI] [PubMed] [Google Scholar]
- 19.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am. J. Respir. Crit. Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. [Google Scholar]
- 21.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 22.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 23.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 2010;54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: Clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 2011;60(Pt. 6):699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredriksson A, Nyström T. Conditional and replicative senescence in Escherichia coli. Curr. Opin. Microbiol. 2006;9:612–618. doi: 10.1016/j.mib.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-Amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodkin-Gal I, Cao S, Chai L, Böttcher T, Kolter R, Clardy J, Losick R. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell. 2012;149:684–692. doi: 10.1016/j.cell.2012.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-Amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCune RM, Feldmann FM, Lambert HP, McDermott W. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J. Exp. Med. 1966;123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCune RM, Feldmann FM, McDermott W. Microbial persistence. II. Characteristics of the sterile state of tubercle bacilli. J. Exp. Med. 1966;123:469–486. doi: 10.1084/jem.123.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avery OT, Dubos R. The protective action of a specific enzyme against type III pneumococcus infection in mice. J. Exp. Med. 1931;54:73–89. doi: 10.1084/jem.54.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: An essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 38.Brötz-Oesterhelt H, Beyer D, Kroll HP, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, Bandow JE, Sahl HG, Labischinski H. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 39.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 40.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: A universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 42.Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461:621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandotra S, Lebron MB, Ehrt S. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6:e1001040. doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 46.Nathan C. Making space for anti-infective drug discovery. Cell Host Microbe. 2011;9:343–348. doi: 10.1016/j.chom.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 47.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 48.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 49.Fernández L, Breidenstein EB, Hancock RE. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 2011;14:1–21. doi: 10.1016/j.drup.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 2007;3:549–556. doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 53.Boshoff HI, Reed MB, Barry CE, III, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burian J, Ramón-García S, Sweet G, Gómez-Velasco A, Av-Gay Y, Thompson CJ. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 2012;287:299–310. doi: 10.1074/jbc.M111.302588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nathan C. Specificity of a third kind: Reactive oxygen and nitrogen intermediates in cell signaling. J. Clin. Invest. 2003;111:769–778. doi: 10.1172/JCI18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies J. Everything depends on everything else. Clin. Microbiol. Infect. 2009;15(Suppl. 1):1–4. doi: 10.1111/j.1469-0691.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Zhang Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 2007;51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhee KY, de Carvalho LP, Bryk R, Ehrt S, Marrero J, Park SW, Schnappinger D, Venugopal A, Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: An unexpected frontier. Trends Microbiol. 2011;19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Myco-bacterium tuberculosis. Antimicrob. Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bryk R, Gold B, Venugopal A, Singh J, Samy R, Pupek K, Cao H, Popescu C, Gurney M, Hotha S, Cherian J, Rhee K, Ly L, Converse PJ, Ehrt S, Vandal O, Jiang X, Schneider J, Lin G, Nathan C. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011;7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 64.Cusumano CK, Pinkner JS, Han Z, Greene SE, Ford BA, Crowley JR, Henderson JP, Janetka JW, Hultgren SJ. Treatment and prevention of urinary tract infection with orally active FimH inhibitors. Sci. Transl. Med. 2011;3:109ra115. doi: 10.1126/scitranslmed.3003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Francolini I, Donelli G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 2010;59:227–238. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 66.Cai W, Wu J, Xi C, Ashe AJ, III, Meyerhoff ME. Carboxyl-ebselen-based layer-by-layer films as potential antithrombotic and antimicrobial coatings. Biomaterials. 2011;32:7774–7784. doi: 10.1016/j.biomaterials.2011.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Epstein AK, Pokroy B, Seminara A, Aizenberg J. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc. Natl. Acad. Sci. U.S.A. 2011;108:995–1000. doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janssens JC, De Keersmaecker SC, De Vos DE, Vanderleyden J. Small molecules for interference with cell-cell-communication systems in Gram-negative bacteria. Curr. Med. Chem. 2008;15:2144–2156. doi: 10.2174/092986708785747580. [DOI] [PubMed] [Google Scholar]
- 69.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2011;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 71.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 72.Silver LL. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE., III PA-824 kills non-replicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nathan C. Microbiology. An antibiotic mimics immunity. Science. 2008;322:1337–1338. doi: 10.1126/science.1167452. [DOI] [PubMed] [Google Scholar]
- 76.Nathan C, Gold B, Lin G, Stegman M, de Carvalho LP, Vandal O, Venugopal A, Bryk R. A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis. 2008;88(Suppl. 1):S25–S33. doi: 10.1016/S1472-9792(08)70034-9. [DOI] [PubMed] [Google Scholar]
- 77.Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D, Ehrt S, Nathan C. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9:21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borysowski J, Gorksi A. Enzybiotics and their potential applications in medicine. In: Villa T, Veiga-Crespo P, editors. Enzybiotics: Antibiotic Enzymes as Drugs and Therapeutics. Wiley; New York: 2010. pp. 1–26. [Google Scholar]
- 79.Fischetti VA. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Casadevall A, Dadachova E, Pirofski LA. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 81.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 82.Bolan GA, Sparling PF, Wasserheit JN. The emerging threat of untreatable gonococcal infection. N. Engl. J. Med. 2012;366:485–487. doi: 10.1056/NEJMp1112456. [DOI] [PubMed] [Google Scholar]
- 83.Hancock RE, Nijnik A, Philpott DJ. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012;10:243–254. doi: 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- 84.Casadevall A, Pirofski LA. Virulence factors and their mechanisms of action: The view from a damage–response framework. J. Water Health. 2009;7(Suppl. 1):S2–S18. doi: 10.2166/wh.2009.036. [DOI] [PubMed] [Google Scholar]
- 85.Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 86.Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van Eeden SJ, Geluk A, Poot A, van der Marel G, Beijersbergen RL, Overkleeft H, Ottenhoff TH, Neefjes J. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- 87.Jayaswal S, Kamal MA, Dua R, Gupta S, Majumdar T, Das G, Kumar D, Rao KV. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA screen. PLoS Pathog. 2010;6:e1000839. doi: 10.1371/journal.ppat.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu K, Koo J, Jiang X, Chen R, Cohen SN, Nathan C. Improved control of tuberculosis and activation of macrophages in mice lacking protein kinase R. PLoS One. 2012;7:e30512. doi: 10.1371/journal.pone.0030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Napier RJ, Rafi W, Cheruvu M, Powell KR, Zaunbrecher MA, Bornmann W, Salgame P, Shinnick TM, Kalman D. Imatinib-sensitive tyrosine kinases regulate mycobacterial pathogenesis and represent therapeutic targets against tuberculosis. Cell Host Microbe. 2011;10:475–485. doi: 10.1016/j.chom.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tobin DM, Vary JC, Jr., Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiang N, Fredman G, Bäckhed F, Oh SF, Vickery T, Schmidt BA, Serhan CN. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. 2012;484:524–528. doi: 10.1038/nature11042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walsh CT, Bassler BL, Nathan CF, O'Brien TF, Rile M, White RJ, Wright GD, Casadevall A, Colwell RR, Hancock REW, McFall-Ngai MJ, Nathan CF, Pirofski L-A, Tzianbos A, Zaller DM. Treating Infectious Diseases in a Microbial World: Report of Two Workshops. National Academies Press; Washington, DC: 2005. pp. 1–63. [Google Scholar]
- 93.Choffnes ER, Relman DA, Mack A. Antibiotic Resistance: Implications for Global Health and Novel Intervention Strategies. National Academies Press; Washington, DC: 2010. p. 474. [PubMed] [Google Scholar]
- 94.Infectious Diseases Society of America (IDSA) Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. Combating antimicrobial resistance: Policy recommendations to save lives. Clin. Infect. Dis. 2011;52(Suppl. 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hallstrom N. Collaboration for Innovation—The Urgent Need for New Antibiotics. ReAct—Action on Antibiotic Resistance; Brussels: 2011. Meeting report—Highlights and conclusions; pp. 1–22. [Google Scholar]
- 96.Nathan C. Antibiotics at the crossroads. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 97.Ambrose PG. Antibiotic bill doesn't GAIN enough ground. Nat. Med. 2011;17:772. doi: 10.1038/nm0711-772. [DOI] [PubMed] [Google Scholar]
- 98.Nathan C. Aligning pharmaceutical innovation with medical need. Nat. Med. 2007;13:304–308. doi: 10.1038/nm0307-304. [DOI] [PubMed] [Google Scholar]
- 99.Mossialos E, Morel CM, Edwards S, Berenson J, Gemmill-Toyama M, Brogan D. Policies and Incentives for Promoting Innovation in Antibiotic Research. World Health Organization; Copenhagen: 2010. p. 195. [Google Scholar]
- 100.Kesselheim AS, Outterson K. Fighting antibiotic resistance: Marrying new financial incentives to meeting public health goals. Health Aff. 2010;29:1689–1696. doi: 10.1377/hlthaff.2009.0439. [DOI] [PubMed] [Google Scholar]
- 101.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 102.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 103.Cogliani C, Goossens H, Greko C. Restricting antimicrobial use in food animals: Lessons from Europe. Microbe. 2011;6:274–279. [Google Scholar]